Introduction
Colorectal cancer is one of the most common
malignant tumors in the gastrointestinal tract (1), including both colon and rectal cancer.
The occurrence of malignant lesions is mainly caused by various
carcinogenic factors such as the environment or heredity of the
intestinal mucosal epithelium (2).
The disease has the third highest occurrence rate in the male
tumors and second highest in the females (3). The incidence rate ranks fourth and the
mortality rate ranks fifth in the malignant tumors of China
(4,5). The incidence of colorectal cancer has
shown a significant upward trend due to factors such as
environment, diet and social pressure (6). Most colorectal cancers are insidious,
not easily identified in the early stage and have an extremely low
diagnostic rate. The majority of patients are already in the
advanced stage when they are diagnosed with colorectal cancer, and
missed the best time for treatment. Colorectal cancer is most
likely to metastasize to the liver, lung, bone and retroperitoneal
lymph nodes. It is already in the advanced stage when distant
metastasis occurs. Therefore, routine detections and early
diagnosis are required for the treatment of colorectal cancer
(7).
RUNX3 is a tumor suppressor gene that has
important regulatory effect on the proliferation, growth and
apoptosis of cells. The development, metastasis and prognosis of
various malignant tumors are related to the expression of
RUNX3 and is expected to become an important indicator for
prognosis and evaluation of tumor invasions (8,9).
Evidence has shown that, RUNX3 can be used as a basis for
judging the degree of malignancy of colon cancer and an indicator
for assisted diagnosis (10).
MicroRNAs (miRNAs), as a class of endogenous non-coding small RNAs,
generally approximately 22 nt in length, can be found in many
organisms such as animals, plants and viruses (11). It has been reported that miRNA, as a
protooncogene or tumor suppressor gene, may be involved in the
development and progression of tumors (12,13).
Related research by Hu et al (14) found that miR-363 can inhibit the
proliferation and metastasis of tumors. Genes showed a low
expression in colorectal cancer, due to its close association with
the occurrence, development and metastasis of colorectal cancer,
therefore, it may be a new target for the gene diagnosis and
treatment in colorectal cancer. However, there are only a few
studies on the association between the expression of miR-363 and
clinicopathological characteristics in colorectal cancer.
The expression of RUNX3 and miR-363 in
colorectal cancer were detected in the present study. The
relationship between the expression levels of RUNX3, miR-363
and the clinicopathological characteristics were studied and the
effect of RUNX3 and miR-363 in the development, progression
and prognosis of colorectal cancer were investigated. In addition,
the value of RUNX3, and miR-363 single diagnosis and two
combined diagnosis in colorectal cancer were compared to provide a
potential theoretical basis to help early clinical diagnosis and
treatment.
Patients and methods
General information
Eighty-five patients who were diagnosed with
colorectal cancer in the First Peoples Hospital of Xiaoshan
Hangzhou (Hangzhou, China) from March 2014 to July 2016 were
collected as the experiment group which included 52 cases of males,
33 cases of females and the mean age was 59.51±8.98 years. There
were 40 cases of colon and 45 cases of rectum. There were also 15
cases of highly differentiated, 52 cases of moderately
differentiated and 18 cases of poorly differentiated. Clinical
stages were: 19 cases in stage I, 29 cases in stage II, 22 cases in
stage III and 15 cases in stage IV. Additionally, 70 healthy
individuals who underwent physical examination during the same
period were included as the control group, comprising 41 males, and
29 females, with a mean age of 58.68±8.81 years.
The inclusion criteria were: i) Patients with
complete clinical and pathological data; ii) not treated with
neoadjuvant chemotherapy, radiotherapy or immunotherapy; iii) all
received tests such as blood tests, urine routine test, liver and
kidney function test electrocardiogram and others; iv) patients
were diagnosed with colorectal cancer in the postoperative
pathology report.
The exclusion criteria were: i) Patients with
autoimmune system defects; ii) non-primary tumor patients; iii)
patients with liver dysfunction or other severe organ disease; iv)
patients who are pregnant or during the period of lactation; v)
patients have mental illness or have had a family history of mental
illness.
This study was approved by the Ethics Committee of
The First Peoples Hospital of Xiaoshan Hangzhou and the
experimental content of the study was described in detail. The
subjects agreed to participate and signed an informed consent.
Blood collection
Peripheral blood (2 ml) was collected from patients
who were on empty stomach in the morning (experiment group), then
loaded into an anticoagulation tube and sent to the laboratory. In
the control group, 2 ml of fasting venous blood (peripheral blood)
was taken in the morning on the day of the physical examination.
After coagulation for 60 min (20–25°C), centrifugation at 1,006.2 ×
g for 10 min at 4°C, the supernatant was collected, avoiding
repeated freezing and thawing.
