Introduction
Lung cancer, one of the most common malignancies and
the leading cause of cancer-associated death in the USA and China
over the past five years, mainly manifests in the form of non-small
cell lung cancer (NSCLC) (1–4). A lack of specific early symptoms often
leads to a delay in the diagnosis and treatment of patients with
NSCLC. Thus, despite notable progress, including minimally invasive
techniques, stereotactic radiotherapy, targeted therapy and
immunotherapy, the five-year survival rate of lung cancer is
estimated to be merely 16% (5). Lung
adenocarcinoma (LUAD) is the major subtype of NSCLC and constituted
~50% of all lung cancer cases (43% in men and 52% in women) in the
USA between 1992 and 2013 (6).
Currently, the mechanism of LUAD initiation and progression is not
entirely clear. Thus, there is a need to identify more effective
biomarkers of this disease.
MicroRNAs (miRs/miRNAs) are a group of
single-stranded, non-protein-coding RNAs that are 19–25 nucleotides
in length, and regulate genes by base-pairing to the
3′-untranslated regions of their target genes (7). Some miRNAs were demonstrated to be
expressed in LUAD tissues and to be associated with survival in
patients (8,9). miR-198-5p was reported to be involved
in various human malignancies (10–12),
such as NSCLC (13). Yang et
al (13) reported that
miR-198-5p inhibits the development of LUAD in vitro and
in vivo by regulating fibroblast growth factor receptor 1;
however, the exact role of miR-198-5p in LUAD and its underlying
mechanism remain poorly understood.
Aiming to discover the clinical value of miR-198-5p
in LUAD, its expression level was detected using RT-qPCR. The
associations between miR-198-5p expression and the
clinicopathological characteristics of patients with LUAD,
including overall survival (OS) time, were analyzed. Furthermore,
meta-analyses based on microarray or miRNA-sequencing data from
Gene Expression Omnibus (GEO), ArrayExpress and The Cancer Genome
Atlas (TCGA) were performed. Finally, the target mRNAs of
miR-198-5p were examined to explore the potential signaling
pathways involved. A summary of the study design is illustrated in
Fig. 1.
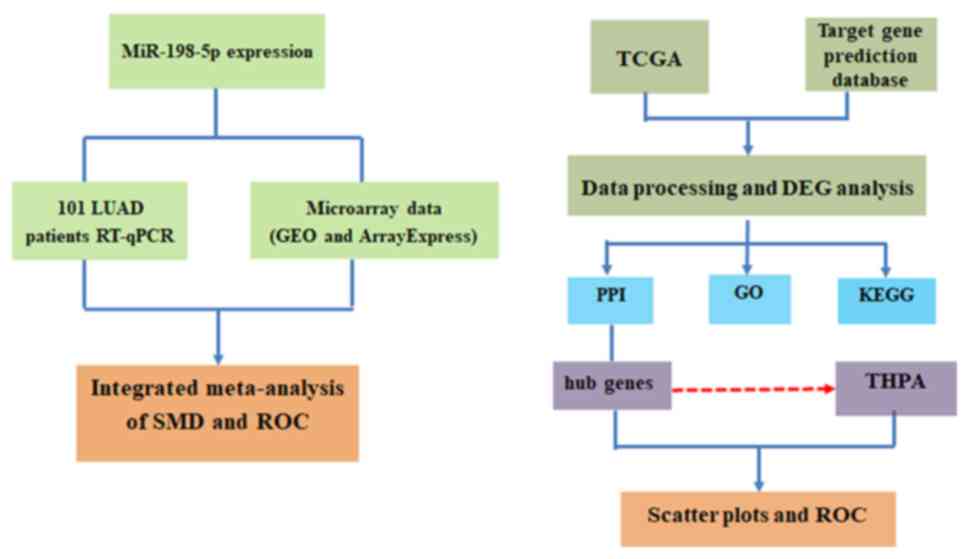 | Figure 1.Flow diagram of the study design.
miR, microRNA; LUAD, lung adenocarcinoma; RT-qPCR, reverse
transcription-quantitative PCR; GEO, Gene Expression Omnibus; SMD,
standard mean difference; ROC, receiver operating characteristic;
TCGA, The Cancer Genome Atlas; DEG, differentially expression gene;
PPI, protein-protein interaction; GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; THPA, The Human Protein
Atlas. |
Materials and methods
Patients
A total of 101 patients with LUAD were enrolled in
the study between January 2014 and December 2016 at The First
Affiliated Hospital of Guangxi Medical University (Nanning, China),
whose Medical Ethics Committee approved this study (approval no.
2015 KY-E-041). Each participant signed an informed consent form.
The inclusion criteria were as follows: i) Diagnosed with LUAD with
complete pathological reports; ii) both tumor and adjacent
non-tumor tissues were available; iii) staging information was
complete; iv) age ≤80 years; and v) home address and telephone
number were available. The exclusion criteria were as follows: i)
Any history of malignancies other than LUAD; and ii) antitumor
therapy before pathological tissues were obtained.
Paraffin-embedded samples of tumor tissues and matched adjacent
normal tissues were collected from each patient. All samples were
fixed with 10% neutral formalin for 16–24 h at room temperature
prior to paraffin embedding. The staging was classified according
to the 7th edition of the UICC-AJCC Tumor-Node-Metastasis staging
system (14). The median patient
follow-up time was 28.4 months. OS time was defined as the period
between pathological diagnosis and death.
RT-qPCR
The miRNeasy FFPE kit (Qiagen GmbH) was used to
extract total RNA from the aforementioned samples, and NanoDrop
2000 (Thermo Fisher Scientific, Inc.) was used to measure the
concentration of RNA. Mir-X™ miRNA qRT-PCR TB Green® Kit
(Takara Bio, Inc.) was used to synthesize cDNA from total RNA.
miR-191-5p was considered as an internal reference due to its
stable expression in lung cancer and normal lung tissues (15). The primer sequence for miR-198-5p was
5′-CAACGGAAUCCCAAAAGCAGCU-3′ and the sequence for miR-191-5p was
5′-GGUCCAGAGGGGAGAUAGGUUC-3′. PCR was performed with an Applied
Biosystems 7900 PCR system (Thermo Fisher Scientific, Inc.) using
SYBR Premix Ex Taq (Takara Bio, Inc.). The reaction conditions of
the PCR were as follows: 98°C for 10 min, followed by 42 cycles of
3 sec at 98°C and 30 sec at 55°C. The 2−ΔΔCq method was
utilized to calculate the relative expression of miR-198-5p
(16).
Database searches
A LUAD-associated microarray search was performed in
the GEO (www.ncbi.nlm.nih.gov/geo) (17) and ArrayExpress (www.ebi.ac.uk/arrayexpress) (18) databases on December 1, 2017. The
following terms were used: [‘lung’ OR ‘pulmonary’ OR ‘respiratory’
OR ‘bronchioles’ OR ‘bronchi’ OR ‘alveoli’ OR ‘pneumocytes’ OR ‘air
way’ (MeSH)] and [‘cancer’ OR ‘carcinoma’ OR ‘tumor’ OR ‘neoplas*
OR malignan* squamous cell carcinoma’ OR ‘adenocarcinoma’ (MeSH)]
or/and [‘MicroRNA’ OR ‘miRNA’ OR ‘MicroRNA’ OR ‘Small Temporal RNA’
OR ‘noncoding RNA’ OR ‘ncRNA’ OR ‘small RNA’ (MeSH)].
Studies that met the following criteria were
included: First, LUAD tissues were available in the LUAD group, and
normal lung tissues were available in the control group. Secondly,
>5 samples were available in each group. Thirdly, the miR-198-5p
level was available for the LUAD and control groups. Fourthly, only
human samples were included. Because the microarray data obtained
from the two databases were identical, GEO was chosen for the
subsequent analyses. The expression data of miR-198-5p were
log2-tranformed for further analysis.
The PubMed (www.ncbi.nlm.nih.gov/pubmed), Web of Science
(clarivate.com/products/web-of-science), Science Direct
(www.sciencedirect.com), Ovid (http://www.ovid.com/), LILACS (lilacs.bvsalud.org/en), Wiley Online Library
(www.onlinelibrary.wiley.com/), EMBASE
(ttuhsc.libguides.com/embase), and
CNKI (cnki.net) databases were also searched for
publications that discussed miR-198-5p expression in LUAD, and only
one relevant article was retrieved (19). In addition, TCGA miRNA data
associated with LUAD and non-cancerous samples from The University
of California Santa Cruz Xena (xena.ucsc.edu)
was downloaded. Two cohorts were obtained, each with <3 samples
available. Therefore, due to an insufficient number of patients
with LUAD or control cases in the miR-198-5p expression data, TCGA
was not included in the investigation.
Data preprocessing and differentially
expressed gene (DEG) analysis
The DESeq R package (version 1.18.1) (20) was employed to process the original
gene expression data of mRNA in LUAD that was obtained from TCGA
(ID: TCGA-LUAD). The DEGs between the LUAD and non-cancer groups
were identified, and upregulated DEGs with a fold change (FC) >2
were subjected to further analysis. As miR-198-5p exhibited a lower
expression level in LUAD tissues, downregulated DEGs were not
selected for subsequent bioinformatics analyses.
Identification of potential target
genes
The potential targets of miR-198-5p were predicted
using miRWalk (version 2.0; zmf.umm.uni-heidelberg.de/mirwalk2), a comprehensive
database that includes 12 online predictive tools: microT-CDS,
microT4, miRanda, miRBridge, miRDB, miRMap, miRNAMap, PICTAR2,
PITA, RNA22, RNAhybrid and TargetScan (21). Genes that appeared in more than two
databases were cross-referenced with the upregulated DEGs
identified in TCGA analysis. The overlapping genes were considered
to be potential target genes of miR-198-5p in LUAD.
