Introduction
Ovarian cancer is one of the common gynecological
tumors, whose incidence and mortality rates are fairly high among
gynecological tumors (1). At
present, studies have focused on genetic detection and targeted
therapy worldwide. Some targeted drugs have satisfactory effects in
treating ovarian cancer and prolonging life-span of patients
(2,3).
It has been recently proven that laminin subunit α3
(LAMA3) plays an important role in the occurrence of various
types of cancer that encode laminin. Moreover, LAMA3 is essential
for the formation and function of basement membrane and plays an
additional role in regulating cell migration and mechanical signal
transduction (4). This gene encodes
an α-subunit and can regulate keratinocyte growth factor, epidermal
growth factor and insulin-like growth factor (5,6).
Currently, research on this gene mostly focuses on gastric cancer,
albeit some studies have focused on breast and prostate cancer
(7). However, there are few studies
on LAMA3 in ovarian cancer. In particular, the
characteristics of this gene in the occurrence and development of
lung cancer remain unclear (8,9).
In this study, the correlation of LAMA3 gene
expression with the onset and prognosis of ovarian cancer was
investigated in order to provide references for clinical diagnosis
and treatment.
Patients and methods
Study subjects
A total of 210 ovarian cancer patients who received
surgical resection in West China Second Hospital (Chengdu, China)
from March 2011 to March 2013 were randomly selected. The patients
were aged 48.34±12.43 years on average, including 65 patients in
stage I, 43 patients in stage II, 53 patients in stage III and 49
patients in stage IV. Another 160 non-ovarian cancer patients who
needed ovariectomy were also selected, and they were aged
49.76±13.45 years on average.
This study was approved by the Ethics Committee of
West China Second Hospital, and patients and their families were
informed of the study purpose and agreed and signed informed
consent.
RNA extraction and qPCR
A comparison of LAMA3 gene expression among
carcinoma tissues, para-carcinoma tissues and non-carcinoma normal
tissues obtained from ovarian cancer patients was carried out via
quantitative polymerase chain reaction (qPCR)
Ovarian cancer, para-carcinoma and non-carcinoma
tissues were ground with liquid nitrogen, and ribonucleic acid
(RNA) was extracted from samples using TRIzol in strict accordance
with the protocol. One microgram RNA was taken and reverse
transcribed into complementary deoxyribonucleic acid (cDNA)
according to instructions of the reverse transcriptase kit. The
concentration of cDNA was adjusted, followed by detection of the
messenger RNA (mRNA) level using the CFX 96 PCR instrument (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) according to instructions of
the SYBR® Premix Ex Taq™ II kit. Reaction conditions are
as follows: 95°C for 2 min, 94°C for 15 sec, 50°C for 25 sec, a
total of 40 cycles. The corresponding primer sequences are shown in
Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene name | Primer sequences |
|---|
| LAMA3 | F:
5′-ACCCAGGCCAAGGACCTGAGG-3′ |
|
| R:
5′-GTGTTGCCCGATTAACATTG-3′ |
| β-actin | F:
5′-GTGGACATCCGCAAAGAC-3′ |
|
| R:
5′-GAAAGGGTGTAACGCAACTA-3′ |
Methylation detection of tissue
samples
Genomic DNA was extracted using the tissue DNA
extraction kit (Tianjin Genetic Biotech, Beijing, China). The
samples were added into the BDS Hypersil C18 column of
high-performance liquid chromatographic instrument (Shimadzu,
Kyoto, Japan) using the microsyringe, followed by elution under low
temperature using a mixed solution of methanol, sodium
pentanesulfonate and triethylamine as the mobile phase at a flow
rate of 1 l/min, ultraviolet wavelength of 273 nm and sensitivity
of 0.01 AUFS. The standard samples of deoxycytidine and
deoxymethylcytosine were used as controls, and the DNA methylation
content in samples was detected. It was measured for 3 times in
each sample and the average was taken.