Experimental instruments and
reagents
TRIzol kit (Shanghai Shenggong Bio Co., Shanghai,
China); DNase I (Shanghai Shenggong Bio Co., Shanghai, China); cDNA
Reverse Transcription kit (Takara Biotechnology, Co., Ltd., Dalian,
China); Ultraviolet spectrophotometer (Beijing Youpu General
Technology Co., Ltd., Beijing, China), and real-time PCR kit
(Beijing Aide Lai Biotechnology Co., Ltd., Beijing, China) were
used in the study. The ABI 7500 real-time PCR detector was
purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Experimental procedures of
RT-qPCR
Total RNA in serum was extracted using TRIzol
reagent according to the protocol. The template RNA with DNase I
(RNA-free) was digested and the contamination of genomic DNA was
eliminated. Ultraviolet spectrophotometer was used for the
determination of purity and concentration, and 1.5% agarose gel
electrophoresis was used for detection of RNA integrity. The RNA
concentration was adjusted to 500 ng/µl. The transcription of RNA
samples was reversed into cDNA by using reverse transcriptase and
was conducted in strict accordance with the protocol. The SYBR
RT-qPCR (Thermo Fisher Scientific, Inc.) system was 20 µl, 2X Ultra
SYBR One-Step RT-qPCR buffer 10 µl, RNA template 2 µl,
nuclease-free water 5.5 µl, 1 µl each of the upstream and
downstream primers and upper enzyme mix 0.5 µl. The RT-qPCR
reaction conditions were: Pre-denatured at 95°C for 10 min,
denatured at 95°C for 15 sec, annealed and extended at 60°C for 1
min, with 40 cycles. The primers in this experiment were designed
by Primer Premier 5.0 (Premier Biosoft International, Palo Alto,
CA, USA) primer design software, generated by Tianjin Saier
Biotechnology Co., Ltd. (Tianjin, China). GAPDH was used as
an internal reference for RUNX3 and U6 was used as an
internal reference for miR-363. The specific primer sequences are
shown in Table I. The above system
configuration is strictly in accordance with the instructions. The
results showed that fluorescence signal in the process of
amplification of the cycle number Cq value started from the number
of cycles corresponding to the inflection point from the background
to the exponential growth phase. The relative expression level of
the target gene RUNX3 mRNA and miR-363 in blood was
calculated by 2−ΔCq (15).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Internal
reference | Upstream
primer | Downstream
primer |
|---|
| RUNX3 |
5′-AGGCAATGACGAGAACTACTCC-3′ |
5′-CGAAGGTCGTTGAACCTGG-3′ |
| GAPDH |
5′-GCACCGTCAAGGCTGAGAAC-3′ |
5′-ATGGTGGTGAAGACGCCAGT-3′ |
| miR-363 |
5′-ACACTCCAGCTGGGAATTGCACGGTATCCA-3′ |
5′-TGGTGTCGTGGAGTCG-3′ |
| U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
5′-AACGCTTCACGAATTTGCGT-3′ |
Observation indices
The clinical basic information between the groups
was compared. The differences of RUNX3 mRNA and miR-363
expression levels between the experiment and control groups were
observed. The correlation between the expression levels of
RUNX3 mRNA and miR-363 and clinical stage and
differentiation was analyzed in accordance with the
clinicopathological features of patients with colorectal
cancer.
Statistical analysis
The statistical analysis of the experimental data
was performed by SPSS 19.0 software system (IBM Corp., Armonk, NY,
USA). The enumeration data are expressed as [n (%)]. Chi-square
test was used for the comparison between groups and mean ± SD was
used to represent the measurement data. Paired t-test was used for
comparison between the groups. One-way analysis of variance (ANOVA)
and LSD post hoc test were used for comparisons between the means
of multiple groups. The correlation between the expression levels
of RUNX3 and miR-363 with the clinical stage and degree of
differentiation was based on the Spearman correlation coefficient.
The sensitivity and specificity of individual and combined tests
were assessed using the receiver operating curve (ROC). The
diagnostic value of RUNX3 and miR-363 combined test of
colorectal cancer was analyzed by binary logistic regression.
P<0.05 was considered to indicate a statistically significant
result.
Results
Comparison of general information
The clinical baseline information in terms of sex,
age, body mass index, smoking status, alcoholic status, diastolic
blood pressure, systolic blood pressure, white blood cell (WBC),
hemoglobin (HB), red blood cell (RBC) count and platelet (PLT)
count were collected from the experiment group and the control
group. There was no difference between the groups (P>0.05)
(Table II).
 | Table II.Comparison of clinical general data
between the groups (mean ± SD)/[n (%)]. |
Table II.
Comparison of clinical general data
between the groups (mean ± SD)/[n (%)].
|
Characteristics | Experiment group
(n=85) | Control group
(n=70) |
χ2/t | P-value |
|---|
| Sex |
|
| 0.109 | 0.74 |
|
Male | 52 (61.18) | 41 (58.67) |
|
|
|
Female | 33 (38.82) | 29 (41.43) |
|
|
| Age (years) | 59.51±8.98 | 58.68±8.87 | 0.576 | 0.57 |
| Body mass index
(kg/m2) | 20.12±2.18 | 20.32±2.09 | 0.579 | 0.56 |
| Smoking status |
|
| 1.127 | 0.29 |
|
Smoking | 45 (52.94) | 43 (61.43) |
|
|
|
Non-smoking | 40 (47.06) | 27 (38.57) |
|
|
| Alcoholic
status |
|
| 0.231 | 0.63 |
|
Alcoholic | 47 (55.29) | 36 (51.43) |
|
|
|
Non-alcoholic | 38 (44.71) | 34 (48.57) |
|
|
| Diastolic blood
pressure (mmHg) | 76.23±12.13 | 77.36±12.45 | 0.570 | 0.57 |
| Systolic blood
pressure (mmHg) | 113.34±18.24 | 117.39±19.35 | 2.321 | 0.98 |
| WBC
(×109/l) | 5.89±3.65 | 6.21±3.51 | 0.553 | 0.58 |
| HB (gm/dl) | 11.87±1.91 | 12.59±2.16 | 1.865 | 0.06 |
| PLT
(×109/l) | 153.67±21.81 | 156.17±22.71 | 0.697 | 0.49 |
| RBC
(1012/l) | 4.76±0.61 | 4.64±0.58 | 1.246 | 0.21 |
Comparison of RUNX3 and miR-363
expression levels between the groups
RT-qPCR was used to detect the expression of
RUNX3 and miR-363. Τhe expression of RUNX3 in the
experiment group was significantly lower than that in the control
group, and the difference between the groups was statistically
significant (P<0.05) (Table
III). The expression level of miR-363 was also significantly
lower than that in the control group (P<0.05) (Table III).
 | Table III.Comparison of the expression levels
of RUNX3 and miR-363 in the groups (mean ± SD). |
Table III.
