Introduction
Liver cancer is one of the most frequently diagnosed
types of cancer and it was identified as a leading cause of
cancer-associated mortality, with an estimated 782,500 new cases
and 745,500 mortalities occurring worldwide during 2012 (1). The incidence and mortality rates of
liver cancer in China account for ~50% of cases worldwide (1). Of the primary liver cancers occurring
worldwide, 70–90% are hepatocellular carcinoma (HCC) (1). At present, surgery remains the
first-line treatment option for HCC (2). However, the early clinical symptoms of
HCC are not obvious and are often overlooked. The majority of
patients with HCC are diagnosed at an advanced stage of the disease
when there are limited therapeutic options available, which results
in a high mortality rate (3).
Therefore, the identification of specific and sensitive biomarkers
for the early diagnosis of HCC is required.
MicroRNAs (miRNAs) are a family of single-stranded
non-coding RNAs, which are typically 20–25 nucleotides in length
and do not encode for any proteins (4). miRNAs can regulate protein expression
at the post-transcriptional level via the negative regulation of
mRNA translation by binding to specific sequences in the 3′
untranslated region of their target mRNAs (4). Notably, certain miRNAs are dysregulated
in cancers and can serve as a critical determinant of cancer
initiation and malignant progression (5). Aberrant expression levels of miRNAs
have been detected in a variety of different types of cancer,
including HCC. Cancer cells may also release miRNAs into the blood
circulation. Previous studies revealed that there are different
levels of miRNA expression between tumors and corresponding
non-cancerous tissues, and that serum miRNAs can discriminate
cancer patients from healthy subjects (6,7).
Circulating miRNAs have been demonstrated to possess high levels of
stability and can easily be obtained from patients with cancer
(8). Furthermore, blood samples have
the advantage of being minimally invasive and can be subjected to
continuous in vitro testing, as well as being highly
reproducible. The discovery of dysregulated miRNAs as potential
circulating biomarkers in the plasma and/or serum may represent a
useful approach for future diagnosis, prognosis and personalized
therapy of patients with cancer (9,10).
In a previous study, aberrant expression of miR-155,
miR-96 and miR-99a between HCC tissues and their matched adjacent
normal liver tissues were validated through miRNA microarray assay
and reverse transcription-quantitative PCR (RT-qPCR) (11). Given that blood samples can be easily
obtained, and it has been demonstrated that miRNAs are stably
maintained in human serum/plasma, it was suggested that they can be
utilized as potential biomarkers for cancer diagnosis and
prognosis. In the present study, 30 HCC tissues and their matched
adjacent normal liver tissues were collected, as well as 30 HCC and
30 healthy serum samples, and the expression levels of miR-155,
miR-96 and miR-99a were measured using RT-qPCR. The diagnostic and
prognostic potential of these miRNAs for HCC were also
investigated.
Materials and methods
Tissue and serum samples
In total, 30 HCC tissues and their matched adjacent
normal liver tissues (≥5 cm from the tumor site) were collected
from patients from the Affiliated Tumor Hospital of Guangxi Medical
University (Nanning, China) between January 2012 and December 2013.
All 30 patients with HCC (age range, 26–67; female to male ratio,
7:23) were given a follow-up examination 58–70 months following
hepatectomy. Overall survival was defined based on the elapsed time
between the time of surgery and mortality. According to the median
cut-off values, 30 patients with HCC were categorized into
high-expression and low-expression of miRNA. In addition, the 30
HCC (age range, 31–70; mean age, 50.7±10.56) and 30 healthy (age
range, 34–69; mean age, 49.8±10.76) blood samples (2 ml) were also
obtained from the Affiliated Tumor Hospital of Guangxi Medical
University between January 2014 and December 2014. There was no
significant difference in the age of the patients observed between
the HCC group and healthy group (P>0.05). All the participants
provided written informed consent. Patients included in this study
were patients with HCC diagnosed by histopathological examination
and according to the National Comprehensive Cancer Network clinical
practice guidelines for oncology. The exclusion criteria were as
follows: i) Patients with HCC who received anticancer therapy prior
to surgery; and ii) Patients with HCC who also suffered from other
severe diseases or malignant tumors simultaneously. Furthermore,
healthy volunteers with autoimmune diseases, severe liver and
kidney diseases, hematologic diseases, and infectious diseases were
excluded from this study. All tissues were diagnosed independently
following liver resection and were snap frozen in liquid nitrogen
and immediately stored at −80°C. Serum samples were centrifuged
immediately at 13,900 × g for 10 min at 4°C after collection and
then stored at −20°C for the future experiments. The present study
was approved by the Ethics Committee of the Affiliated Tumor
Hospital of Guangxi Medical University. The Tumor-Node-Metastasis
(TNM) stage was determined according to the 6th edition of the TNM
Classification of Malignant Tumor. The clinical characteristics of
the included participants are summarized in Table I.
 | Table I.Demographic and clinical
characteristics of the patients with hepatocellular carcinoma and
30 healthy subjects included in the present study. |
Table I.