Gene Ontology (GO), Kyoto Encyclopedia
of Genes and Genomes (KEGG) and protein-protein interaction (PPI)
maps
To discern the biological attributes of the putative
target genes, GO (22) and KEGG
enrichment analyses (23) were
conducted using online functional annotation tools from the
Database for Annotation, Visualization and Integrated Discovery
(version 6.7; david.ncifcrf.gov) (24) and were completed using the
ClusterProfiler R package with default threshold (P<0.05)
(version 3.4.1; bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
(25). The functional network graph
of the selected genes was further visualized. The PPI maps of the
putative mRNAs were constructed using the Search Tool for the
Retrieval of Interacting Genes/Proteins database (version 10.0;
version10.string-db.org) with default
settings.
Validation of the hub genes of
miR-198-5p in LUAD based on TCGA and The Human Protein Atlas
(THPA)
In the PPI network, genes were selected whose edges
(the number of the protein-protein interactions) ranked among the
top 10, and were considered the hub genes of miR-198-5p in LUAD. If
several genes had the same edges, the genes with higher
logFC values were selected. Scatter plots and receiver
operating characteristic (ROC) curves were generated to show the
expression of each hub gene, based on TCGA. The insufficient data
related to miR-198-5p expression in TCGA restricted the analysis.
Subsequently, the expression data of the hub genes at the protein
level were downloaded from THPA (http://www.proteinatlas.org/), an open source database
that provides data on the expression of proteins in a variety of
human tissues. The expression of the hub genes was analzyed
according to the staining intensity in THPA.
Statistical analysis
The quantitative data of miR-198-5p expression are
presented as the mean ± standard deviation. Student's t-test for
independent samples, paired sample t-test, and one-way analysis of
variance with Fisher's least significance difference post hoc test
were performed in SPSS (version 19.0; IBM Corp.) to analyze the
association between miR-198-5p expression and various
clinicopathological characteristics. P<0.05 was considered to
indicate a statistically significant difference.
The sensitivity and specificity of mir-198-5p, as a
diagnostic marker, was assessed by generating ROC curves, and
calculating the area under the curve (AUC) using SPSS version
19.0.
The Kaplan-Meier method was used to estimate the OS
rate of patients with LUAD, and the log-rank test was used to
calculate the differences between groups. To evaluate the survival
variables, univariate and multivariate (if applicable) Cox
regression analyses were performed.
To evaluate the differential expression of
miR-198-5p between LUAD tissues and normal tissues, data from GEO
and ArrayExpress were subjected to a continuous variable
meta-analysis in Stata 12 (StataCorp LP) and the results are
presented as the overall standardized mean difference (SMD) and a
95% confidence interval (CI). The fixed effects model was used
first. Heterogeneity across studies was assessed using the
χ2 test of Q and the I2 statistic. The
existence of heterogeneity was defined as P<0.05 or
I2>50%, indicating that the random effects model was
more suitable. The source of heterogeneity was detected by a
sensitivity analysis, as described previously (26). Begg's test was employed to assess
publication bias with the criterion of P<0.05. Based on the
included microarray, the ROC curves were plotted. Subsequently, a
diagnostic meta-analysis was conducted with MetaDiSc 1.4
(http://www.hrc.es/investigacion/metadisc_en.htm).
Heterogeneity was detected in this model by a meta-regression and a
threshold effect analysis. Finally, the continuous variable
meta-analysis was repeated following the inclusion of the in-house
RT-qPCR data for comparison. The data from both the in-house
RT-qPCR analysis and a previously published study that investigated
miR-198-5p expression in LUAD-associated pleural effusion (19) were included and the diagnostic
meta-analysis was subsequently repeated.
Results
Expression of miR-198-5p and diverse
clinicopathological characteristics in LUAD
The clinicopathological characteristics of 101
patients with LUAD are listed in Table
I. According to the results of the in-house RT-qPCR, the
expression of miR-198-5p was lower in LUAD tissues than in
corresponding adjacent non-tumor lung tissues (4.469±2.495 vs.
5.301±2.502, respectively; P=0.015; AUC, 0.599; Fig. 2A). However, an AUC value of 0.599
indicated that the performance of the ROC curve was not
satisfactory. Subsequently, the expression of miR-198-5p in 101
LUAD tissues was compared for various clinicopathological factors.
As shown in Table I, the samples
from patients <60 years of age had lower expression of
miR-198-5p compared with those from patients ≥60 years of age
(3.732±1.781 vs. 4.972±2.787, respectively; P=0.008; AUC, 0.621;
Fig. 2B). The expression of
miR-198-5p was also lower in tissues with vascular invasion
compared with those without vascular invasion (3.6774±1.9411 vs.
4.819±2.642, respectively; P=0.033; AUC, 0.638; Fig. 2C), and samples from patients at
Tumor-Node-Metastasis stages III–IV compared with those at stages
I–II (3.939±2.069 vs. 5.154±2.838, respectively; P=0.019; AUC,
0.637; Fig. 2D). Moreover, a
decrease in miR-198-5p expression was observed in samples with
lymph node metastasis compared with those without lymph node
metastasis (4.008±2.156 vs. 5.042±2.781, respectively; P=0.038;
AUC, 0.622; Fig. 2E).
 | Table I.Association between microRNA-198-5p
expression and the clinicopathological features in LUAD. |
Table I.
Association between microRNA-198-5p
expression and the clinicopathological features in LUAD.
| Clinicopathological
feature | n | Mean ± SD | Statistical
valuea | P-value |
|---|
| Tissue |
|
| −2.477 | 0.015 |
|
LUAD | 101 | 4.469±2.495 |
|
|
|
Non-tumor | 101 | 5.301±2.502 |
|
|
| Sex |
|
| −0.091 | 0.928 |
|
Male | 56 | 4.448±2.103 |
|
|
|
Female | 45 | 4.494±2.937 |
|
|
| Age |
|
| −2.727 | 0.008 |
|
<60 | 41 | 3.732±1.781 |
|
|
|
≥60 | 60 | 4.972±2.787 |
|
|
| Smoking state |
|
| −1.147 | 0.258 |
| No | 26 | 3.769±1.754 |
|
|
|
Yes | 18 | 4.439±2.106 |
|
|
| Size |
|
| 0.689 | 0.493 |
| ≤3
cm | 53 | 4.632±2.619 |
|
|
| >3
cm | 48 | 4.288±2.365 |
|
|
| EGFR mutation |
|
| −0.096 | 0.924 |
|
Wild-type | 20 | 3.835±1.968 |
|
|
|
Mutation | 13 | 3.900±1.792 |
|
|
| EGFR
amplification |
|
| 0.348 | 0.730 |
| No | 21 | 3.948±2.009 |
|
|
|
Yes | 12 | 3.708±1.679 |
|
|
| Vascular
invasion |
|
| 2.159 | 0.033 |
| No | 70 | 4.819±2.642 |
|
|
|
Yes | 31 | 3.677±1.941 |
|
|
| TNM stage |
|
| 2.391 | 0.019 |
|
I–II | 44 | 5.154±2.838 |
|
|
|
III–IV | 57 | 3.939±2.069 |
|
|
| LNM |
|
| 2.105 | 0.038 |
| No | 45 | 5.042±2.781 |
|
|
|
Yes | 56 | 4.008±2.156 |
|
|
| EGFR protein
expression |
|
| 0.091 | 0.928 |
|
Low | 22 | 3.882±1.994 |
|
|
|
High | 11 | 3.818±1.691 |
|
|
| MET expression |
|
| −0.510 | 0.614 |
|
Low | 20 | 3.725±1.808 |
|
|
|
High | 13 | 4.069±2.023 |
|
|
|
Gradingb |
|
| 0.448 | 0.640 |
| I | 17 | 4.979±3.073 |
|
|
| II | 61 | 4.328±2.242 |
|
|
|
III | 23 | 4.465±2.495 |
|
|
The entire cohort was divided into two groups (low
and high mirR-198-5p expression) according to the median expression
level of miR-198-5p in the LUAD tissues (median, 3.60). The
survival rate of patients with high miR-198-5p expression was
higher than those with low expression (P<0.001; Fig. 2F). The univariate analysis revealed
that the expression level of miR-198-5p served as an independent
prognostic factor for OS time (P<0.001; Table II). The multivariate analysis was
not performed because other clinicopathological factors did not
appear to be effective as prognostic predictors.
 | Table II.Univariate analysis based on the
follow-up of patients with lung adenocarcinoma. |
Table II.
Univariate analysis based on the
follow-up of patients with lung adenocarcinoma.
|
| Overall survival
time (months) |
|---|
|
|
|
|---|
| Variable | P-value | HR | Lower limit | Upper limit |
|---|
| Sex |
| Male
vs. female | 0.456 | 0.779 | 0.404 | 1.502 |
| Age |
| <60
vs. ≥60 | 0.901 | 0.956 | 0.470 | 1.945 |
| Size |
| ≤3 cm
vs. 3 cm | 0.318 | 1.396 | 0.725 | 2.691 |
| Vascular
invasion |
| No vs.
yes | 0.860 | 1.059 | 0.562 | 1.995 |
| TNM stage |
| I–II
vs. III–IV | 0.081 | 1.955 | 0.921 | 4.153 |
| LNM |
| No vs.
yes | 0.389 | 1.351 | 0.681 | 2.683 |
| Grade |
| II vs.
I | 0.699 | 1.213 | 0.456 | 3.233 |
| III vs.
I | 0.698 | 1.108 | 0.660 | 1.860 |
| II vs.