Comparison of the expression levels
of RUNX3 and miR-363 in the groups (mean ± SD).
| Item | No. of cases | RUNX3 | miR-363 |
|---|
| The experiment
group | 85 | 0.93±0.38 | 0.47±0.21 |
| The control
group | 70 |
2.18±0.87a |
1.57±0.68a |
| t |
| 11.94 | 19.77 |
| P-value |
| <0.001 | <0.001 |
Association between the expression of
RUNX3 and miR-363 with clinicopathological characteristics in
patients with colorectal cancer
The clinical information of 85 patients with colon
cancer was collected for comparison. Some relevant data showed that
the expression level of RUNX3 in the blood of patients with
colorectal cancer was not significantly associated with sex, age
and tumor location (P>0.05). However, it was associated with
tumor size, degree of differentiation, lymph node metastasis, depth
of invasion and clinical stages (P<0.05). The expression level
of miR-363 was not associated with sex, age tumor location and
tumor size (P>0.05). It was correlated with the degree of
differentiation, lymph node metastasis, depth of invasion and
clinical stages (P<0.05) (Table
IV).
 | Table IV.Association between RUNX3,
miR-363 and clinicopathological characteristics (mean ± SD). |
Table IV.
Association between RUNX3,
miR-363 and clinicopathological characteristics (mean ± SD).
| Characteristic | No. of cases | RUNX3 | F/t | P-value | miR-363 | F/t | P-value |
|---|
| Sex |
|
| 0.699 | 0.49 |
| 1.379 | 0.17 |
|
Male | 52 | 0.98±0.37 |
|
| 0.64±0.21 |
|
|
|
Female | 33 | 0.92±0.41 |
|
| 0.58±0.17 |
|
|
| Age (years) |
|
| 0.729 | 0.47 |
| 1.111 | 0.27 |
|
<50 | 39 | 0.95±0.39 |
|
| 0.57±0.19 |
|
|
|
≥50 | 46 | 1.02±0.48 |
|
| 0.62±0.22 |
|
|
| Tumor
locations |
|
| 1.776 | 0.08 |
| 1.837 | 0.07 |
|
Colon | 40 | 0.93±0.35 |
|
| 0.54±0.17 |
|
|
|
Rectum | 45 | 1.08±0.42 |
|
| 0.61±0.18 |
|
|
| Tumor sizes
(cm) |
|
| 2.346 | 0.02 |
| 0.708 | 0.48 |
|
<5 | 57 | 1.13±0.54 |
|
| 0.51±0.19 |
|
|
| ≥5 | 28 |
0.86±0.40c |
|
| 0.48±0.17 |
|
|
|
Differentiation |
|
| 17.91 | <0.001 |
| 13.55 | <0.001 |
| Highly
differentiated | 15 | 1.43±0.49 |
|
| 0.63±0.23 |
|
|
| Medium
differentiated | 52 |
1.03±0.31a |
|
|
0.47±0.17a |
|
|
| Poorly
differentiated | 18 |
0.75±0.16a,b |
|
|
0.32±0.10a,b |
|
|
| Lymph node
metastasis or non-metastasis |
|
| 3.809 | <0.001 |
| 4.340 | <0.001 |
|
Metastasis | 49 | 0.79±0.38 |
|
| 0.43±0.14 |
|
|
|
Non-metastasis | 36 |
1.20±0.61c |
|
|
0.59±0.20c |
|
|
| Infiltration
depth |
|
| 4.849 | <0.001 |
| 4.807 | <0.001 |
|
T1+T2 | 28 | 1.18±0.43 |