Single nucleotide polymorphism (SNP)
typing of rs12373237 via multiple PCR
The primer sequences at the SNP site and its TaqMan
probe sequences were designed using Oligo7.0 (Table II), and primers were synthesized by
Sangon, Shanghai. One microliter DNA solution and 1.2 µl primer
solution were prepared (including 0.4 µl forward primers, 0.4 µl
reverse primers and 0.4 µlprobe primers) and added into the
pre-prepared 17.8 µl TransStart Probe PCR SuperMix (Beijing
TransGen Biotech Co., Ltd.) vibrated slightly, mixed evenly and
placed into the CFX96 fluorescence quantitative PCR instrument
(Bio-Rad Laboratories, Inc.). Three repeated wells were set for
each sample, diethylpyrocarbonate (DEPC)-treated water was used as
the negative control, and the positive plasmid containing this
sequence (synthesized by Sangon, Shanghai, China) was used as the
positive control. In terms of genotype determination, the genotype
close to the abscissa was wild-type homozygote, which when close to
ordinate was mutant-type homozygote, and when close to 45° line was
heterozygote.
 | Table II.rs12373237 primer sequences. |
Table II.
rs12373237 primer sequences.
| SNP | Primer sequence | Probe sequence |
|---|
| rs12373237 | F:
5′-ATGATAGCGATTCTCCTCCTC-3′ | FAM:
5′-ACATAGGTCTACTTTTTTGG-3′ |
|
| R:
5′-AACAGGTCAGCGGGAACCTTG-3′ | VIC:
5′-ACATAGGTCCACTTTTTTGG-3′ |
Immunohistochemistry
Immunohistochemical detection of laminin expression
was carried out as previously described (10). Criteria for positive cells in stained
samples: i) There are brown yellow particles, and ii) the staining
intensity is higher than that of non-specific staining in the field
of view. The tumor cells and interstitial endothelial cells were