III | 0.754 | 0.892 | 0.438 | 1.817 |
| miR-198-5p
expression |
| High
vs. low | <0.001 | 0.272 | 0.133 | 0.555 |
Meta-analysis based on LUAD microarray
data
The expression pattern and diagnostic value of
miR-198-5p was evaluated by a meta-analysis based on microarray
data from the GEO datasets. A total of 483 samples from six GEO
datasets (in which bodily fluid samples were not available)
including GSE14936 (27), GSE25508
(28), GSE48414 (29), GSE51853 (30), GSE63805 (31) and GSE7419 were included to generate
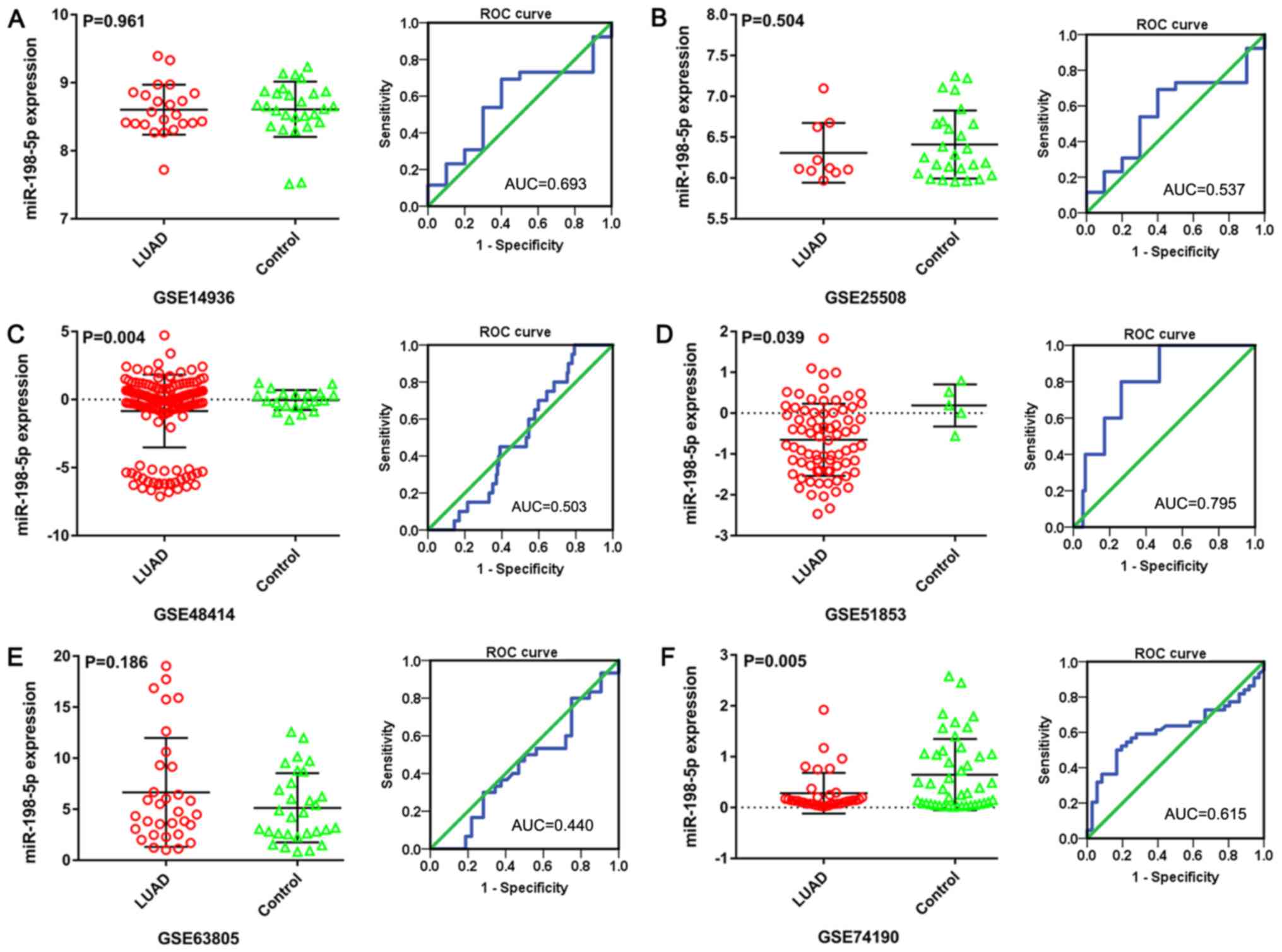
scatter plots and corresponding ROC curves (Fig. 3). No difference in miR-198-5p
expression between LUAD and non-tumor lung tissues was observed in
dataset GSE14936 (8.604±0.368 vs. 8.609±0.406, respectively;
P=0.961; AUC, 0.693; Fig. 3A) and
dataset GSE25508 (6.308±0.366 vs. 6.409±0.416, respectively;
P=0.504; AUC, 0.537; Fig. 3B).
miR-198-5p showed a lower expression in LUAD than in non-tumor lung
tissues in both the GSE48414 (−0.844±2.673 vs. −0.040±0.731,
respectively; P=0.004; AUC, 0.503; Fig.
3C) and GSE51853 (−0.652±0.882 vs. −0.189±0.516, respectively;
P=0.039; AUC, 0.795; Fig. 3D)
datasets. For the GSE63805 dataset, miR-198-5p expression in LUAD
was similar to that in non-tumor lung tissues (6.642±5.328 vs.
5.132±3.397, respectively; P=0.186; AUC, 0.440; Fig. 3E). In contrast, the data from the
GSE74190 dataset showed that miR-198-5p level was lower in LUAD
than in non-tumor lung tissues (0.281±0.400 vs. 0.646±0.702,
respectively; P=0.005; AUC, 0.615; Fig.
3F).
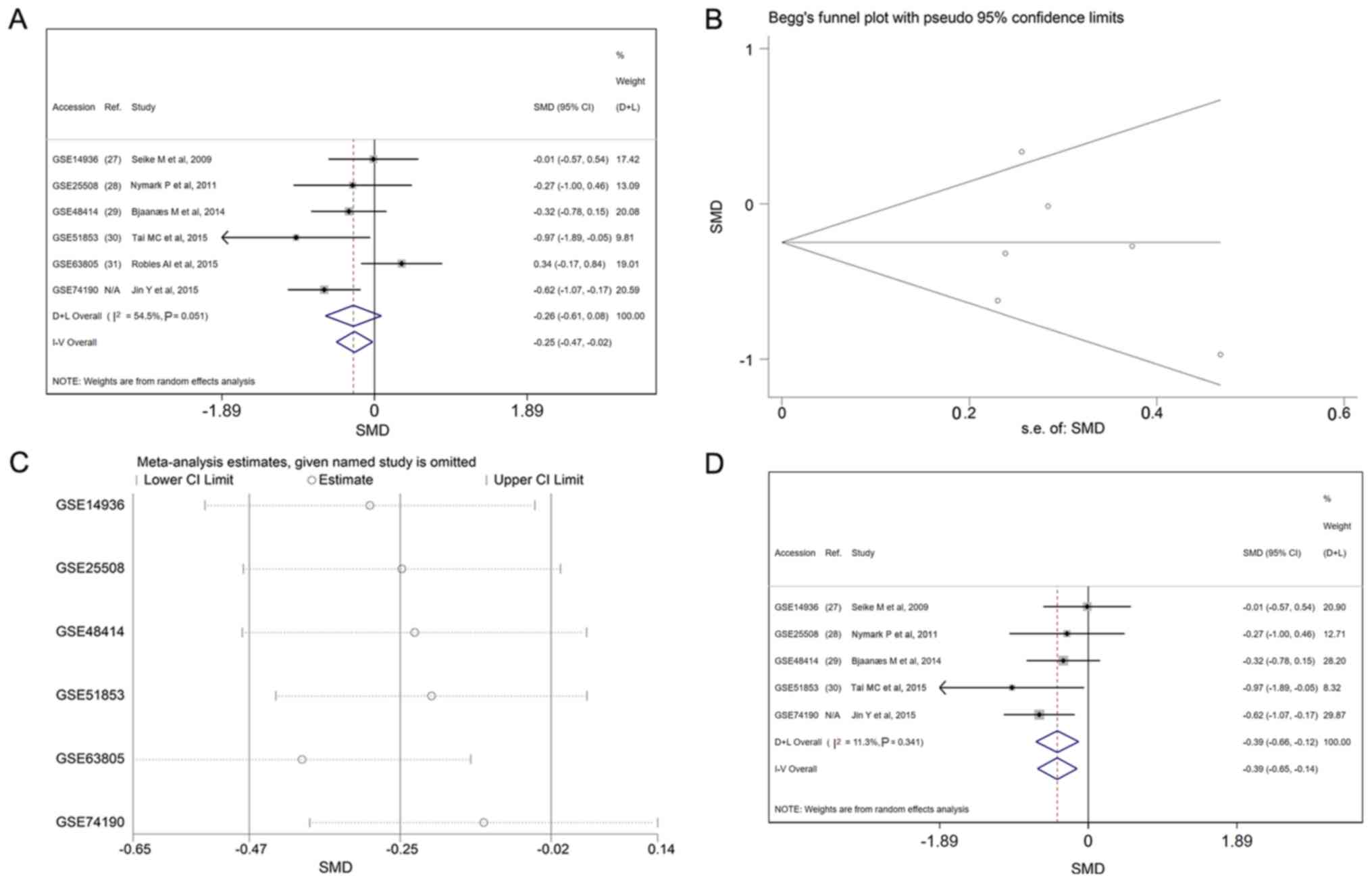
miR-198-5p expression is illustrated in forest plots
(Fig. 4A). An I2 value of
54.5% indicated the existence of heterogeneity across studies. The
combined effect size was −0.25 (95% CI, −0.47 to −0.02) in the
fixed effects models and −0.26 (95% CI, −0.61 −0.08) in the random
effects models. The funnel plots revealed no publication bias in
this meta-analysis (P>0.05; Fig.