Demographic and clinical
characteristics of the patients with hepatocellular carcinoma and
30 healthy subjects included in the present study.
| Characteristic | Tissue sample, n
(%) | Serum sample from
patients, n (%) | Serum sample from
healthy controls, n (%) |
|---|
| Sex |
|
|
|
| Male | 23 (76.7) | 18 (60.0) | 16 (53.3) |
|
Female | 7 (23.3) | 12 (40.0) | 14 (46.7) |
| Age |
|
|
|
| ≤50
years | 17 (56.7) | 16 (53.3) | 15 (50.0) |
| >50
years | 13 (43.3) | 14 (46.7) | 15 (50.0) |
| AFP |
|
|
|
| >20
ng/ml | 18 (60.0) | 18 (60.0) | 0 (0) |
| ≤20
ng/ml | 12 (40.0) | 12 (40.0) | 30 (100) |
| Tumor size |
|
|
|
| ≤5
cm | 12 (40.0) | 11 (36.7) | – |
| >5
cm | 18 (60.0) | 19 (63.3) | – |
| Liver
cirrhosis |
|
|
|
|
Present | 12 (40.0) | 22 (73.3) | – |
|
Absent | 18 (60.0) | 8 (26.7) | – |
|
Differentiation |
|
|
|
| Well +
moderate | 10 (33.3) | 20 (66.7) | – |
|
Poor | 20 (66.7) | 10 (33.3) | – |
| TNM stage |
|
|
|
|
I+II | 17 (56.7) | 12 (40.0) | – |
|
III+IV | 13 (43.3) | 18 (60.0) | – |
Total RNA extraction from tissues
Total RNA was isolated from all test tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The isolated total
RNA was then purified using a NucleoSpin® RNA clean-up
kit (Macherey-Nagel GmbH and Co.), and the quantity and quality of
total RNA were determined by using a NanoDrop 2000
spectrophotometer (version 1.6.198; Thermo Fisher Scientific, Inc.)
and agarose gel electrophoresis. RNA was stored at −80°C for
subsequent RT-qPCR analysis.
Total RNA extraction from serum
Total RNA of serum was extracted using miRNeasy
Serum/Plasma kit (Qiagen GmbH) according to the manufacturer's
protocol. The NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.) was used to measure the quality of total RNA, and
agarose gel electrophoresis was used to ensure the integrity of the
total RNA. RNA was stored at −80°C for subsequent RT-qPCR
analysis.
RT-qPCR
For the miRNA RT-qPCR, 1 µg RNA was reverse
transcribed to cDNA using a miScript II RT kit (Qiagen GmbH)
according to the manufacturer's protocol. RT-PCR assays were
performed as previously described (11). RT-qPCR was performed using an Agilent
MX 3000 real-time PCR system (Agilent Technologies, Inc.) with the
miScript SYBR-Green PCR kit (Qiagen GmbH) and reactions were
performed in triplicate. U6 was used as an endogenous control to
normalize the expression level of miRNA extracted from the tissue
samples, and miR-16 was used as an endogenous control for the serum
samples (12). The thermocycling
conditions were the following: Initial denaturation for 95°C for 15
min, followed by 40 cycles of 94°C for 15 sec, annealing at 55°C
for 30 sec and extension at 70°C for 30 sec. The primers used in
the present study were purchased from Qiagen GmbH. The sequences
for the primers used were as follows: miR-155 forward,
5′-CCTTTGCTGGAATGGACAAGAAC-3′; miR-96 forward,
5′-TTGGGTGAAATATATTGTGCGTCTC-3′; miR-99a forward,
5′-GAGTCCTGGACACCCAACTACAAG-3′; miR-16 forward,
5′-UAGCAGCACGUAAAUAUUGGCG-3′; U6 forward,
5′-GCACCGTCAAGGCTGAGAAC-3′; miScript universal reverse primer,
5′-AGCCGAAGTGAGCCACTGAA-3′. Relative levels of the three miRNAs
were calculated using the 2−∆∆Cq method (13).
Serum detection of α-fetoprotein
(AFP)
Serum AFP level was quantitatively measured using
electrochemiluminescence immunoassay analyzer kit (Cobas e 601
module; Roche Diagnostics) according to the manufacturer's
protocol. Normal reference values for AFP were 0–20 ng/ml.
Statistical analysis
One-way ANOVA followed by Student-Newman-Keuls post
hoc was used to assess differences in the miRNA expression levels
between the group of normal and pathological samples. Student's
t-test was used to compare the ages of the two groups. The receiver
operating characteristic (ROC) curve was applied to assess the
diagnostic values of the three miRNAs investigated. Sensitivity and
specificity levels were obtained according to the Youden's index
(sensitivity + specificity-1) (9).