observed and positioned. Five different fields of view (×400) were
selected to observe each sample section. The relative expression
level was: The number of positive cells >50% (3 points), 25–49%
(2 points), 1–24% (1 point) and <1% (0 point). The staining
intensity was: deep staining (3 points), moderate staining (2
points) and light staining (1 point). The two scores were added up
as a basis of determining the expression level, and a total score
≤3 points indicated a low expression, while >3 points indicated
high expression.
Statistics of 5-year survival status of patients via
follow-up. Patients were followed up via telephone or visit every
month, the recovery condition of patients was inquired, the number
of recurrence and death was recorded, and the 5-year
recurrence-free survival rate and overall survival rate were
calculated.
Statistical analysis
The Statistical Product and Service Solutions (SPSS)
19.0 software was used for data processing. Measurement data were
expressed as mean ± SD. The t-test and one-way analysis of variance
and LSD post hoc test were used for the comparison of means.
Chi-square test was used for enumeration data. Hardy Weinberg
equilibrium law was used for genotype distribution analysis.
Survival analysis was determined using the Kaplan-Meiersurvival
curve and log-rank test. P<0.05 indicates that the difference
was statistically significant.
Results
LAMA3 gene expression level in ovarian
patients
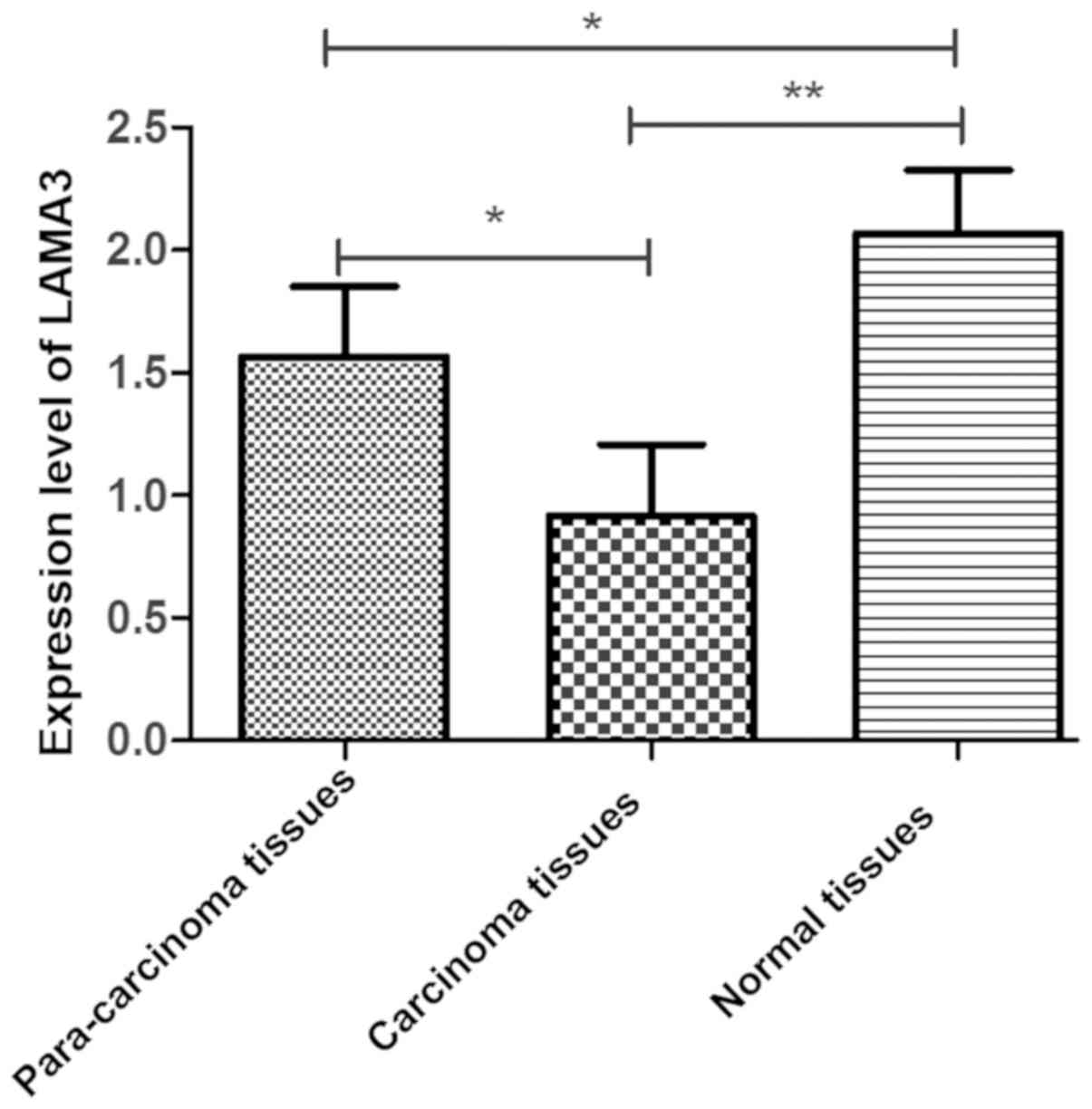
Comparison of LAMA3 gene expression level
among carcinoma tissues, para-carcinoma tissues and non-carcinoma
normal tissues in ovarian cancer patients was carried out. The