4B). Subsequently, a sensitivity analysis was performed to
detect the source of heterogeneity. Each dataset was removed in
turn and the meta-analysis was repeated to recalculate the
heterogeneity. Upon removal of the GSE63805 dataset, the value of
I2 sharply decreased to 11.3%. No such decrease in
I2 was observed when any other of the datasets was
removed (Fig. 4C). GSE63805 was thus
identified as the source of heterogeneity. The forest plot was then
regenerated based on the fixed effects model, following the
elimination of GSE63805 (Fig.
4D).
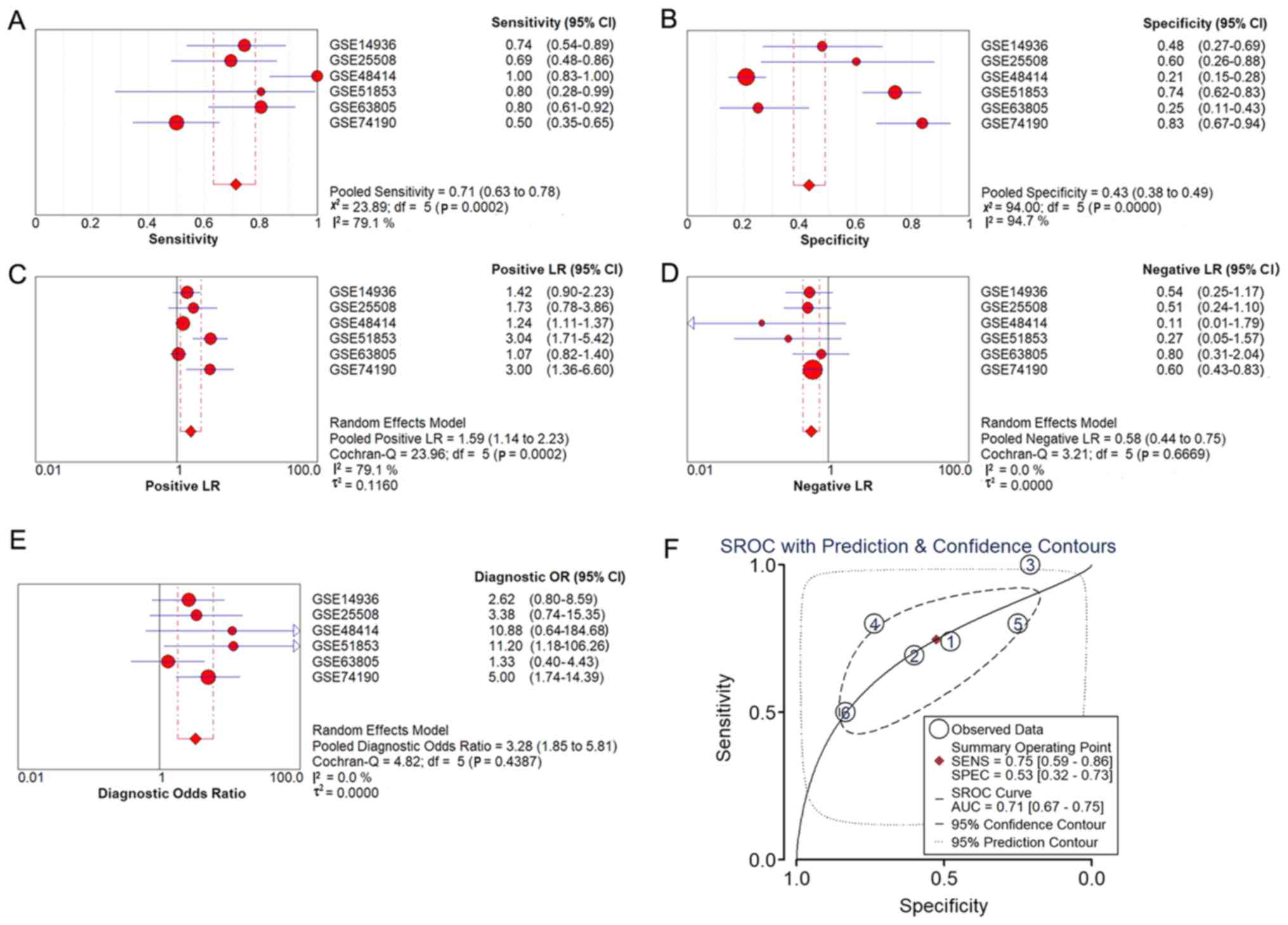
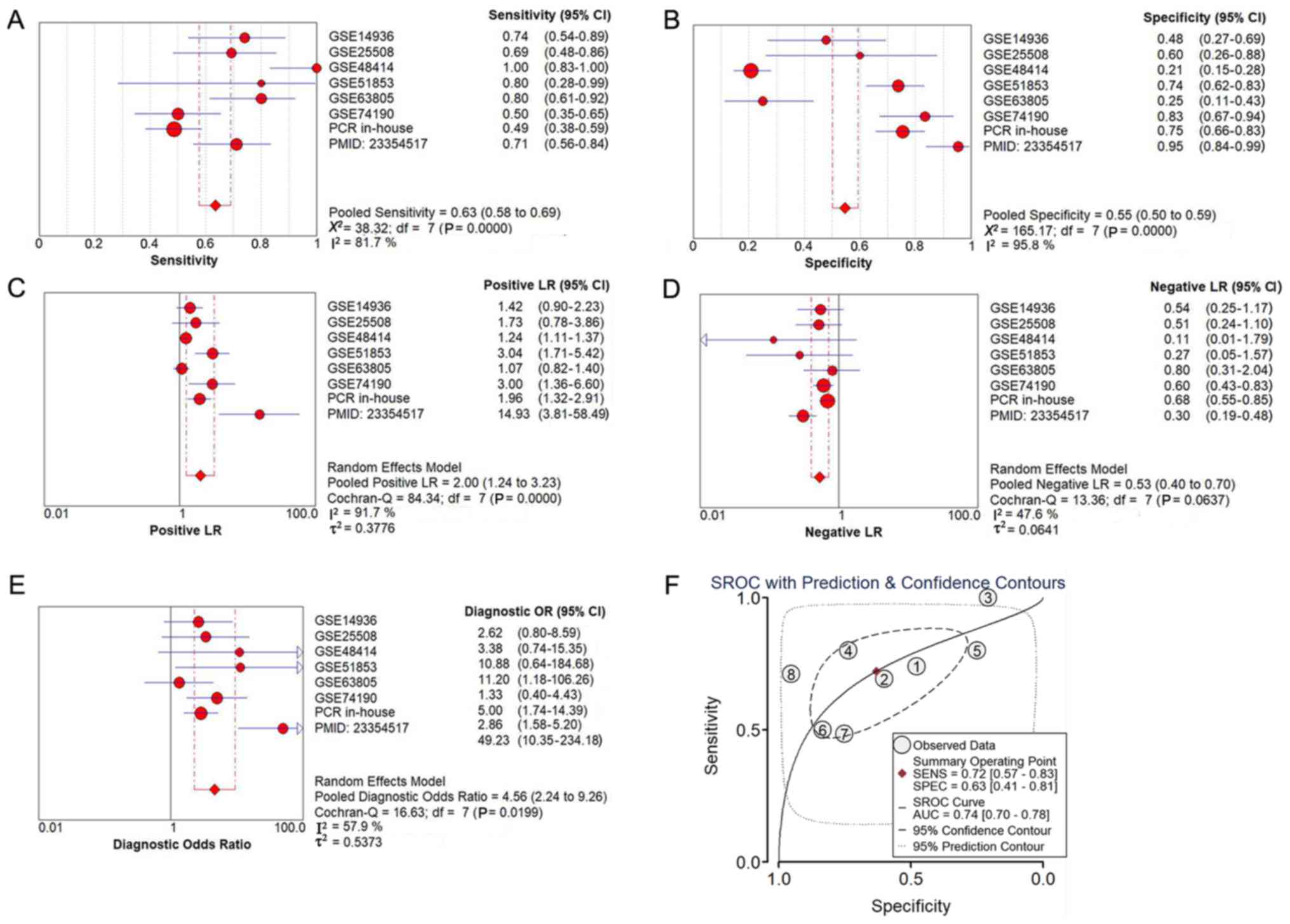
The diagnostic meta-analysis and the summarized ROC
curve, based on the six included microarray datasets, are displayed
in Fig. 5. The pooled sensitivity,
specificity, positive likelihood ratio (PLR), negative likelihood
ratio (NLR) and diagnostic odds ratio (DOR) were 0.71 (95% CI,
0.63–0.78), 0.43 (95% CI, 0.38–0.49), 1.59 (95% CI, 1.14–2.23),
0.58 (95% CI, 0.44–0.75) and 3.28 (95% CI, 1.85–5.81),
respectively, and the AUC was 0.71.