The ROC curves of the combinations were constructed using binary
logistic regression analysis. Survival curves were plotted using
the Kaplan-Meier method and compared using the log-rank test.
Statistical analysis was performed with SPSS software (version
17.0; SPSS, Inc.), and P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miRNAs in tissue and
serum of patients with HCC
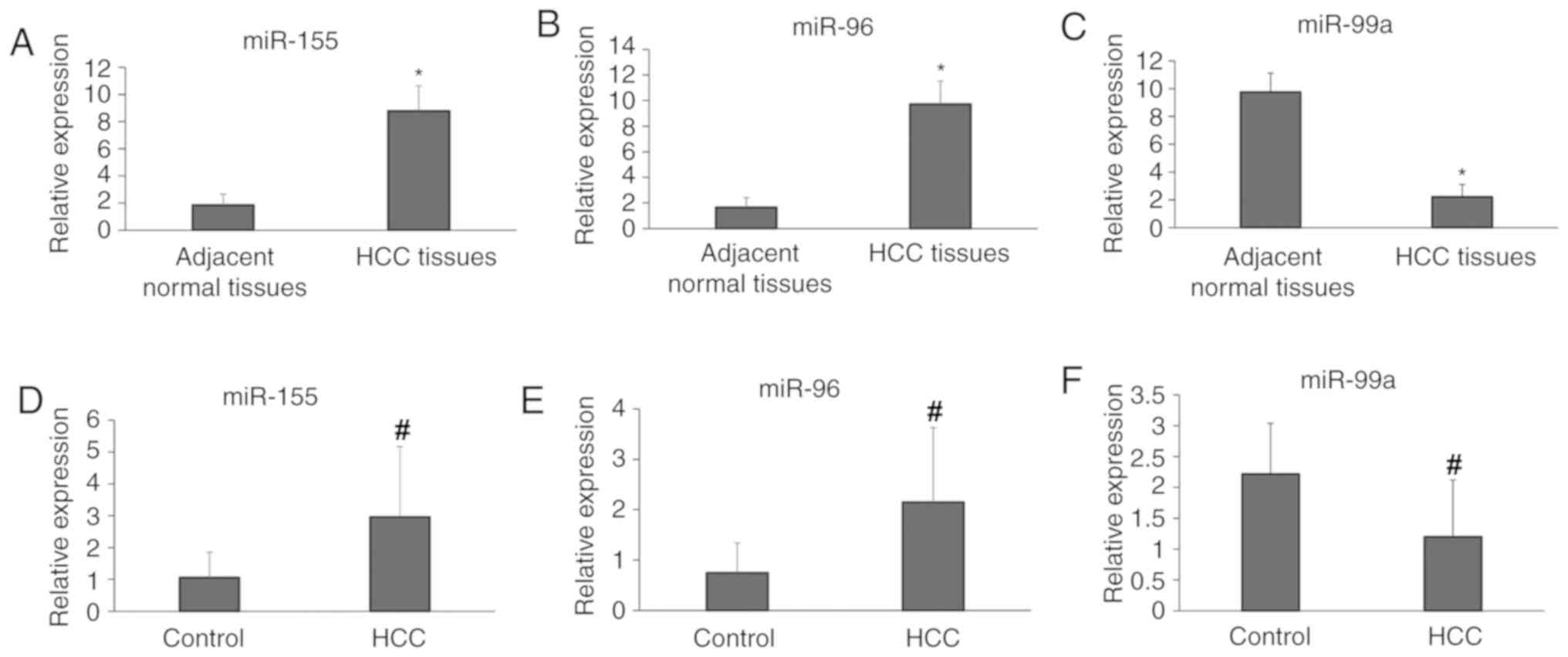
In the present study, the expression levels of
miR-155, miR-96 and miR-99a were analyzed in 30 HCC tissues and
their matched adjacent normal liver tissues. The present results
indicated that compared with the adjacent normal tissues, the
expression levels of miR-155 and miR-96 in HCC tissues were
significantly upregulated, whereas miR-99a was significantly
downregulated (P<0.05; Fig.
1A-C). Consistent with the results from tissue samples, the
expression levels of miR-155 and miR-96 were also identified to be
upregulated in the sera of patients with HCC compared with healthy
controls. In addition, the serum levels of miR-99a were
significantly downregulated in patients with HCC compared with
healthy controls (P<0.05; Fig.
1D-F).