expression level of LAMA3 was lower in carcinoma tissues
than those in normal and para-carcinoma tissues (P<0.05).
Moreover, the expression level of LAMA3 was lower in
para-carcinoma tissues than that in normal tissues (P<0.05)
(Fig. 1).
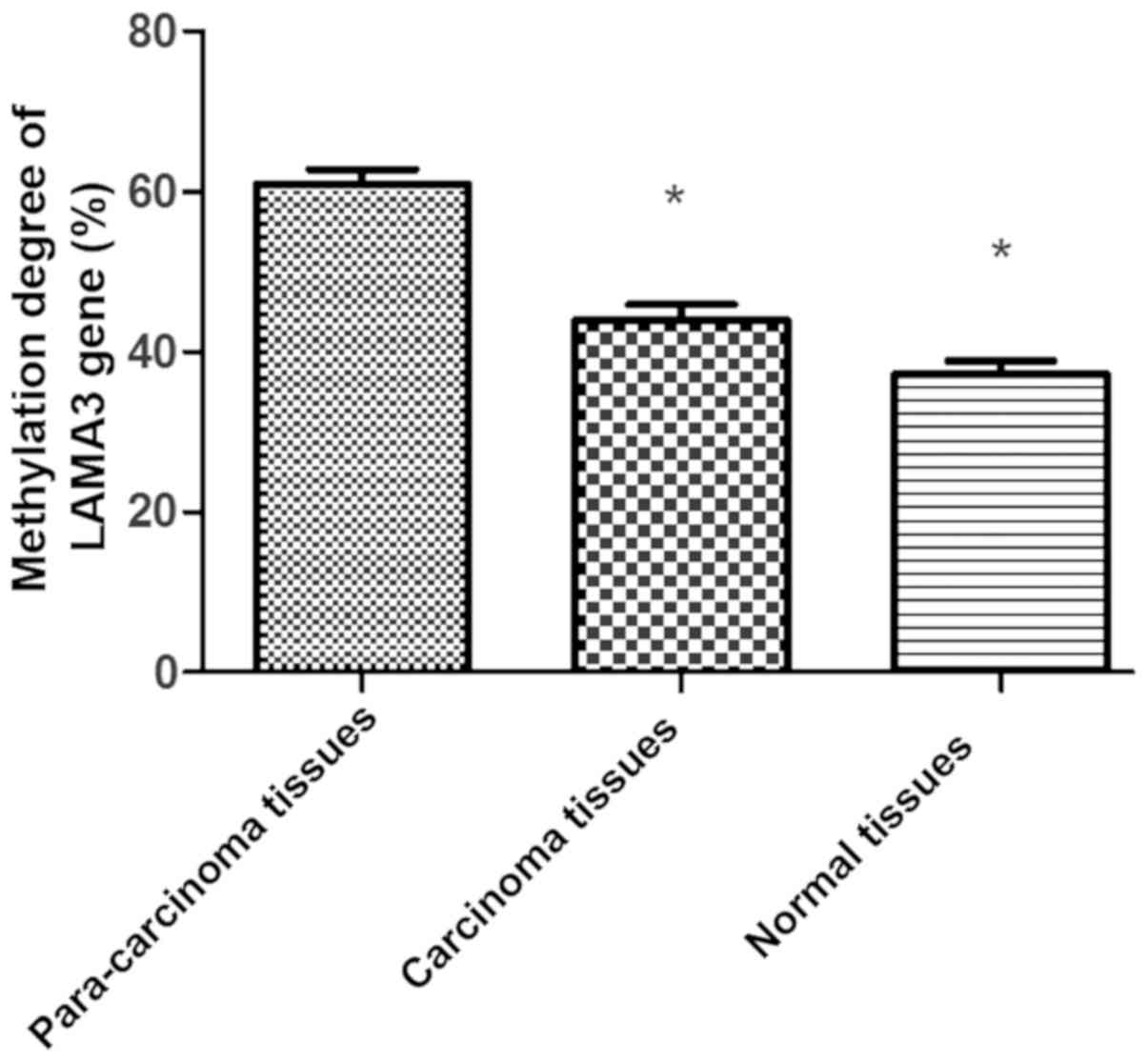
Comparison of methylation degree of
LAMA3 gene
The methylation degree was lower in carcinoma
tissues and normal tissues than that in para-carcinoma tissues
(P<0.05) (Fig. 2).
Correlation between LAMA3 gene
polymorphism and onset of ovarian cancer
In ovarian cancer patients, the distribution
frequency of TT and TC genotypes of rs12373237 had no statistically
significant differences compared with that in control group
(P>0.05), but there was a statistically significant difference
in the CC genotype (P<0.05) (Table
III). The genotype distribution in both groups met the
Hardy-Weinberg equilibrium law (Pcontrol =0.31,
Pobservation =0.35). The odds ratio (OR) was 1.195 in
the dominant model (TC+CC/TT) (P=0.532), and 4.333 in the recessive
model (CC/TT+TC) (P=0.028). In the co-dominant model, the OR was
0.967 in TC/TT and 4.43 in CC/TT (P=0.09) (Table IV).
 | Table III.Distribution frequency of rs12373237
(n, %). |
Table III.
Distribution frequency of rs12373237
(n, %).
|
| Observation group
(n=210) | Control group
(n=160) |
|
|
|---|
|
|
|
|
|
|
|---|
| Genotype | N | % | N | % | χ2
test | P-value |
|---|
| TT | 125 |
59.5 |
102 | 63.8 |
0.391 | 0.532 |
| TC | 64 |
30.5 | 54 | 33.7 |
0.235 | 0.628 |
| CC | 21 | 10 | 4 | 2.5 | 4.8 | 0.028 |
| T | 314 |
74.8 |
258 | 80.6 |
0.971 | 0.324 |
| C | 106 |
25.2 | 62 | 19.4 |
|
|
 | Table IV.Disease risks in different models. |
Table IV.
Disease risks in different models.