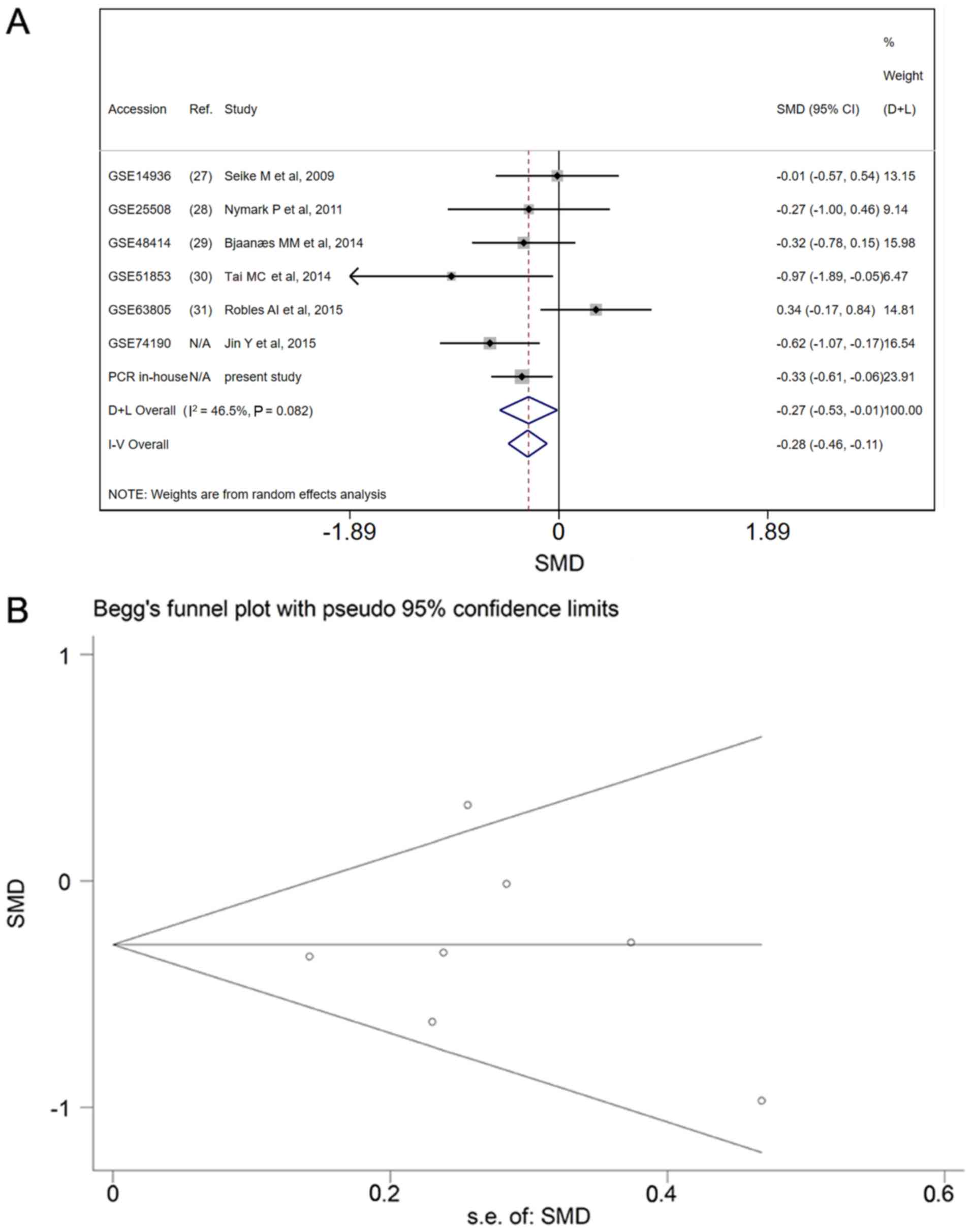
Meta-analysis of miR-198-5p expression
based on patients recruited for the present study and GEO LUAD
data
The miR-198-5p expression data from the patients
recruited for the present study were added to the data from the GEO
datasets and the meta-analysis was repeated to determine the
expression pattern in LUAD. Based on the fixed effects model, the
combined effect size was −0.28 (95% CI, −0.46–0.11) with
I2=46.5% (Fig. 6A). No
publication bias was identified (P>0.05; Fig. 6B). Upon adding of the data from the
present study and data that from a published article
(PMID:23354517) (19), the pooled
sensitivity, specificity, PLR, NLR and DOR were 0.62 (95% CI,
0.56–0.58), 0.51 (95% CI, 0.46–0.56), 1.67 (95% CI, 1.18–2.36),
0.64 (95% CI, 0.54–0.76) and 3.07 (95% CI, 2.03–4.64), respectively
(Fig. 7A-E), and the AUC was 0.74
(Fig. 7F).
GO and KEGG pathway analysis and
construction of a PPI network
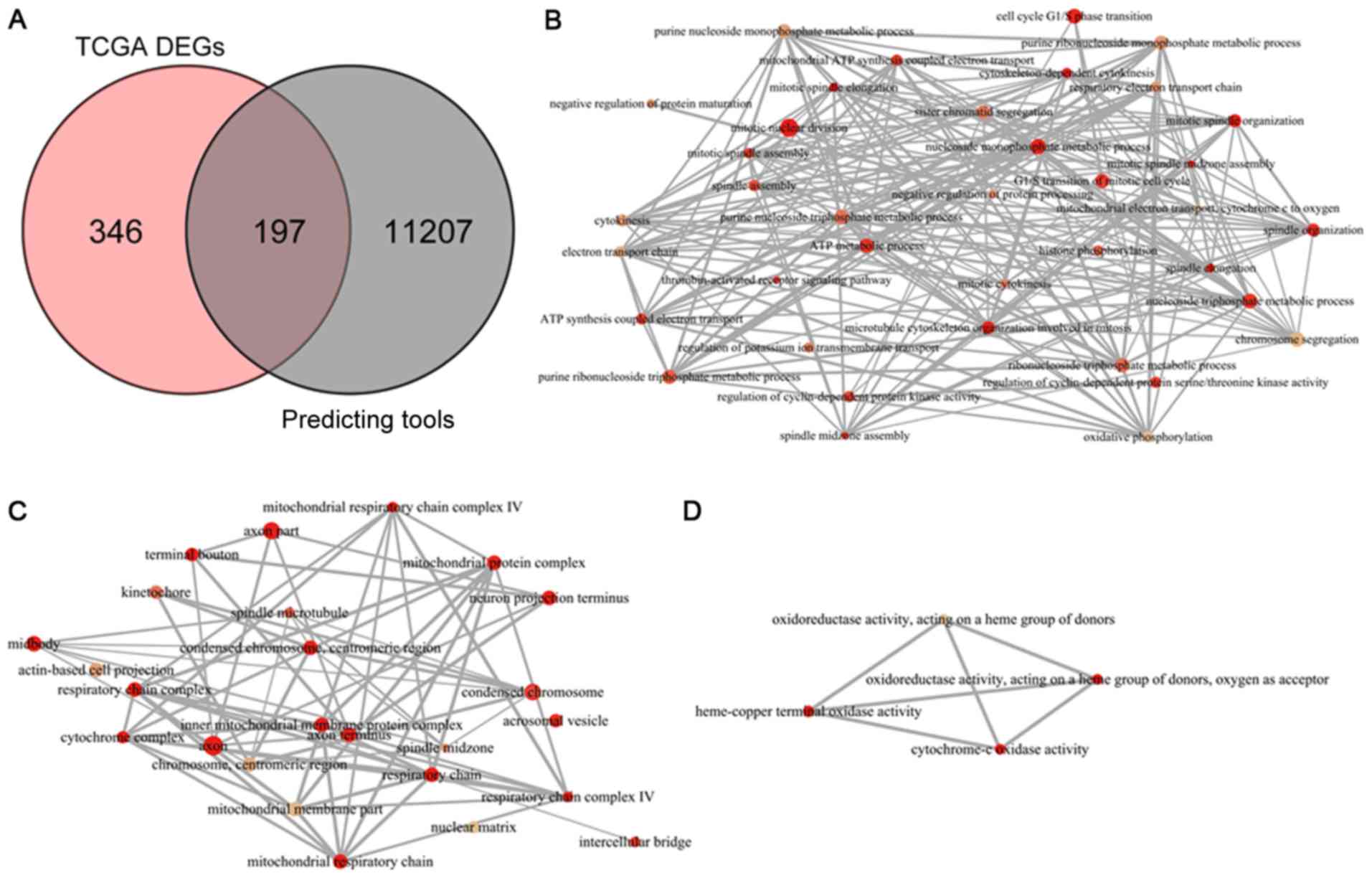
Overall, 197 overlapping genes were identified as
potential targets of miR-198-5p in LUAD (Fig. 8A). The GO enrichment analysis
included three categories: Biological process (BP; Fig. 8B), cellular component (CC; Fig. 8C) and molecular function (MF;
Fig. 8D). The top five terms of each
category are listed in Table III.