Diagnostic value of the combination of
the serum levels of miRNAs and AFP in patients with HCC and healthy
controls
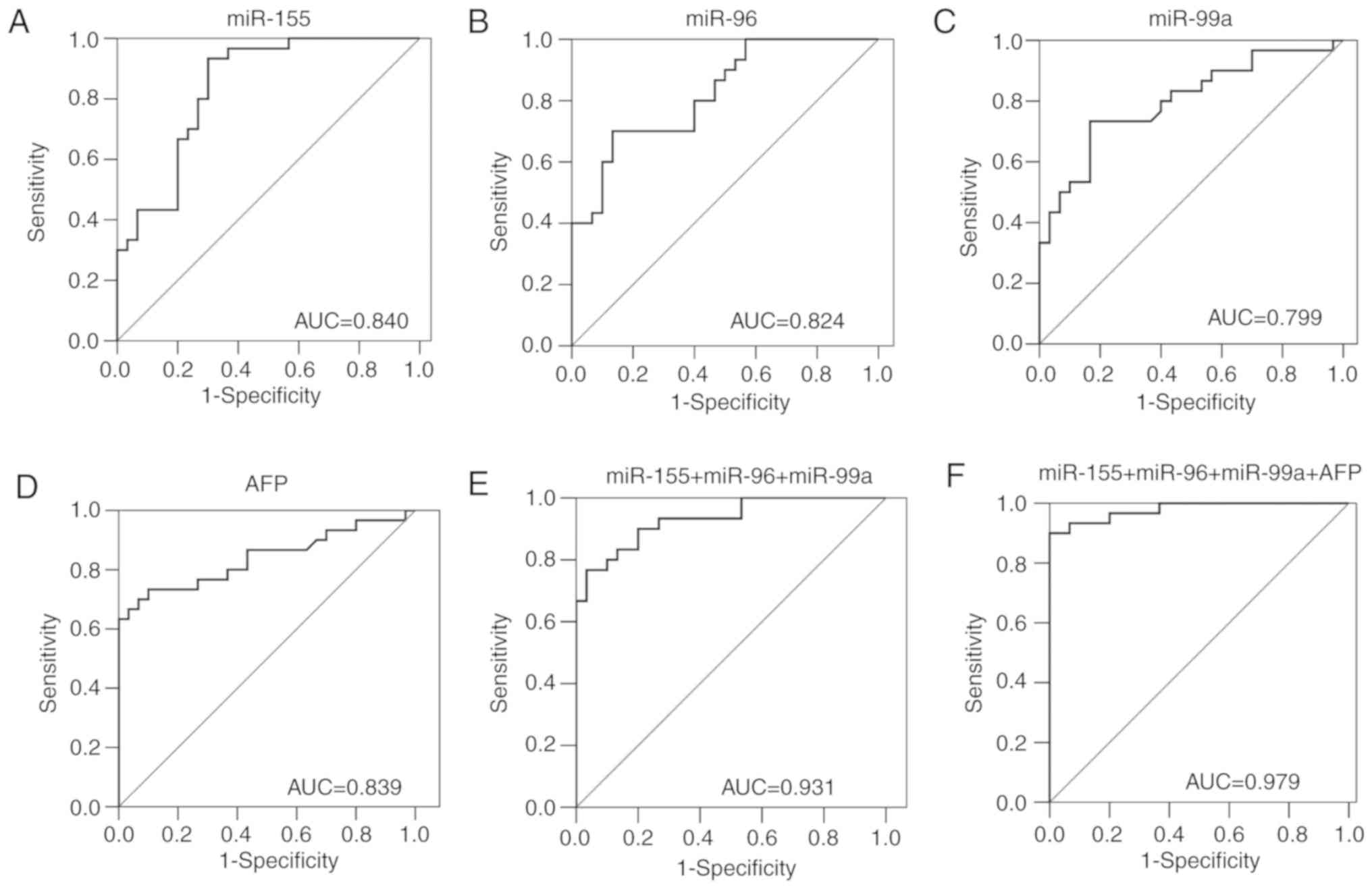
To determine whether the serum levels of miR-155,
miR-96, miR-99a and AFP exhibited diagnostic value in HCC, the ROC
curves were plotted to analyze their diagnostic sensitivity,
specificity and area under the curve (AUC) (Fig. 2). The AUC of the ROC analyses for
four tested biomarkers in HCC were: i) miR-155, 0.840; ii) miR-96,
0.824; iii) miR-99a, 0.799; and iv) AFP, 0.839 (Table II). When miR-155, miR-96 and miR-99a
were combined to form a panel, the combination had a higher
accuracy than each miRNA separately in discriminating patients with
HCC from healthy controls [AUC, 0.931; 95% CI, 0.870–0.992;
sensitivity, 76.7%; specificity, 96.7%]. The addition of AFP to the
combination of the aforementioned miRNAs resulted in the highest
diagnostic accuracy (AUC, 0.979; 95% CI, 0.903–0.999; sensitivity,
90.0%; specificity, 100.0%).
 | Table II.AUC and the corresponding 95% CI of
serum microRNA and AFP in patients with hepatocellular carcinoma
compared with healthy controls. |
Table II.