| Model | Genotype | Observation group
(n=210) | Control group
(n=160) | OR (95% CI) | P-value |
|---|
| Dominant model | TT | 125 | 102 | 1 | 0.532 |
|
| TC+CC | 85 | 58 | 1.195
(0.732–1.532) |
|
| Recessive model | TT+TC | 189 | 156 | 1 | 0.028 |
|
| CC | 21 |
4 | 4.333
(2.321–7.786) |
|
| Co-dominant
model | TT | 125 | 102 | 1 | 0.09 |
|
| TC | 64 | 54 | 0.967
(0.657–1.214) |
|
|
| CC | 21 |
4 | 4.43
(2.451–7.698) |
|
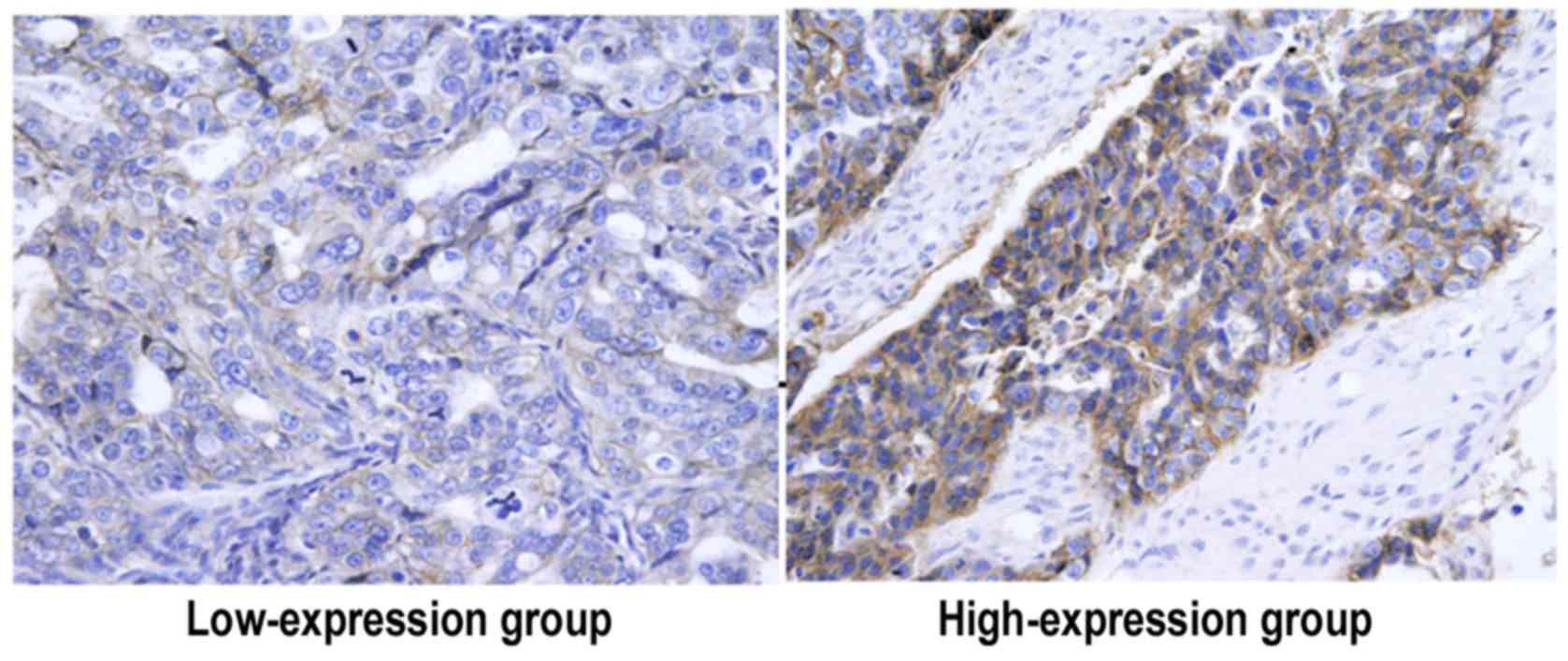
Expression of laminin
According to the immunohistochemical detection,
there were 134 cases (63.8%) in high-expression group, which was
more than that in low-expression group (76 cases, 36.2%)
(P<0.05) (Fig. 3).