|
| 0.60±0.17 |
|
|
|
T3+T4 | 57 |
0.81±0.27c |
|
|
0.44±0.13c |
|
|
| Clinical
stages |
|
| 8.896 | <0.001 |
| 9.498 | <0.001 |
| I +
II | 48 | 1.48±0.63 |
|
| 0.69±0.21 |
|
|
| III +
IV | 37 |
0.53±0.13c |
|
|
0.30±0.15c |
|
|
Correlation between the expression
levels of RUNX3 and miR-363 with clinical stages and
differentiation
The expression of RUNX3 and miR-363 was
detected by RT-qPCR. Correlation between the expression levels of
RUNX3 and miR-363 with the degree of differentiation were
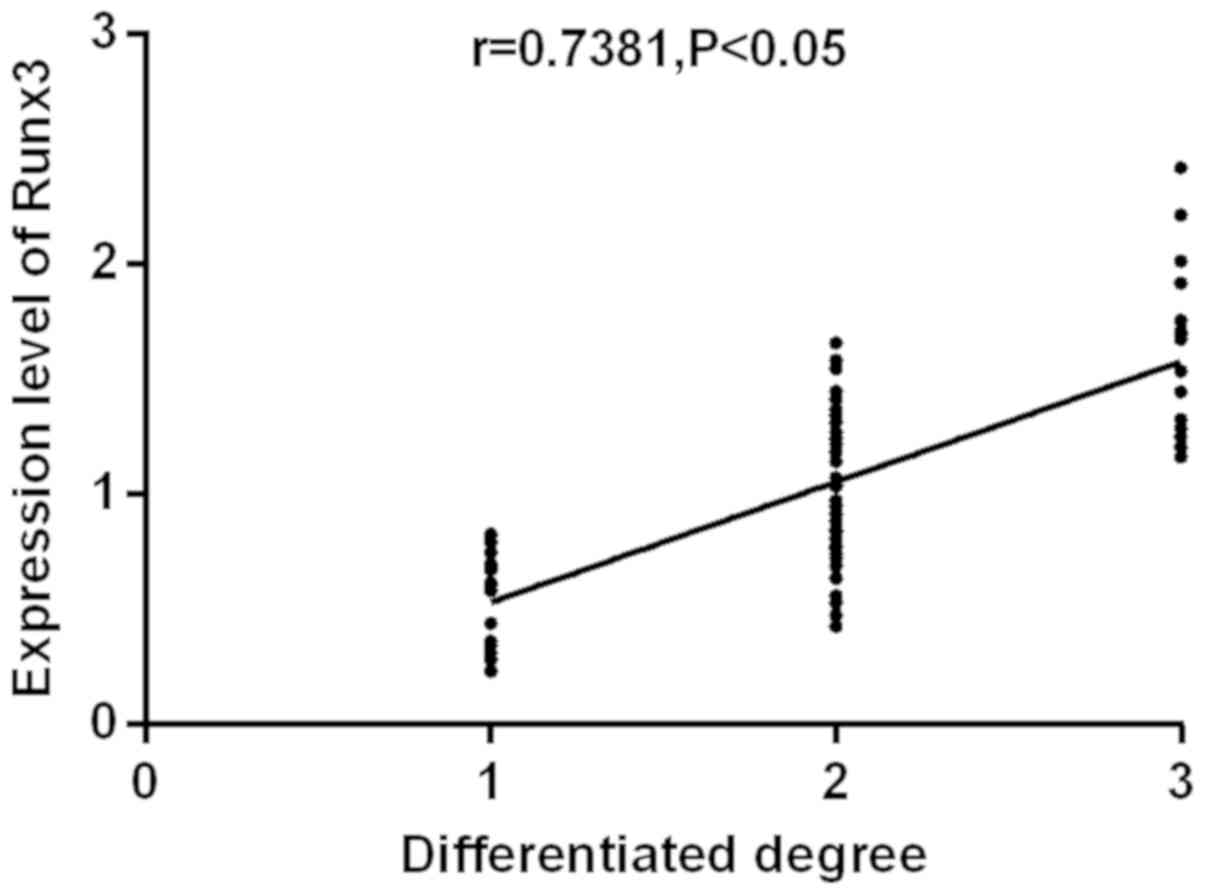
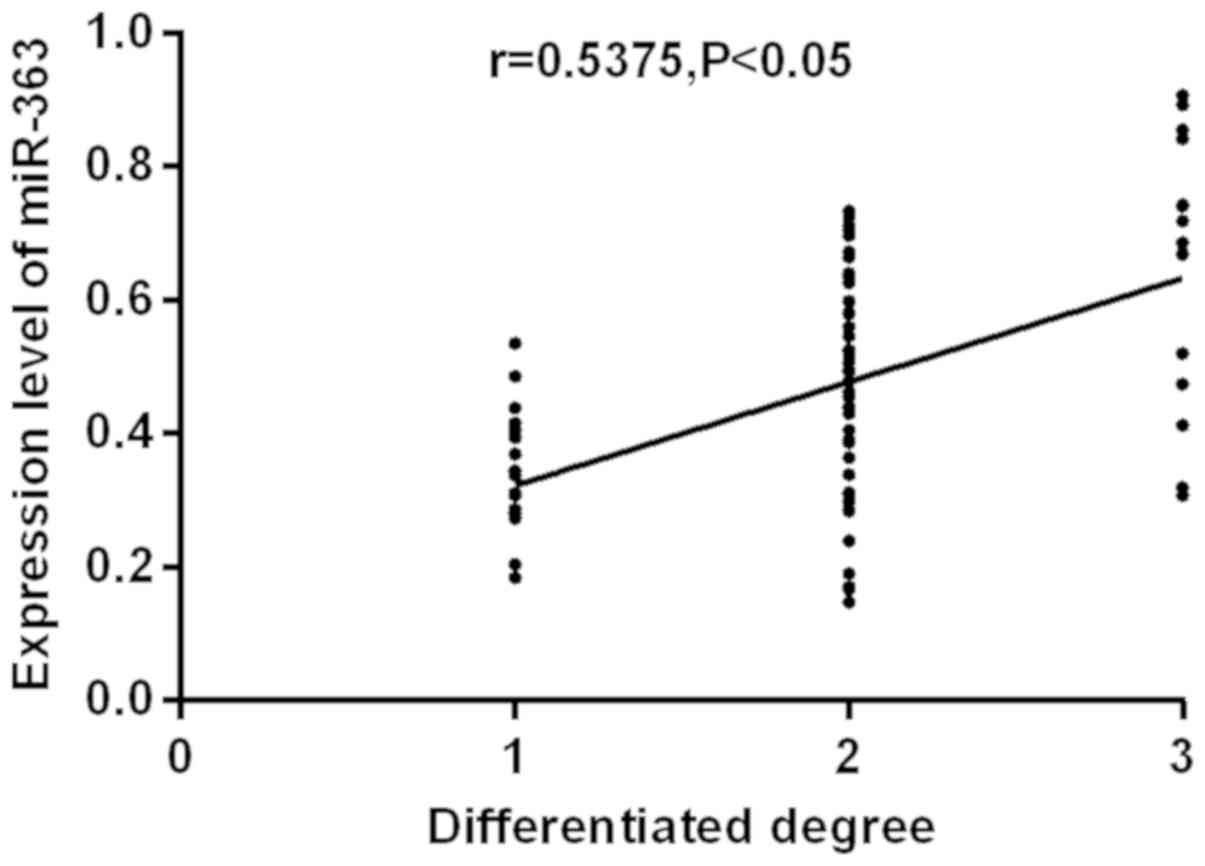
analyzed (Figs. 1 and 2). RUNX3 and miR-363 were
significantly positively correlated with the degree of
differentiation (r=0.7381, r=0.5375; P<0.05). The higher the
degree of differentiation, the higher the expression level of
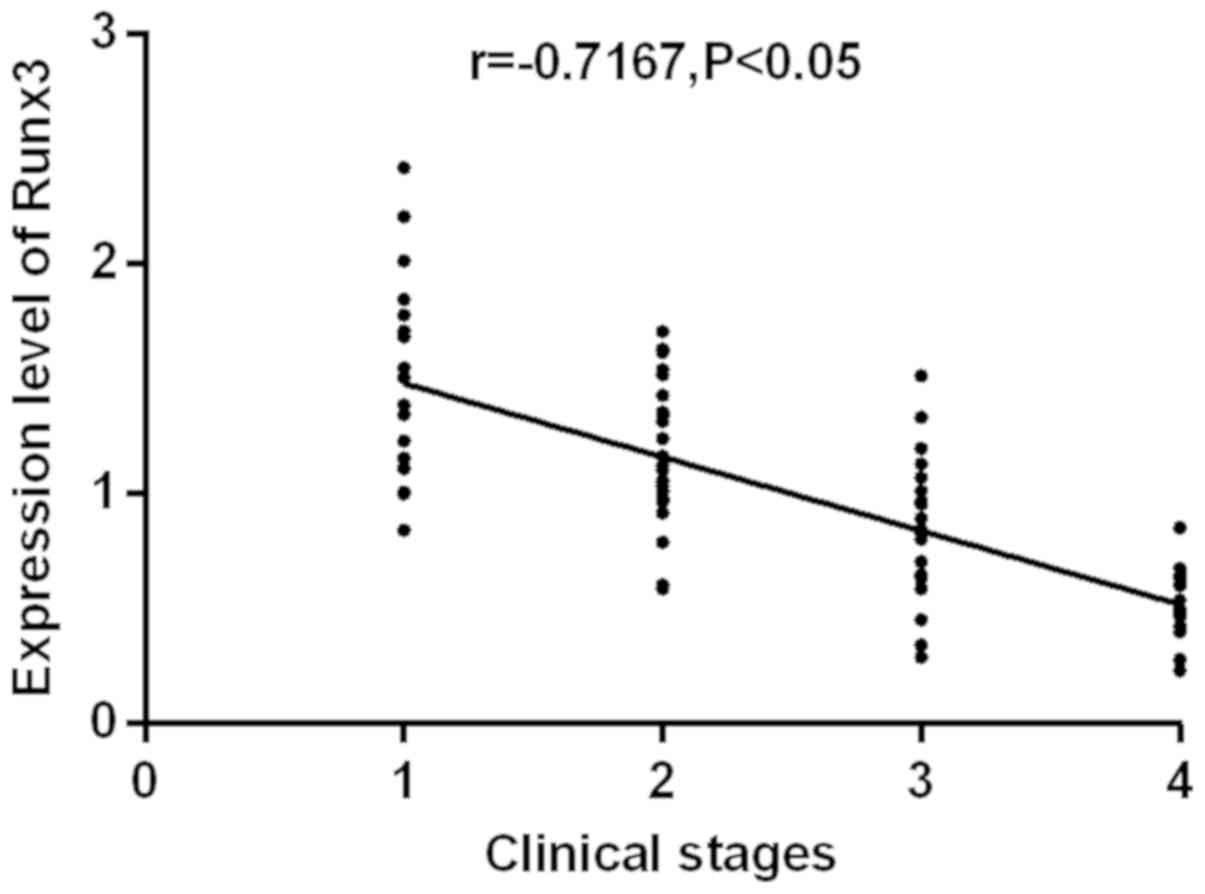
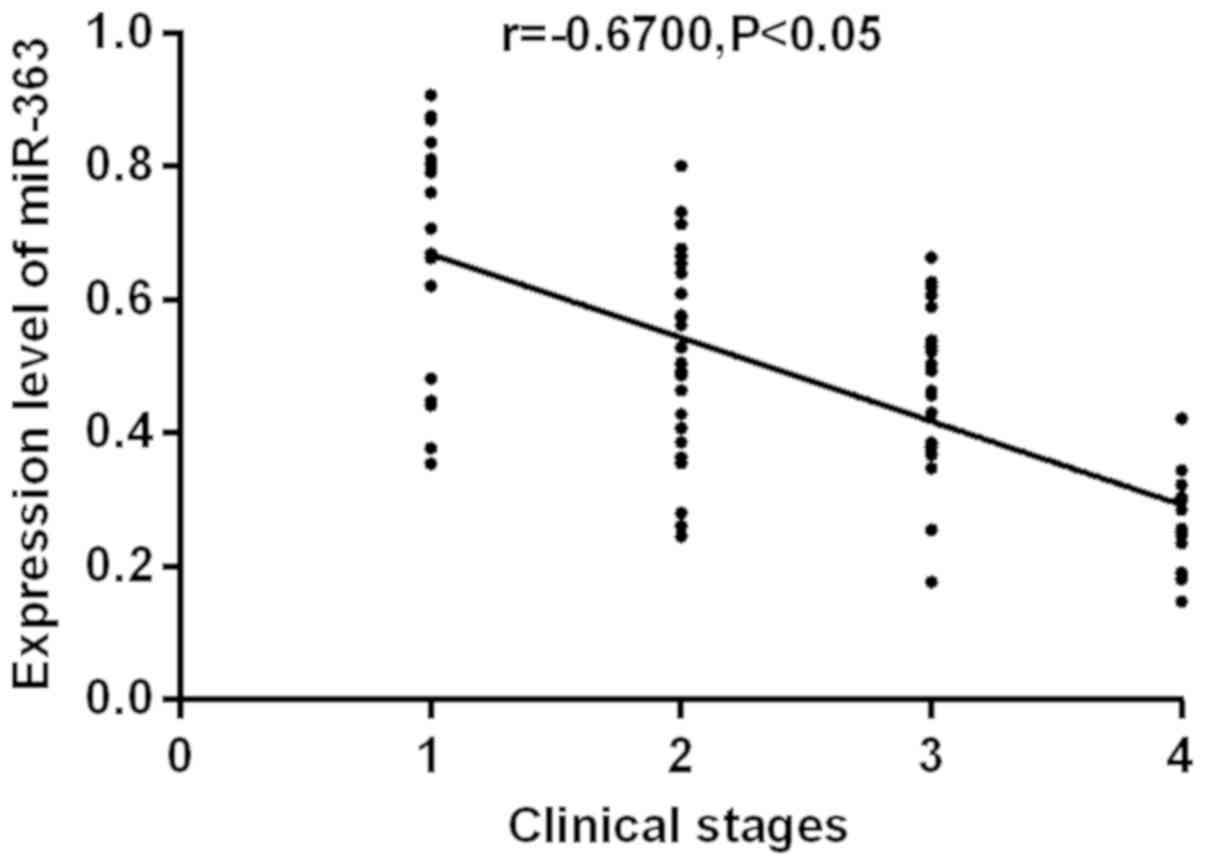
RUNX3 and miR-363. The correlation between the expression
levels of RUNX3 and miR-363 with clinical stages were
analyzed (Figs. 3 and 4). RUNX3 and miR-363 were
significantly negatively correlated with clinical stages
(r=−0.7167, −0.6700; P<0.05). As clinical staging continues to
increase, the expression levels of RUNX3 and miR-363
gradually decrease.