AUC and the corresponding 95% CI of
serum microRNA and AFP in patients with hepatocellular carcinoma
compared with healthy controls.
| Tumor markers | Sensitivity, % | Specificity, % | AUC | SE | 95% CI | P-value |
|---|
| miR-155 | 93.3 | 70.0 | 0.840 | 0.051 | 0.739–0.941 | <0.001 |
| miR-96 | 70.0 | 86.7 | 0.824 | 0.052 | 0.722–0.927 | <0.001 |
| miR-99a | 73.3 | 83.3 | 0.799 | 0.058 | 0.687–0.912 | <0.001 |
| AFP | 66.7 | 96.7 | 0.839 | 0.054 | 0.733–0.946 | <0.001 |
| miR-155 + miR-96 +
miR-99a | 76.7 | 96.7 | 0.931 | 0.031 | 0.870–0.992 | <0.001 |
| miR-155 + miR-96 +
miR-99a + AFP | 90.0 | 100.0 | 0.979 | 0.015 | 0.903–0.999 | <0.001 |
Association between miRNA expression
levels and prognosis
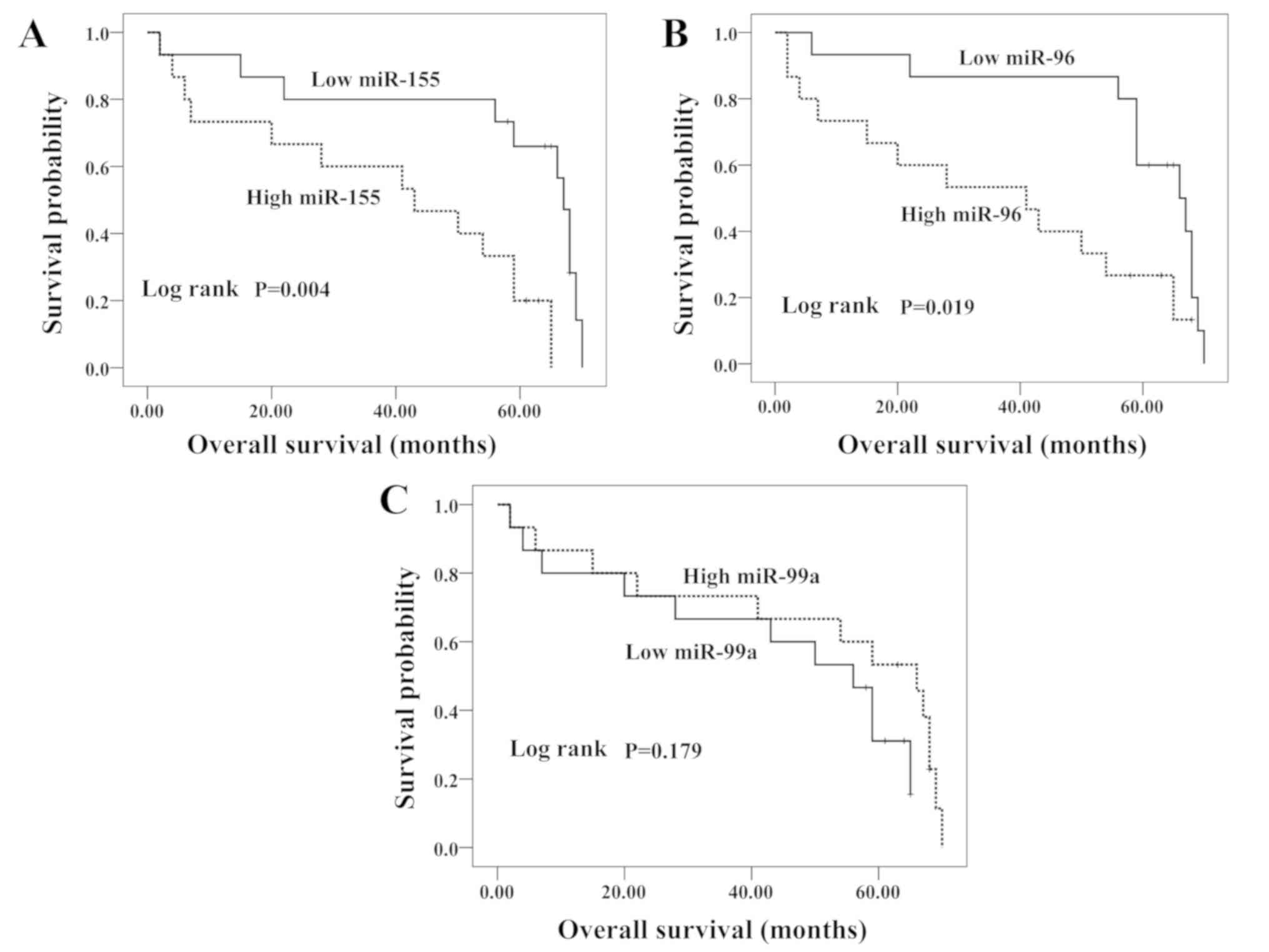
In order to evaluate the possible prognostic value
of miR-155, miR-96 and miR-99a, an overall survival analysis for
patients with HCC was performed. Kaplan-Meier curves were plotted
between the high or low miRNA expression levels in HCC tissues and
overall survival. The median survival time of patients with HCC was
58.5 months (Fig. 3). Kaplan-Meier
analysis revealed that high levels of miR-155 and miR-96 were
significantly associated with the decreased overall survival time
of patients with HCC (P=0.004 and P=0.019, respectively),
indicating that miR-155 and miR-96 may possess prognostic value for
patients with HCC. In contrast, the patients with low miR-99a
expression had slightly reduced survival times compared with those
with high miR-99a. However, these results were not statistically
significant (P=0.179).