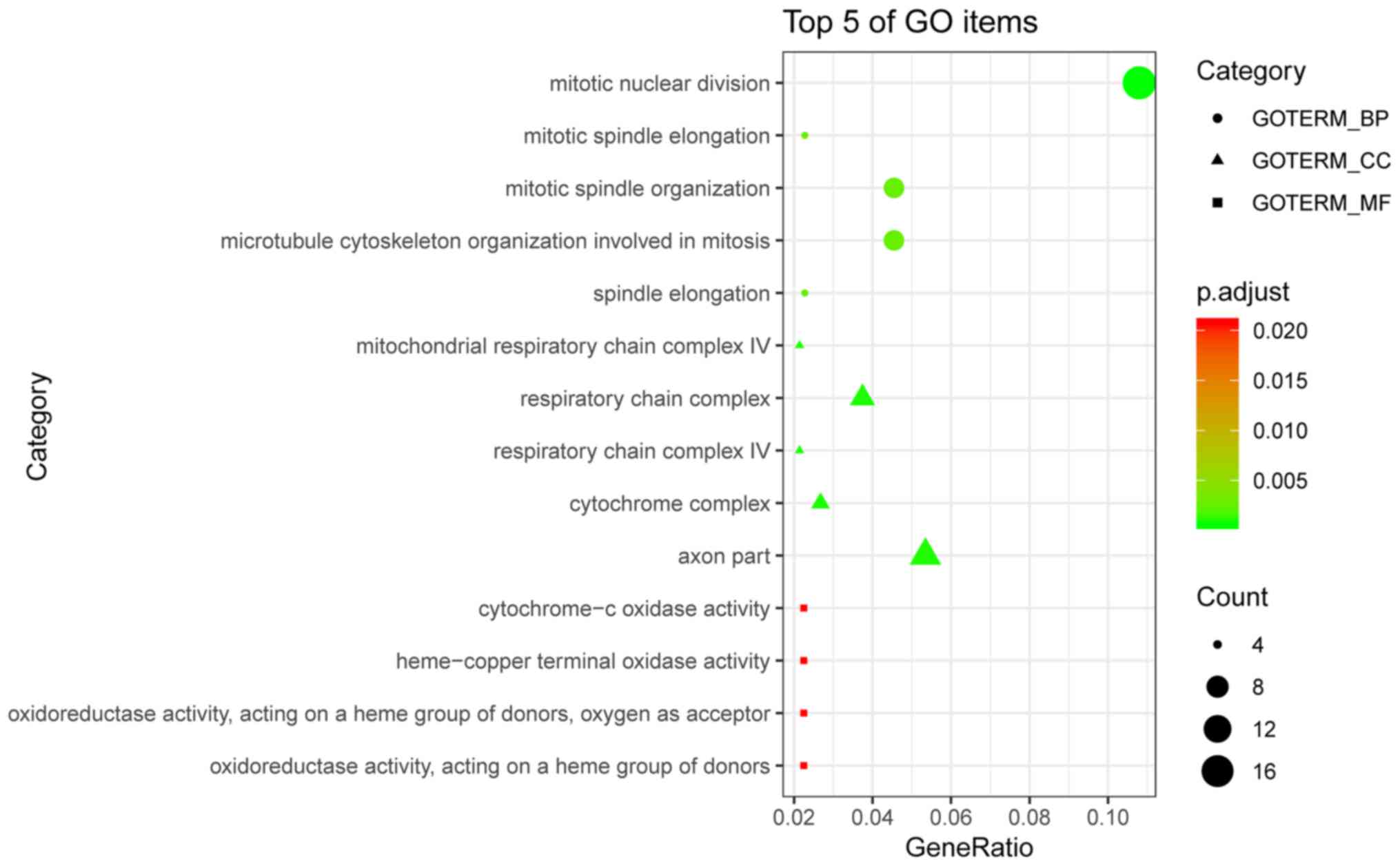
The bubble plot in Fig. 9 displays
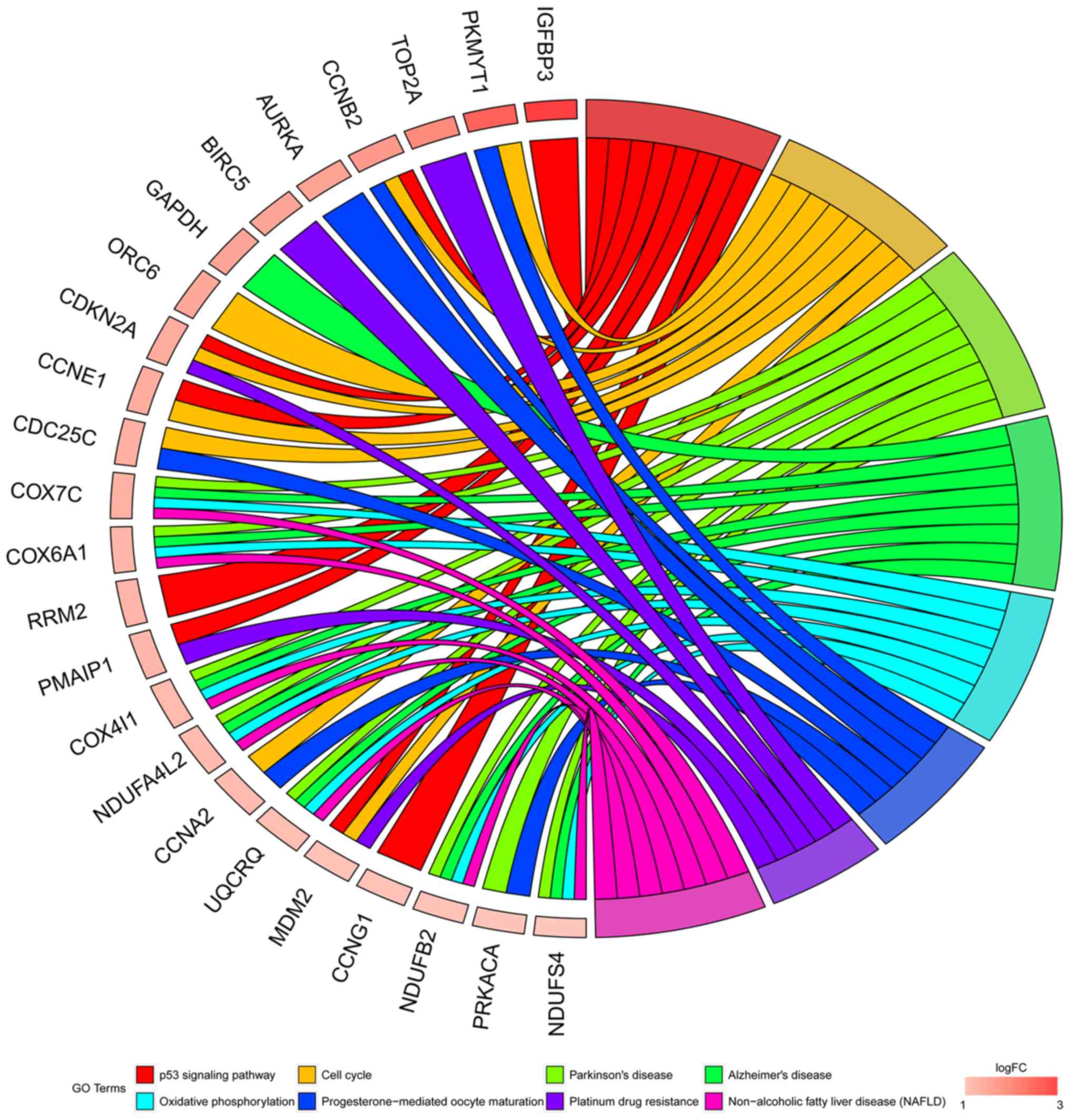
the enriched GO terms. In the KEGG pathway analyses, the potential
targets of miR-198-5p were most associated with the p53 signaling
pathway (P=1.42×10−6; Table
IV and Fig. 10). Other enriched
terms included ‘cell cycle’, ‘Parkinson's disease’, ‘Alzheimer's
disease’, ‘oxidative phosphorylation’, ‘progesterone-mediated
oocyte maturation’, ‘platinum drug resistance’, and ‘non-alcoholic
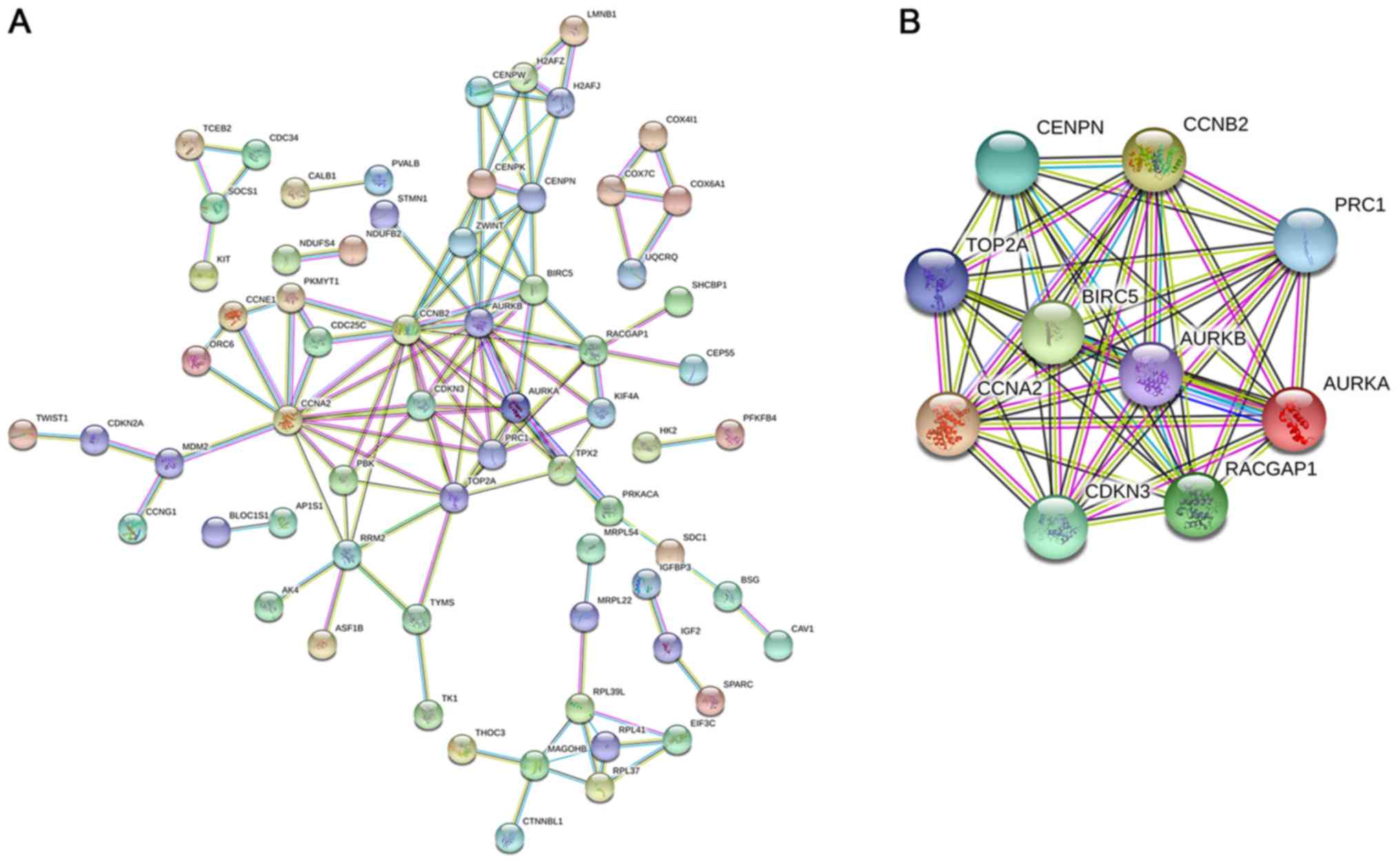
fatty liver disease (NAFLD)’. The PPI network is shown in Fig. 11A with 66 representative nodes (ones
with the largest number of edges) from 197 hub genes and the
corresponding 123 edges and a PPI enrichment
P=1.55×10−14. The selected hub genes were
G2/mitotic-specific cyclin-B2 (CCNB2), Aurora kinase B (AURKB),
cyclin-A2 (CCNA2), DNA topoisomerase 2-α (TOP2A), Aurora kinase A
(AURKA), baculoviral IAP repeat-containing protein 5 (BIRC5),
centromere protein N (CENPN), Rac GTPase-activating protein 1
(RACGAP1), protein regulator of cytokinesis 1 (PRC1) and
cyclin-dependent kinase inhibitor 3 (CDKN3) (Fig. 11B).
 | Table III.Five most enriched terms in BP, CC
and MF based on the GO analysis. |
Table III.