Comparison of the value of RUNX3 and
miR-363 single test and two combined tests for the diagnosis of
colorectal cancer
The comparison of the diagnostic value in this part
was taken from the participation in the discussion between the
experiment group and the normal control group. Then the
sensitivity, specificity and Youden indices in the single and
combined tests were compared. Sensitivity levels from high to low
from the combined test was (88.57%), miR-363 (85.71%) and
RUNX3 (78.57%). The specificity level from high to low in
the combined test was (96.47%), miR-363 (95.29%) and RUNX3
(80.0%). The results suggest that RUNX3+miR-363 combined
test has the highest sensitivity and specificity levels. Among
them, Youden had the largest indices in the combined test. For the
screening of colorectal cancer, the larger the Youden index, the
better the detection and the higher the authenticity (Table V).
 | Table V.Comparison of the value of
RUNX3 and miR-363 single test and two combined tests for
colorectal cancer diagnosis. |
Table V.
Comparison of the value of
RUNX3 and miR-363 single test and two combined tests for
colorectal cancer diagnosis.
| Test | Sensitivity | Specificity | Youden indices |
|---|
| RUNX3 | 78.57% | 80.00% | 0.59 |
| miR-363 | 85.71% | 95.29% | 0.81 |
|
RUNX3+miR-363 | 88.57% | 96.47% | 0.85 |
Evaluation of RUNX3 and miR-363 single
test and combined test in the diagnosis of colorectal cancer
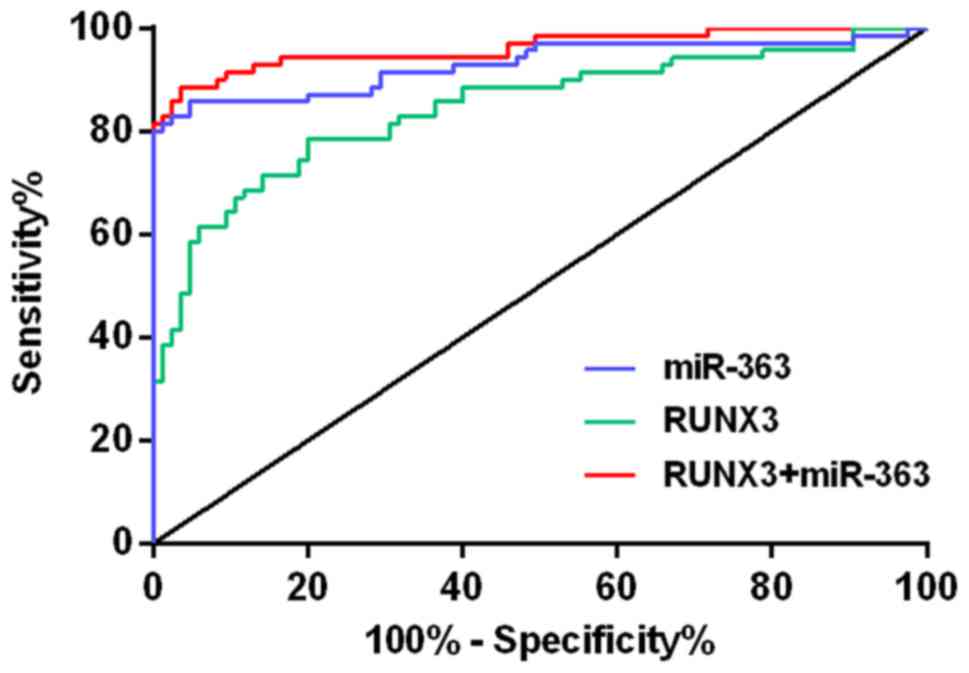
ROC curves were plotted based on their sensitivity
and specificity levels for both single and combined tests. In the
area under the ROC curve (AUC) the larger the AUC, the greater the
diagnostic value. Combination diagnosis of AUC is greater than
single diagnosis. The optimal thresholds for RUNX3 and
miR-363 are 1.367 and 0.752, respectively, the diagnostic
efficiency is the highest at this time-point (Table VI and Fig. 5).
 | Table VI.Evaluation of RUNX3 and
miR-363 single test and two combined tests for the diagnosis of
colorectal cancer. |
Table VI.
Evaluation of RUNX3 and
miR-363 single test and two combined tests for the diagnosis of
colorectal cancer.
|
|
|
|
| 95% Confidence
interval |
|---|
|
|
|
|
|
|
|---|
| Detection
method | Optimal critical
value | AUC | P-value | Upper limit | Lower limit |
|---|
| RUNX3 | 1.367 | 0.845 | <0.001 | 0.780 | 0.909 |
| miR-363 | 0.752 | 0.930 | <0.001 | 0.883 | 0.976 |
|
RUNX3+miR-363 | – | 0.961 | <0.001 | 0.930 | 0.992 |
Discussion
Colorectal cancer has become the third most common
cancer in the world. It is the fourth malignant tumor that causes
human death (16). It has a high
morbidity and mortality rates and there is a significant
heterogeneity among individual patients (17). The main prevention and treatment of
colorectal cancer is to control its incidence and reduce its
mortality rates. Therefore, it is very necessary to find a
sensitive and specific index to judge the risk of occurrence,
development, metastasis and recurrence of colorectal cancer
patients, thus an individualized treatment plan has to be developed
(18). Currently, pathogenesis of
colorectal cancer is not yet fully understood. There are some
reports showing that its carcinogenesis may originate from
intestinal mucosal epithelial cells, a series of genetic
alterations are involved in the sequence changes of ‘abdominal
epithelial dysplasia - adenoma - carcinoma’ (19).
RUNX3, as a tumor suppressor gene, a member
of the RUNX3 family of transcription factors, it has an
important effect in cell differentiation and apoptosis, and cell
cycle regulation (20). An important
reason for the downregulation of RUNX3 expression was due to
methylation of CpG islands in the RUNX3 promoter region. Its
anticancer mechanism is mainly related to the TGF-β pathway which
induces growth inhibition and apoptosis (21,22).