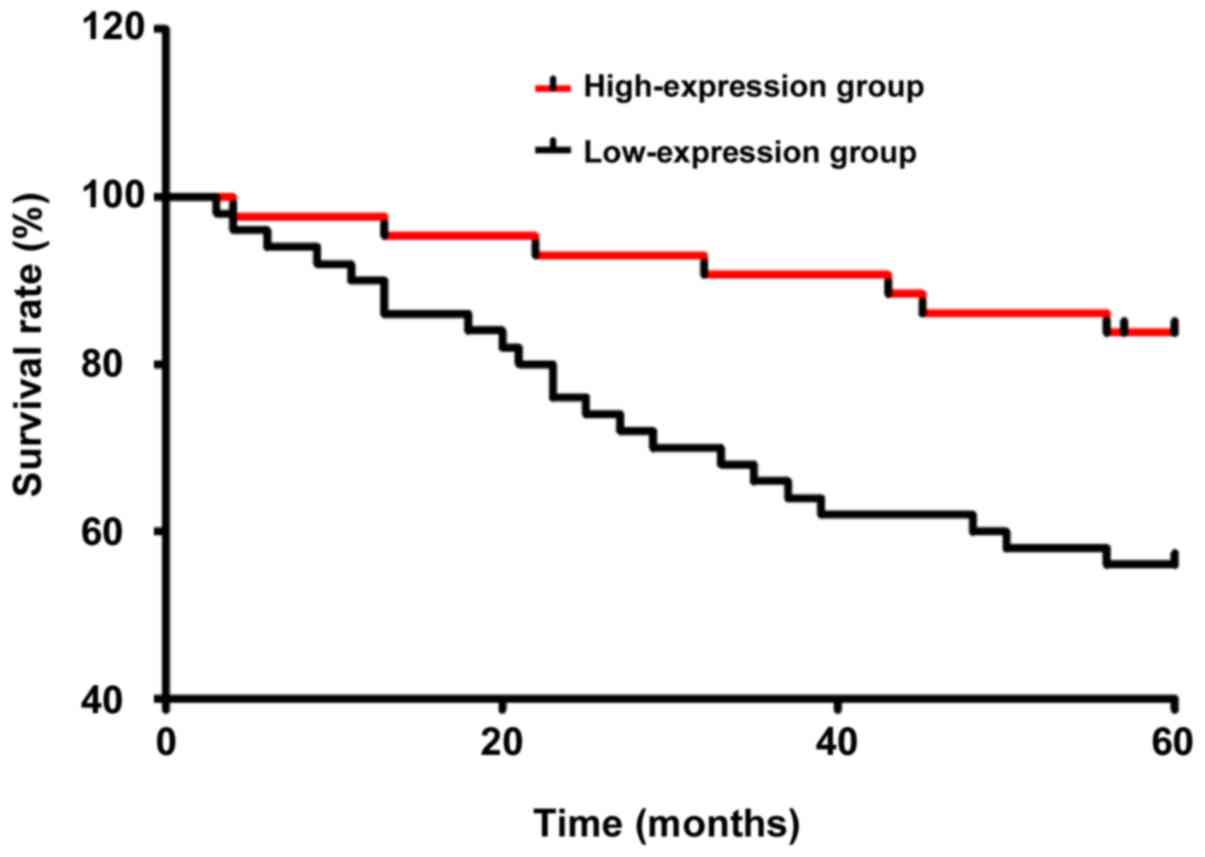
Five-year survival curve
According to the analysis of 5-year survival rate,
the recurrence-free survival rate and overall survival rate in
LAMA3 high-expression group were significantly higher than those in
LAMA3 low-expression group (P<0.05) (Table V). The Kaplan-Meier survival curves
and log-rank test in both groups are shown in Fig. 4.
 | Table V.Comparison of survival rate between
low-expression group and high-expression group [n (%)]. |
Table V.
Comparison of survival rate between
low-expression group and high-expression group [n (%)].
| Group | n | Recurrence-free
survival rate | Overall survival
rate |
|---|
| High-expression
group | 134 | 91 (67.9) | 116 (86.6) |
| Low-expression
group | 76 | 27 (35.5) | 44
(57.9) |
| χ2
test |
| 13.92 | 39.84 |
| P-value |
| <0.001 | <0.001 |
Discussion
At present, with the development of molecular
diagnosis and individualized treatment technique for tumors, the
research emphasis of ovarian cancer, one of the
frequently-occurring gynecological tumors, has been gradually
transferred to the gene and molecular levels (11,12). The
pathogenesis of ovarian cancer is complex, and the external
environmental stimulus, gene damage and emergence of tumor stem
cells are important theories of its onset (13,14).
Surgical resection is the most effective therapeutic method for
ovarian cancer currently, which, however, will lead to various
sequelae, such as obesity, hypertension, heart disease and
osteoporosis (15). In terms of drug
therapy, chemotherapeutic drugs applied clinically are harmful to
the body's immune system, and the emerging cell therapy has
obtained a certain effect only in in vitro experiments and
animal experiments, but its anti-solid tumor effect in the human
body is still unsatisfactory (16,17).
Therefore, studying the pathogenesis to clarify the correlation
between gene and onset is of great significance in the prevention
of disease and development of targeted drugs.
LAMA3 encodes laminin and widely exists in
each organ of human, which can regulate cell attachment and
migration during embryonic development through interaction with
other extracellular matrix components and binding to cells via
high-affinity receptor, thus indirectly promoting and inhibition of
the growth of tumor cells (18).