Five most enriched terms in BP, CC
and MF based on the GO analysis.
| Category | ID | Description | Count, n | P-value |
|---|
| GOTERM_BP | GO:0007067 | Mitotic nuclear
division | 19 |
2.49×10−7 |
| GOTERM_BP | GO:0000022 | Mitotic spindle
elongation | 4 |
3.47×10−6 |
| GOTERM_BP | GO:0007052 | Mitotic spindle
organization | 8 |
4.62×10−6 |
| GOTERM_BP | GO:1902850 | Microtubule
cytoskeleton organization involved in mitosis | 8 |
4.62×10−6 |
| GOTERM_BP | GO:0051231 | Spindle
elongation | 4 |
5.16×10−6 |
| GOTERM_CC | GO:0005751 | Mitochondrial
respiratory chain complex IV | 4 |
5.09×10−6 |
| GOTERM_CC | GO:0098803 | Respiratory chain
complex | 7 |
8.48×10−6 |
| GOTERM_CC | GO:0045277 | Respiratory chain
complex IV | 4 |
1.01×10−5 |
| GOTERM_CC | GO:0070069 | Cytochrome
complex | 5 |
1.08×10−5 |
| GOTERM_CC | GO:0033267 | Axon part | 10 |
1.41×10−5 |
| GOTERM_MF | GO:0004129 | Cytochrome-c
oxidase activity | 4 |
1.70×10−4 |
| GOTERM_MF | GO:0015002 | Heme-copper
terminal oxidase activity | 4 |
1.70×10−4 |
| GOTERM_MF | GO:0016676 | oxidoreductase
activity, acting on a Heme group of donors, oxygen as acceptor | 4 |
1.70×10−4 |
| GOTERM_MF | GO:0016675 | Oxidoreductase
activity, acting on a heme group of donors | 4 |
1.96×10−4 |
 | Table IV.KEGG pathway enrichment of the
potential target genes of microRNA-198-5p in lung
adenocarcinoma. |
Table IV.
KEGG pathway enrichment of the
potential target genes of microRNA-198-5p in lung
adenocarcinoma.
| Category | ID | Description | Count, n | P-value |
|---|
| KEGG_PATHWAY | hsa04115 | p53 signaling
pathway | 8 |
1.42×10−6 |
| KEGG_PATHWAY | hsa04110 | Cell cycle | 8 |
1.21×10−4 |
| KEGG_PATHWAY | hsa05012 | Parkinson's
disease | 8 |
3.10×10−4 |
| KEGG_PATHWAY | hsa05010 | Alzheimer's
disease | 8 |
1.07×10−3 |
| KEGG_PATHWAY | hsa00190 | Oxidative
phosphorylation | 7 |
1.12×10−3 |
| KEGG_PATHWAY | hsa04914 |
Progesterone-mediated oocyte
maturation | 6 |
1.2×10−3 |
| KEGG_PATHWAY | hsa01524 | Platinum drug
resistance | 5 |
1.86×10−3 |
| KEGG_PATHWAY | hsa04932 | Non-alcoholic fatty
liver disease (NAFLD) | 7 |
2.15×10−3 |
Validation of the hub genes using TCGA
and THPA
According to data from TCGA, the expression of all
the selected hub genes was higher in the LUAD group than in the
control group (all P<0.001), and the AUCs for CCNB2, AURKB,
CCNA2, TOP2A, AURKA, BIRC5, CENPN, RACGAP1, PRC1 and CDKN3 were
0.968, 0.962, 0.965, 0.985, 0.947, 0.959, 0.830, 0.871, 0.971 and
0.936, respectively (Fig. 12). The
expression of the hub genes at the protein level, obtained from
THPA, is displayed in Fig. 13.
Eight of the ten hub genes were upregulated in LUAD tissues,
whereas the expression data of CENPN and CDKN3 were not available
in the THPA database.
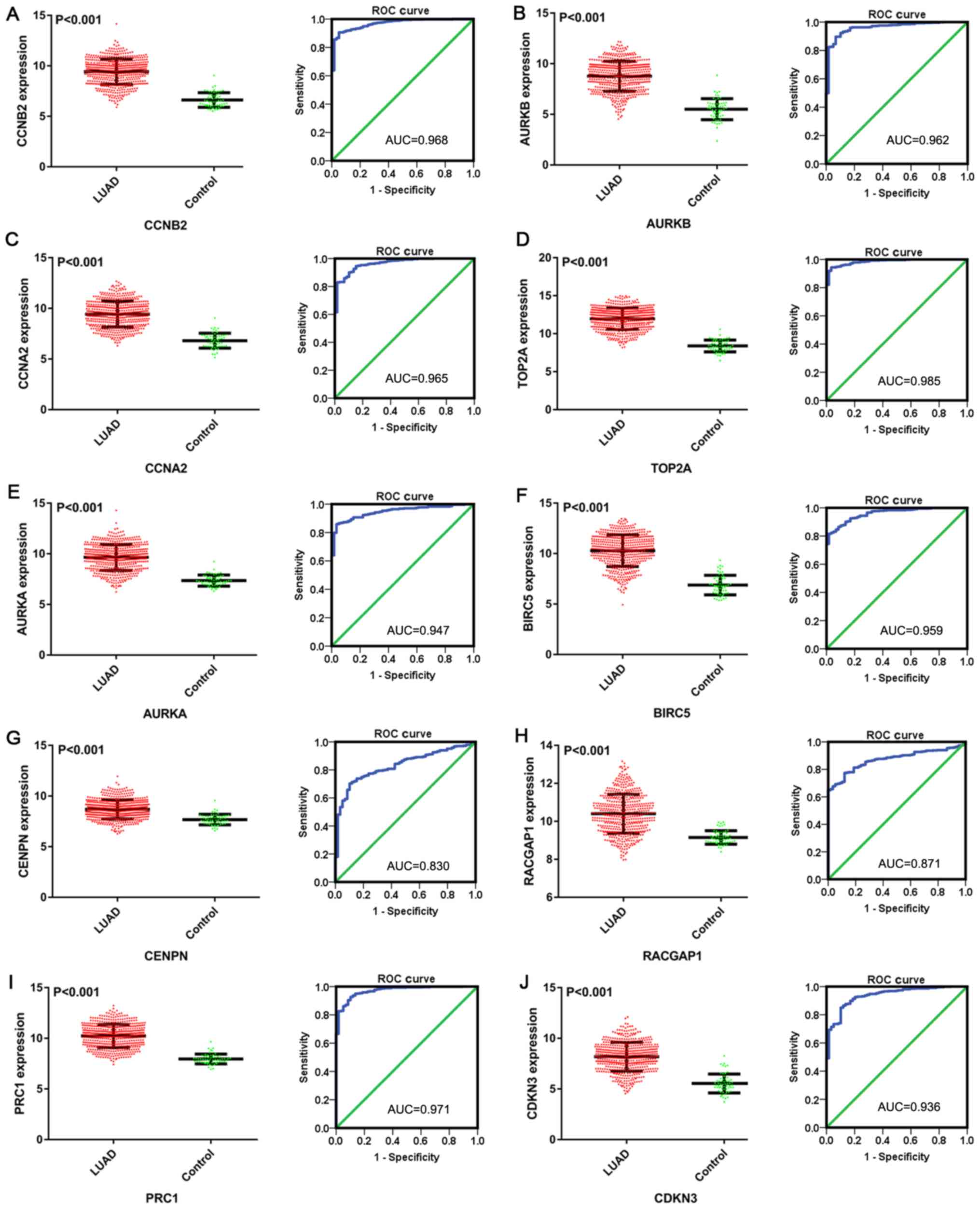 | Figure 12.Scatter plots and ROC curves of 10
hub genes of microRNA-198-5p in LUAD based on data from The Cancer
Genome Atlas. (A) CCNB2; (B) AURKB; (C) CCNA2; (D) TOP2A; (E)
AURKA; (F) BIRC5; (G) CENPN; (H) RACGAP1; (I) PRC1; and (J) CDKN3.