However, the TGF-β/Smad signaling pathway is involved in the
development and progression of colorectal cancer (23). Studies have shown that (24) RUNX3 gene may be a key target
for TGF-β signaling pathways, participated in the negative
regulation of epithelial cells by the TGF-β pathway, promoter
region methylation and heterozygosity loss was the main mechanism.
miR-363 is a novel small molecule RNA that has an important effect
in the occurrence and development of some tumors. It was first
discovered that the expression of miR-363 was in the head and neck
squamous cell carcinoma of lymph node metastasis, and miR-363 has a
low expression in tissues. Furthermore, it also showed a low
expression level in some highly invasive tumor cell lines. Further
studies have found that miR-363 affects the invasion and metastasis
of tumor cells mainly through targeted inhibition of podoplanin
(PDPN) (25,26).
In the present study, by comparing the information
of the general clinical baseline, it was found that there was no
difference between the experiment group and the control group. The
interference from other factors on the experimental results were
excluded, to ensure reliable results in this research. The data
showed that the expression level of RUNX3 in the blood of
the experiment group was lower than the control group. Ku et
al (10) used RT-qPCR to detect
cancer cell lines in colon cancer patients. It was found that
approximately half of the colon cancer cell lines had no expression
or decreased expression of RUNX3. The expression level of
miR-363 in the blood of the experiment group was also significantly
lower than the control group (P<0.05). Xu et al (27) detected the expression of miR-363 in
colorectal cancer tissues by RT-qPCR. Compared with normal
colorectal epithelial cell lines, miR-363 expression was
significantly downregulated in six colorectal cancer cell lines.
The differences were statistically significant (P<0.05) and
consistent with the results of this research. The study used
peripheral blood, which is easier to obtain than tissue. Through
the research of the relationship between the expression of
RUNX3 and miR-363 with clinicopathological features in
patients with colorectal cancer, the results have shown that
RUNX3 was associated with tumor size, degree of
differentiation, lymph node metastasis, depth of invasion and
clinical stages (P<0.05), which is consistent with the results
of Watanabe et al (28).
However, the results of Mu et al (29) found that RUNX3 expression was
not associated with tumor size, differentiation and histological
types. There are some differences between this research and the
results of Watanabe et al, so further research is required.
We should be aware of that with the development of colorectal
cancer, the expression level of RUNX3 gradually decreases.
In the development of colorectal cancer, the loss of RUNX3
expression presents a cumulative process. Therefore, RUNX3
can be used as a new marker to judge the clinical stages of
colorectal cancer. Moreover, Soong et al (20) found that the positive expression of
RUNX3 in the nucleus of colon cancer tissues is related to
the stage of disease. miR-363 is associated with differentiation,
lymph node metastasis, depth of invasion and clinical stages
(P<0.05). Also studies have shown, the comparison of miR-363
among different pathological differentiation, TNM stages and lymph
node metastasis in patients with colorectal cancer, the difference
was statistically significant and consistent with our research
(14). It has been shown that the
clinical stage and differentiation of cervical cancer are related
to miR-363 and that there is scarce researches on colorectal cancer
(30). Therefore, miR-363 requires a
deeper study on colorectal cancer. The results of this study showed
that RUNX3 and miR-363 are significantly positively
correlated with the degree of differentiation. Both genes have an
important effect in the occurrence and development of colorectal
cancer. However, RUNX3 and miR-363 showed a significant
negative correlation with clinical stage. As the disease worsens,
the expression levels of RUNX3 and miR-363 are
downregulated. This indicates that RUNX3 and miR-363 can be
used to determine the severity of patients condition. Based on
relevant reports, there is scarce research on the diagnosis of
colorectal cancer with RUNX3 and miR-363 and no research was
found on the two combined diagnosis. The present study compared the
diagnostic significance of RUNX3 and miR-363 single and
combined tests in colorectal cancer, it was found that the
sensitivity and specificity levels of the combined diagnosis of
RUNX3 and miR-363 were higher than the single test. Also the
combined test has the largest Youden indices, for the screening of
colorectal cancer, the larger the Youden index, the better the
detection effect and the higher the authenticity. By plotting the
ROC curve, the AUC shows that the combined diagnosis is larger than
the single diagnosis, and the larger the AUC, the greater the
diagnostic value. The above two points indicate that the value of
the combined test of RUNX3 and miR-363 is higher than the
single test.
In this study, the expression levels of RUNX3
and miR-363 in the blood of patients were studied from various
aspects, which provides a reference for clinical research. However,
there is a lack of follow-up in patients after surgery, so the
lifestyle and prognosis of patients need to be explored in future
studies. Also the exact mechanism of action of RUNX3 and
miR-363 in the development of colorectal cancer remains to be
further studied. Searches have shown that there only exist a few
relevant documents on the combined tests of RUNX3 and
miR-363, so this aspect requires strengthening in subsequent
research.
In summary, the expression of RUNX3 gene and
miR-363 in serum of patients with colorectal cancer is lower than
normal people, the low expression of RUNX3 and miR-363 is
closely related to various biological behavior of colorectal cancer
and shows potential as a reference for the evaluation of patients
with colorectal cancer, and to provide a guidance for the treatment
of colorectal cancer. Combined test of RUNX3 and miR-363 has
an important significance in the diagnosis and treatment evaluation
of colorectal cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors contributions
GL conceived and designed the study, collected the
patients data, analyzed and interpreted the patient data regarding
the colorectal cancer, wrote the manuscript, and read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Peoples Hospital of Xiaoshan Hangzhou (Hangzhou, China).
Signed informed consents were obtained from the patients or the
guardians.
Patient consent for publication
Not applicable.
Competing interests
The author declares no competing interests.
References
|
1
|
Wong MC, Lam AT, Li DK, Lau JT, Griffiths
SM and Sung JJ: Factors associated with practice of colorectal
cancer screening among primary care physicians in a Chinese
population: A cross-sectional study. Cancer Epidemiol. 33:201–206.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matsuoka K, Watanabe N and Nakamura K:
O-glycosylation of a precursor to a sweet potato vacuolar protein,
sporamin, expressed in tobacco cells. Plant J. 8:877–889. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng R, Zeng H, Zhang S, Chen T and Chen
W: National estimates of cancer prevalence in China, 2011. Cancer
Lett. 370:33–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang XQ, Tang ZX, Yu D, Cui SJ, Jiang YH,
Zhang Q, Wang J, Yang PY and Liu F: Epithelial but not stromal
expression of collagen alpha-1(III) is a diagnostic and prognostic
indicator of colorectal carcinoma. Oncotarget. 7:8823–8838.