Studies have found that the expression level of LAMA3 is related to
the occurrence of gastric cancer, and its high expression can
inhibit the occurrence of gastric cancer (19). In this study, it was also found that
the expression level of LAMA3 in ovarian cancer was lower
than those in para-carcinoma tissues and non-carcinoma tissues,
which was similar to the research report on gastric cancer. Svoboda
et al (20) also analyzed the
liver cancer tissues and surrounding normal tissues using the C-DNA
microarray and RT-PCR, and found that the expression of
LAMA3 gene is upregulated in liver cancer tissues,
indicating that the expression of LAMA3 gene has a
consistent correlation with a variety of cancers, including ovarian
cancer, and they share a common signaling pathway in molecular
mechanism.
It was also found in this study that the methylation
degree was lower in para-carcinoma tissues and normal tissues than
that in carcinoma tissues, suggesting that the higher the
methylation degree is, the higher the probability of ovarian cancer
will be. Shukla et al (21)
found that LAMA3 is expressed in benign breast tissues and deleted
in breast cancer tissues, and the LAMA3 gene expression in
breast cancer cell lines is restored after demethylation,
indicating that methylation may be a mechanism of expression
silencing of LAMA3 gene. According to the gene polymorphism
study, it has been proved that the base structure and number of the
same gene are different in different people, and such a difference
will lead to changes in the translation level, thereby affecting
the physical development and metabolism related to the gene.
Rs12373237 is a point mutation in the intron of LAMA3 gene,
and it has been reported that its site mutation is certainly
related to the cardiovascular and cerebrovascular diseases and
chronic renal diseases. This study showed that the CC homozygous
mutation of rs12373237 had a high correlation with the onset of
ovarian cancer. The analysis suggests that the CC mutation may
reduce the expression level of LAMA3 gene and the production
amount of laminin.
In this study, the 5-year recurrence-free survival
rate and overall survival rate of the patients were compared, and
it was found that both rates in LAMA3 high-expression group
were significantly higher than those in low-expression group,
indicating that the expression level of LAMA3 gene also
affects the postoperative recovery and therapeutic effect.
In conclusion, the expression level and base
mutation of LAMA3 gene can change the level of laminin,
which have a certain influence on the onset and prognosis of
ovarian cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LT and PW were responsible for PCR and methylation
detection of tissue samples. QW and LZ collected and analyzed
general data of patients. LT wrote the manuscript. LZ helped with
follow-up analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
West China Second Hospital (Chengdu, China) and informed consents
were signed by the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xu Y, Cheng L, Dai H, Zhang R, Wang M, Shi
T, Sun M, Cheng X and Wei Q: Variants in Notch signalling pathway
genes, PSEN1 and MAML2, predict overall survival in Chinese
patients with epithelial ovarian cancer. J Cell Mol Med.
22:4975–4984. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oliveira-Ferrer L, Goswami R, Galatenko V,
Ding Y, Eylmann K, Legler K, Kürti S, Schmalfeldt B and
Milde-Langosch K: Prognostic impact of CEACAM1 in node-negative
ovarian cancer patients. Dis Markers. 2018:67142872018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rhyasen GW, Yao Y, Zhang J, Dulak A,
Castriotta L, Jacques K, Zhao W, Gharahdaghi F, Hattersley MM, Lyne
PD, et al: BRD4 amplification facilitates an oncogenic gene
expression program in high-grade serous ovarian cancer and confers
sensitivity to BET inhibitors. PLoS One. 13:e02008262018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Castro BG, Dos Reis R, Cintra GF, Sousa
MM, Vieira MA and Andrade CE: Predictive factors for surgical
morbidities and adjuvant chemotherapy delay for advanced ovarian
cancer patients treated by primary debulking surgery or interval
debulking surgery. Int J Gynecol Cancer. 28:1520–1528. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saxena P, Severi L, Santucci M, Taddia L,
Ferrari S, Luciani R, Marverti G, Marraccini C, Tondi D, Mor M, et
al: Conformational propensity and biological studies of proline
mutated LR peptides inhibiting human thymidylate synthase and
ovarian cancer cell growth. J Med Chem. 61:7374–7380. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shu C, Yan D, Mo Y, Gu J, Shah N and He J:
Long noncoding RNA lncARSR promotes epithelial ovarian cancer cell
proliferation and invasion by association with HuR and miR-200
family. Am J Cancer Res. 8:981–992. 2018.PubMed/NCBI
|
|
7
|
Coogan PF, Rosenberg L, Palmer JR, Strom
BL, Zauber AG and Shapiro S: Statin use and the risk of breast and
prostate cancer. Epidemiology. 13:262–267. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen P, Fang X, Xia B, Zhao Y, Li Q and Wu
X: Long noncoding RNA LINC00152 promotes cell proliferation through
competitively binding endogenous miR-125b with MCL-1 by regulating
mitochondrial apoptosis pathways in ovarian cancer. Cancer Med.