Discussion
miRNAs have been reported to be involved in a series
of biological processes, in particular in the occurrence and
progression of tumors. Accumulating evidence suggest that numerous
miRNAs are aberrantly expressed in tumor tissues, blood and cells,
and dysregulation of these miRNAs is connected with the occurrence,
progression and prognosis of cancer (14). Certain miRNAs serve oncogenic roles,
with some of these being upregulated in patients with cancer;
whereas other miRNAs are downregulated, acting as tumor suppressors
(6). However, all these miRNAs are
highly associated with the carcinogenesis, progression and
prognosis of patients with cancer. Thus, it is possible that many
miRNAs could serve as biomarkers for the diagnosis and prognosis of
various types of cancer, including HCC (7). Based on previous microarray assay and
RT-PCR data (11), miR-155, miR-96
and miR-99a were selected as candidate miRNAs for the diagnosis and
prognosis of HCC.
miR-155 acts as an oncogenic miRNA, which has been
found to be upregulated in various types of tumor, such as breast
cancer (15), gastric cancer
(16), lung cancer (17), colorectal cancer (18) and other solid malignancies. Bašová
et al (15) indicated that
there was a positive correlation between miR-155 levels and the
pathogenesis of early breast cancer relapse, particularly at the
time of post-surgery. These previous data suggested that oncogenic
miRNAs (miR-155 and miR-24) in serum may enable not only the
monitoring of early breast cancer, but may also become a highly
valuable tool for the detection of relapse in patients with early
breast cancer (15). The expression
levels of miR-155 were significantly increased in gastric cancer
tissues, and the upregulation of miR-155 promoted proliferation and
migration of gastric cancer cells (16). A previous study has demonstrated that
miR-155 exerts an oncogenic role in non-small cell lung cancer
(17). It has also been demonstrated
that an enhanced expression level of miR-155 is correlated with a
higher frequency of distant metastases in patients with colorectal
cancer (18). Furthermore, targeting
miR-155-5p may be a useful therapeutic strategy against colon
cancer metastasis (19). A previous
study (20) revealed that the
expression levels of miR-155 were highly upregulated in HCC
tissues, and that miR-155 may be involved in the tumorigenesis of
HCC, which is consistent with the results of the present study.
The miR-99 family consists of three members,
miR-99a, miR-99b and miR-100. Recent studies demonstrated that the
miR-99 family had potential functions as tumor suppressors, which
may affect tumor cell growth, invasion and migration (21). Dysregulation of the miR-99 family
members has been reported to serve an important role in multiple
types of cancer (22–25). The expression levels of miR-99a,
miR-99b and miR-100 were all significantly decreased in glioma
tissues compared with non-neoplastic brain tissues. Further in
vitro experiments indicated that the overexpression of miR-99a,
miR-99b and miR-100 markedly suppressed cellular migration and
invasion in glioma cells (21). In
HCC, oral squamous cell carcinoma (OSCC) and non-small cell lung
cancer, miR-99a also functions as a tumor suppressor (22–24). A
previous study revealed that the low-level expression of miR-99a
was correlated with poor tumor-free survival and overall survival
for patients with HCC (25). For
miR-99b, a previous study demonstrated that miR-99b-5p is expressed
significantly less in liver metastasis lesions compared with paired
primary cancer and can function as a tumor-suppressive microRNA in
liver metastasis of colorectal cancer (26). He et al (27) revealed that the expression levels of
miR-99b-3p were decreased in the tissues of patients with OSCC, and
validated the suppressive role of miR-99b-3p in OSCC via glycogen
synthase kinase 3 β downregulation. In the present study, the
expression levels of miR-99a in HCC tissues and serum were
significantly downregulated compared with healthy controls, in line
with previous studies (22,25).