2016.PubMed/NCBI
|
|
6
|
Zhang ZJ, Zheng ZJ, Kan H, Song Y, Cui W,
Zhao G and Kip KE: Reduced risk of colorectal cancer with metformin
therapy in patients with type 2 diabetes: A meta-analysis. Diabetes
Care. 34:2323–2328. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhong LL, Chen HY, Cho WC, Meng XM and
Tong Y: The efficacy of Chinese herbal medicine as an adjunctive
therapy for colorectal cancer: A systematic review and
meta-analysis. Complement Ther Med. 20:240–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim HJ, Park J, Lee SK, Kim KR, Park KK
and Chung WY: Loss of RUNX3 expression promotes cancer-associated
bone destruction by regulating CCL5, CCL19 and CXCL11 in non-small
cell lung cancer. J Pathol. 237:520–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Estécio MR, Maddipoti S, Bueso-Ramos C,
DiNardo CD, Yang H, Wei Y, Kondo K, Fang Z, Stevenson W, Chang KS,
et al: RUNX3 promoter hypermethylation is frequent in leukaemia
cell lines and associated with acute myeloid leukaemia inv(16)
subtype. Br J Haematol. 169:344–351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ku JL, Kang SB, Shin YK, Kang HC, Hong SH,
Kim IJ, Shin JH, Han IO and Park JG: Promoter hypermethylation
downregulates RUNX3 gene expression in colorectal cancer cell
lines. Oncogene. 23:6736–6742. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and
Li X: MiR-363-3p inhibits the epithelial-to-mesenchymal transition
and suppresses metastasis in colorectal cancer by targeting Sox4.
Biochem Biophys Res Commun. 474:35–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cohen SJ, Cohen RB and Meropol NJ:
Targeting signal transduction pathways in colorectal cancer - more
than skin deep. J Clin Oncol. 23:5374–5385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seymour MT, Maughan TS, Ledermann JA,
Topham C, James R, Gwyther SJ, Smith DB, Shepherd S, Maraveyas A,
Ferry DR, et al FOCUS Trial Investigators; National Cancer Research
Institute Colorectal Clinical Studies Group, : Different strategies
of sequential and combination chemotherapy for patients with poor
prognosis advanced colorectal cancer (MRC FOCUS): A randomised
controlled trial. Lancet. 370:143–152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Croitoru ME, Cleary SP, Di Nicola N, Manno
M, Selander T, Aronson M, Redston M, Cotterchio M, Knight J, Gryfe
R, et al: Association between biallelic and monoallelic germline
MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst.
96:1631–1634. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Soong R, Shah N, Peh BK, Chong PY, Ng SS,
Zeps N, Joseph D, Salto-Tellez M, Iacopetta B and Ito Y: The
expression of RUNX3 in colorectal cancer is associated with disease
stage and patient outcome. Br J Cancer. 100:676–679. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan SH, Ida H, Lau QC, Goh BC, Chieng WS,
Loh M and Ito Y: Detection of promoter hypermethylation in serum
samples of cancer patients by methylation-specific polymerase chain
reaction for tumour suppressor genes including RUNX3. Oncol Rep.
18:1225–1230. 2007.PubMed/NCBI
|
|
22
|
Smith E, De Young NJ, Pavey SJ, Hayward
NK, Nancarrow DJ, Whiteman DC, Smithers BM, Ruszkiewicz AR,
Clouston AD, Gotley DC, et al: Similarity of aberrant DNA
methylation in Barretts esophagus and esophageal adenocarcinoma.
Mol Cancer. 7:752008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nishio M, Sakakura C, Nagata T, Komiyama
S, Miyashita A, Hamada T, Kuryu Y, Ikoma H, Kubota T, Kimura A, et
al: RUNX3 promoter methylation in colorectal cancer: Its
relationship with microsatellite instability and its suitability as
a novel serum tumor marker. Anticancer Res. 30:2673–2682.
2010.PubMed/NCBI
|
|
24
|
Kang KA, Kim KC, Bae SC and Hyun JW:
Oxidative stress induces proliferation of colorectal cancer cells
by inhibiting RUNX3 and activating the Akt signaling pathway. Int J
Oncol. 43:1511–1516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan
M, Wu X and Chen W: Dysregulated miR-363 affects head and neck
cancer invasion and metastasis by targeting podoplanin. Int J
Biochem Cell Biol. 45:513–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo X, Burwinkel B, Tao S and Brenner H:
MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer
Epidemiol Biomarkers Prev. 20:1272–1286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu X, Wu X, Wu S, Jiang Q, Liu H, Chen R
and Sun Y: Study on miR-490-5p and miR-363 as novel biomarkers for
the diagnosis of colorectal cancer. Zhonghua Wei Chang Wai Ke Za
Zhi. 17:45–50. 2014.(In Chinese). PubMed/NCBI
|
|
28
|
Watanabe T, Kobunai T, Ikeuchi H, Yamamoto
Y, Matsuda K, Ishihara S, Nozawa K, Iinuma H, Kanazawa T, Tanaka T,
et al: RUNX3 copy number predicts the development of UC-associated
colorectal cancer. Int J Oncol. 38:201–207. 2011.PubMed/NCBI
|
|
29
|
Mu WP, Wang J, Niu Q, Shi N and Lian HF:
Clinical significance and association of RUNX3 hypermethylation
frequency with colorectal cancer: A meta-analysis. OncoTargets
Ther. 7:1237–1245. 2014. View Article : Google Scholar
|
|
30
|
Tsuji S, Kawasaki Y, Furukawa S, Taniue K,
Hayashi T, Okuno M, Hiyoshi M, Kitayama J and Akiyama T: The
miR-363-GATA6-Lgr5 pathway is critical for colorectal
tumourigenesis. Nat Commun. 5:31502014. View Article : Google Scholar : PubMed/NCBI
|