7:4530–4541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong SS, Zhang MX, Zhang M, Yu Y, Chen J,
Zhang XY and Xu CJ: Follicle-stimulating hormone peptide-conjugated
nanoparticles for targeted shRNA delivery lead to effective gro-α
silencing and antitumor activity against ovarian cancer. Drug
Deliv. 25:576–584. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ahmad F, Shah K, Umair M, Irfanullah JA,
Khan S, Muhammad D, Basit S, Wakil SM, Ramzan K, et al: Novel
autosomal recessive LAMA3 and PLEC variants underlie junctional
epidermolysis bullosa generalized intermediate and epidermolysis
bullosa simplex with muscular dystrophy in two consanguineous
families. Clin Exp Dermatol. 43:752–755. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Wang H, Wang Y, Dong W, Jiang Z
and Yang G: Substrate-mediated gene transduction of LAMA3 for
promoting biological sealing between titanium surface and gingival
epithelium. Colloids Surf B Biointerfaces. 161:314–323. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reimer A, Schwieger-Briel A, He Y, Leppert
J, Schauer F, Kiritsi D, Schneider H, Ott H, Bruckner-Tuderman L
and Has C: Natural history and clinical outcome of junctional
epidermolysis bullosa generalized intermediate due to a LAMA3
mutation. Br J Dermatol. 178:973–975. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gostyńska KB, Yan Yuen W, Pasmooij AM,
Stellingsma C, Pas HH, Lemmink H and Jonkman MF: Carriers with
functional null mutations in LAMA3 have localized enamel
abnormalities due to haploinsufficiency. Eur J Hum Genet. 25:94–99.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pesch M, König S and Aumailley M: Targeted
disruption of the Lama3 gene in adult mice is sufficient to induce
skin inflammation and fibrosis. J Invest Dermatol. 137:332–340.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Arora E, Masab M, Jindal V, Riaz I, Gupta
S and Varadi G: Role of poly adenosine diphosphate ribose
polymerase inhibitors in advanced stage ovarian cancer. Cureus.
10:e26852018.PubMed/NCBI
|
|
16
|
Srinivas KG, Mirza AA, Swamy S, Amarendra
S and Kodaganur G: Metachronous synchronous sternal and colonic
metastasis with asymptomatic colo-colic fistula from carcinoma
ovary rare presentation of ovarian cancer. Indian J Surg Oncol.
8:615–618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stone ML, Chiappinelli KB, Li H, Murphy
LM, Travers ME, Topper MJ, Mathios D, Lim M, Shih IM, Wang TL, et
al: Epigenetic therapy activates type I interferon signaling in
murine ovarian cancer to reduce immunosuppression and tumor burden.
Proc Natl Acad Sci USA. 114:E10981–E10990. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stemmler S, Parwez Q, Petrasch-Parwez E,
Epplen JT and Hoffjan S: Association of variation in the LAMA3
gene, encoding the alpha-chain of laminin 5, with atopic dermatitis
in a German case-control cohort. BMC Dermatol. 14:172014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lincoln V, Cogan J, Hou Y, Hirsch M, Hao
M, Alexeev V, De Luca M, De Rosa L, Bauer JW, Woodley DT, et al:
Gentamicin induces LAMB3 nonsense mutation readthrough and restores
functional laminin 332 in junctional epidermolysis bullosa. Proc
Natl Acad Sci USA. 115:E6536–E6545. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Svoboda M, Hlobilová M, Marešová M,
Sochorová M, Kováčik A, Vávrová K and Dolečková I: Comparison of
suction blistering and tape stripping for analysis of epidermal
genes, proteins and lipids. Arch Dermatol Res. 309:757–765. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shukla DK, Gupta D, Aggarwal A and Kumar
D: A case report of newly diagnosed epithelial ovarian carcinoma
presenting with spontaneous tumor lysis syndrome and its successful
management with rasburicase. Indian J Med Paediatr Oncol.
38:360–362. 2017. View Article : Google Scholar : PubMed/NCBI
|