miR-96 belongs to the miR-183 family, which is
located in the human chromosome 7q32.2. miR-96 is highly expressed
in various types of cancer and is a well-known oncogenic miRNA
(28–30). A previous study demonstrated that
miR-96 was significantly upregulated in breast cancer tissues and
that it could promote breast tumorigenesis both in vitro and
in vivo (28). Yoshino et
al (29) demonstrated that
miR-96 was overexpressed in bladder cancer tissue. The upregulation
of miR-96 in tissues, serum and serum exosomes has also been
observed in patients with lung cancer (30). Furthermore, exosomal miR-96 could act
as a serum biomarker for identifying malignant lung cancer
(30). Iwai et al (31) reported that miR-96-5p expression
levels were markedly upregulated in primary HCC tumors compared
with paired non-tumorous tissues, and suggested that miR-96-5p
inhibits HCC cell apoptosis by targeting caspase-9 mRNA. A previous
study showed that serum miR-96 levels discriminated patients with
HCC from patients with chronic hepatitis B with an AUC of 0.803,
and suggested that serum miR-96 is a promising biomarker for
patients with HCC (32). Another
previous study indicated that upregulation of miR-96 confers an
oncogenic function in HCC dissemination, which could promote cell
invasion and serve as a potential prognostic factor for patients
with HCC (33). The present study
suggested that miR-96 expression was upregulated in tissues and
sera of patients with HCC, and its upregulation was associated with
reduced overall survival time.
In the present study, it was identified that miR-155
and miR-96 were upregulated in tissues and sera of patients with
HCC, whereas miR-99a level was decreased. In addition, the results
revealed that circulating miR-155, miR-96 and miR-99a, and the
combination of the three miRNAs could serve as valuable biomarkers
for the diagnosis of HCC with AUCs of 0.840, 0.824, 0.799 and
0.931, respectively. At present, AFP is one of most frequently used
biomarkers for HCC (7). The addition
of AFP to the combination of miR-155, miR-96 and miR-99b offered a
higher accuracy of HCC diagnosis. The main limitation of the
present study is the small sample size. Further studies using a
larger cohort of patients are therefore required to validate the
diagnostic value of these miRNAs.
In summary, the present study highlighted the
potential role of serum-circulating miR-155, miR-96 and miR-99b
levels for the diagnosis of HCC. The combination of these miRNAs
with AFP could potentially improve the sensitivity and specificity
for HCC diagnosis compared with using either miRNAs or AFP alone.
In addition, elevated expression levels of miR-155 and miR-96 were
identified to be associated with poor survival times in patients
with HCC. Collectively, the present results suggested that miR-155
and miR-96 can act as prospective prognosis predictors of HCC.
Acknowledgements
The authors would like to thank Dr Dev Sooranna,
Imperial College London (London, UK), for assisting with the
preparation of the manuscript.
Funding
The current study was supported by The National
Natural Science Foundation of China (grant no. 81760535) and The
Natural Science Foundation of Guangxi (grant no. 2017GXNSFAA198155)
and the Scientific Research and Technical Development Project of
Qingxiu District (grant no. 2016057).
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SN wrote the manuscript, designed the study and
interpreted the data. HL performed the statistical analysis and
interpreted the results. BG performed the experiments. WW, JL and
AY were involved in the sample collection and data analysis. LZ was
involved in obtaining the funds for the present study, designing
the study and critically revising the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of the Affiliated Tumor Hospital of Guangxi Medical
University (Nanning, China). Written informed consent was obtained
from each participant included within the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Margini C and Dufour JF: The story of HCC
in NAFLD: From epidemiology, across pathogenesis, to prevention and
treatment. Liver Int. 36:317–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang YF and Ho M: Humanization of
high-affinity antibodies targeting glypican-3 in hepatocellular
carcinoma. Sci Rep. 6:338782016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nagy ZB, Wichmann B, Kalmár A, Galamb O,
Barták BK, Spisák S, Tulassay Z and Molnár B: Colorectal adenoma
and carcinoma specific miRNA profiles in biopsy and their
expression in plasma specimens. Clin Epigenetics. 9:222017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
137:1679–1690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oka S, Furukawa H, Shimada K, Hashimoto A,
Komiya A, Fukui N, Tsuchiya N and Tohma S: Plasma miRNA expression
profiles in rheumatoid arthritis associated interstitial lung
disease. BMC Musculoskelet Disord. 18:212017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Zhang X, Zhang Z, Chang J, Wang Z,
Wu Z, Wang C, Sun Z, Ge X, Geng R, et al: Plasma microRNA-based
signatures to predict 3-year postoperative recurrence risk for
stage II and III gastric cancer. Int J Cancer. 141:2093–2102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao B, Ning S, Li J, Liu H, Wei W, Wu F,
Tang Y, Feng Y, Li K and Zhang L: Integrated analysis of
differentially expressed mRNAs and miRNAs between hepatocellular
carcinoma and their matched adjacent normal liver tissues. Oncol
Rep. 34:325–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song J, Bai Z, Han W, Zhang J, Meng H, Bi
J, Ma X, Han S and Zhang Z: Identification of suitable reference
genes for qPCR analysis of serum microRNA in gastric cancer
patients. Dig Dis Sci. 57:897–904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bašová P, Pešta M, Sochor M and Stopka T:
Prediction potential of serum miR-155 and miR-24 for relapsing
early breast cancer. Int J Mol Sci. 18(pii): E21162017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y,
Ying G and Ba Y: MicroRNA-155 promotes gastric cancer growth and
invasion by negatively regulating transforming growth factor-β
receptor 2. Cancer Sci. 109:618–628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu F, Song D, Wu Y, Liu X, Zhu J and Tang
Y: MiR-155 inhibits proliferation and invasion by directly
targeting PDCD4 in non-small cell lung cancer. Thorac Cancer.
8:613–619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS
and Zhou T: Upregulation of microRNA-155 promotes the migration and
invasion of colorectal cancer cells through the regulation of
claudin-1 expression. Int J Mol Med. 31:1375–1380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Al-Haidari A, Algaber A, Madhi R, Syk I
and Thorlacius H: MiR-155-5p controls colon cancer cell migration
via post-transcriptional regulation of human antigen R (HuR).
Cancer Lett. 421:145–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu X, Wen H, Jing L, Yang Y, Wang W, Liang
X, Nan K, Yao Y and Tian T: MicroRNA-155-5p promotes hepatocellular
carcinoma progression by suppressing PTEN through the PI3K/Akt
pathway. Cancer Sci. 108:620–631. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang M, Guo Y, Wu J, Chen F, Dai Z, Fan
S, Li P and Song T: Roles of microRNA-99 family in human glioma.
Onco Targets Ther. 9:3613–3619. 2016.PubMed/NCBI
|
|
22
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan B, Fu Q, Lai L, Tao X, Fei Y, Shen J,
Chen Z and Wang Q: Downregulation of microRNA 99a in oral squamous
cell carcinomas contributes to the growth and survival of oral
cancer cells. Mol Med Rep. 6:675–681. 2012.PubMed/NCBI
|
|
24
|
Yin H, Ma J, Chen L, Piao S, Zhang Y,
Zhang S, Ma H, Li Y, Qu Y, Wang X and Xu Q: MiR-99a enhances the
radiation sensitivity of non-small cell lung cancer by targeting
mTOR. Cell Physiol Biochem. 46:471–481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Jin H, Liu H, Lv S, Wang B, Wang
R, Liu H, Ding M, Yang Y, Li L, et al: MiRNA-99a directly regulates
AGO2 through translational repression in hepatocellular carcinoma.
Oncogenesis. 3:e972014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li W, Chang J, Wang S, Liu X, Peng J,
Huang D, Sun M, Chen Z, Zhang W, Guo W and Li J: miRNA-99b-5p
suppresses liver metastasis of colorectal cancer by down-regulating
mTOR. Oncotarget. 6:24448–24462. 2015.PubMed/NCBI
|
|
27
|
He K, Tong D, Zhang S, Cai D, Wang L, Yang
Y, Gao L, Chang S, Guo B, Song T, et al: miRNA-99b-3p functions as
a potential tumor suppressor by targeting glycogen synthase
kinase-3β in oral squamous cell carcinoma Tca-8113 cells. Int J
Oncol. 47:1528–1536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hong Y, Liang H, Uzair-Ur-Rehman, Wang Y,
Zhang W, Zhou Y, Chen S, Yu M, Cui S, Liu M, et al: miR-96 promotes
cell proliferation, migration and invasion by targeting PTPN9 in
breast cancer. Sci Rep. 6:374212016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yoshino H, Seki N, Itesako T, Chiyomaru T,
Nakagawa M and Enokida H: Aberrant expression of microRNAs in
bladder cancer. Nat Rev Urol. 10:396–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B,
Xie Y, Ye Y and Liu J: Circulating exosomal microRNA-96 promotes
cell proliferation, migration and drug resistance by targeting
LMO7. J Cell Mol Med. 21:1228–1236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iwai N, Yasui K, Tomie A, Gen Y, Terasaki
K, Kitaichi T, Soda T, Yamada N, Dohi O, Seko Y, et al: Oncogenic
miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in
hepatocellular carcinoma. Int J Oncol. 53:237–245. 2018.PubMed/NCBI
|
|
32
|
Chen Y, Dong X, Yu D and Wang X: Serum
miR-96 is a promising biomarker for hepatocellular carcinoma in
patients with chronic hepatitis B virus infection. Int J Clin Exp
Med. 8:18462–18468. 2015.PubMed/NCBI
|
|
33
|
Leung WK, He M, Chan AW, Law PT and Wong
N: Wnt/β-catenin activates MiR-183/96/182 expression in
hepatocellular carcinoma that promotes cell invasion. Cancer Lett.
362:97–105. 2015. View Article : Google Scholar : PubMed/NCBI
|