ROC, receiver operating characteristic; LUAD, lung adenocarcinoma,
CCNB2, G2/mitotic-specific cyclin-B2; AURKB, Aurora kinase B;
CCNA2, cyclin-A2; TOP2A, DNA topoisomerase 2-α; AURKA, Aurora
kinase A; BIRC5, baculoviral IAP repeat-containing protein 5;
CENPN, centromere protein N; RACGAP1, Rac GTPase-activating protein
1; PRC1, protein regulator of cytokinesis 1; CDKN3,
cyclin-dependent kinase inhibitor 3. |
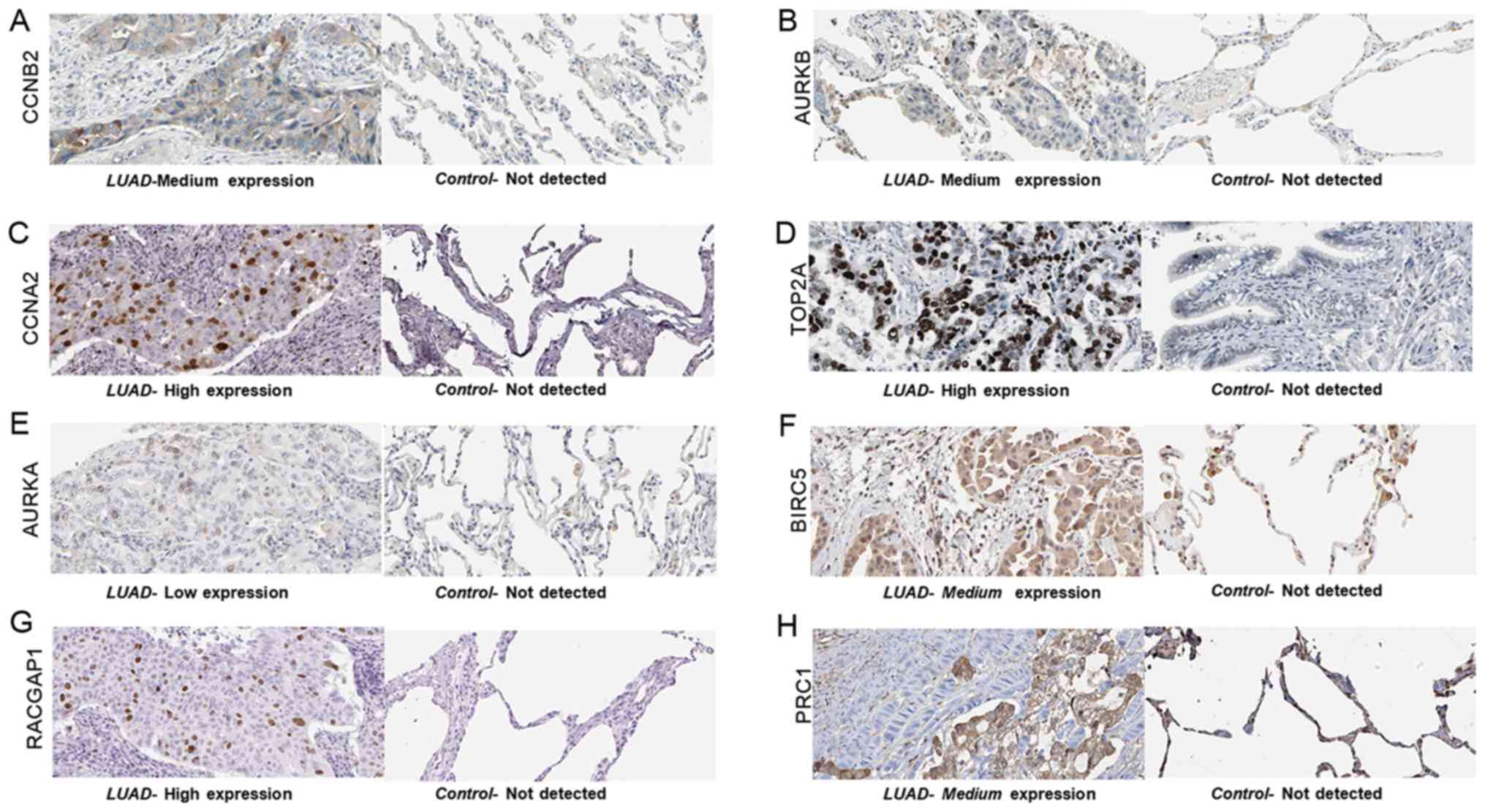 | Figure 13.The expression pattern of the hub
genes in LUAD tissues and adjacent normal lung tissues (control),
obtained from The Human Protein Atlas database. (A) CCNB2 protein
was moderately expressed in LUAD tissue. (B) Medium expression of
AURKB was observed in LUAD tissue. (C) CCNA2 was highly expressed
in LUAD tissue. (D) TOP2A was highly expressed in LUAD tissue. (E)
AURKA expression was low in LUAD. (F) Medium expression of BIRC5 in
LUAD tissue. (G) High expression of RACGAP1 in LUAD tissue. (H)
PRC1 protein staining was medium in LUAD tissues. For all proteins,
no staining was observed in normal lung tissue. LUAD, lung
adenocarcinoma, CCNB2, G2/mitotic-specific cyclin-B2; AURKB, Aurora
kinase B; CCNA2, cyclin-A2; TOP2A, DNA topoisomerase 2-α; AURKA,
Aurora kinase A; BIRC5, baculoviral IAP repeat-containing protein
5; RACGAP1, Rac GTPase-activating protein 1; PRC1, protein
regulator of cytokinesis 1. |
Discussion
In the present study, the expression of miR-198-5p
in LUAD was determined by in-house RT-qPCR on samples from 101
patients. Subsequently, a continuous variable meta-analysis based
on microarray data was performed to validate this expression
pattern. The results of the RT-qPCR and the microarray data were
then integrated into a cohort of 584 cases, which enabled direct
comparison. Comprehensive bioinformatics analyses were performed to
explore the underlying pathways of miR-198-5p in LUAD. In addition,
the expression data of the potential targets of miR-198-5p were
acquired from TCGA and THPA. By comprehensively analyzing the
results of the in-house RT-qPCR, microarray data mining and
meta-analysis, it was found that miR-198-5p was differentially
expressed in LUAD tissues and non-tumor lung tissues. Moreover, the
downregulation of miR-198-5p in LUAD tissues was associated with
multiple clinicopathological characteristics and served as an
independent prognostic factor. In contrast, the potential target
genes of miR-198-5p that were predicted by online tools were
overexpressed in LUAD, according to TCGA and THPA data.
It has been previously observed that miR-198-5p is
involved in multiple physiological processes, and its dysfunction
can lead to various diseases. This miR was upregulated in both
glomerular and tubulointerstitial regions of patients with lupus
nephritis, indicating its participation in autoimmunity (32). In the tissues of fetuses with
anencephaly, miR-198-5p was highly expressed and participated in
the pathogenesis of anencephaly by regulating genes in a protein
interaction network (33); however,
downregulation of miR-198-5p was observed in respiratory syncytial
virus infection and preeclampsia (34,35). In
Parkinson's disease, the expression level of miR-198-5p has been
reported to be low and this mRNA is involved in several
neurodegenerative pathways (36).
Its upregulation in several malignant diseases, including
esophageal cancer (37), multiple
myeloma (38), pancreatic ductal
adenocarcinoma (39), retinoblastoma
(40) and tongue squamous cell
carcinoma (41) was also reported.
Moreover, the expression of miR-198-5p was negatively correlated
with patient survival (37–39); however, miR-198-5p inhibits the
development of certain types of cancer (42). In certain studies, downregulation of
miR-198-5p was observed in prostate cancer (11), osteosarcoma (42), glioblastoma (43), as well as in certain malignancies of
the digestive system, such as colorectal cancer (44), gastric cancer (45), pancreatic cancer (46) and hepatocellular carcinoma (47).
miR-198-5p was reported to enhance chemotherapy
sensitivity in glioblastoma by targeting
O6-methylguanine-DNA methyltransferase (43) or by directly inhibiting cell
proliferation and migration (12),
vital processes for tumorigenesis. In addition, there is evidence
indicating that miR-198-5p participates in anti-tumor immunity,
since its overexpression in the CD8+ T cells of patients
with renal cell carcinoma leads to immune dysfunction via the
targeting of Janus kinase 3 and myeloid cell leukemia 1 (48).
In a previous study, decreased expression of
miR-198-5p was observed in lung squamous cell carcinoma tissues
(SMD, −0.34; 95% CI, −0.71–0.04) (49). For LUAD, miR-198-5p initially
received attention due to its differential expression between
benign pleural effusion and LUAD-associated malignant pleural
effusion (LA-MPE). Han et al (19,50)
reported decreased expression of miR-198-5p in LA-MPE, indicating
its diagnostic potential for this condition. It was observed that
miR-198-5p inhibits lung cancer cellular proliferation and induced
apoptosis by regulating the expression of fibroblast growth factor
receptor 1 (13).
The chemotherapeutic sensitivity of human LUAD cell
line A549 was demonstrated to be positively regulated by miR-198-5p
(13); however, miR-198-5p
expression and the prognosis of patients with LUAD, in terms of OS,
were not found to be associated. The in-house RT-qPCR performed in
the present study revealed that miR-198-5p was downregulated in
LUAD tissues. The continuous variable meta-analysis, which was
based on microarray data, showed the same trend. Following the
inclusion of the RT-qPCR data to the microarray data, no
heterogeneity or publication bias was observed. Considering the
unsatisfactory AUCs in the diagnostic meta-analysis, the diagnostic
value of miR-198-5p in LUAD was not well-established. In addition,
all the samples used for continuous variable meta-analysis were
obtained from lung tissues rather than bodily fluids such as blood
and urine, limiting the use of miR-198-5p as a diagnostic marker
for LUAD. Surgery is the major mode of therapy for patients with
early-stage (TNM stages I–II) LUAD, whereas patients with
advanced-stage (TNM stages III–IV) LUAD rely mainly on other
treatments, such as radiotherapy, chemotherapy, molecular targeted
therapy and immunotherapy (51).
Thus, the patients were divided into two groups (those at early
stage and those at advanced stage) and lower expression of
miR-198-5p was observed in samples from patients at stages III–IV
compared with those at stages I–II.
Lung cancer is considered to be an age-associated
disease, and multiple age-associated miRNAs have been reported to
be expressed aberrantly in lung cancer (52). In the present study, the level of
miR-198-5p was associated with age, whereas age and the prognosis
of patients were not associated. Whether the prognosis of patients
with LUAD was affected by other clinicopathological factors is
worthy of further investigation. miR-198-5p expression was also
associated with blood vessel invasion and lymph node metastasis in
patients with LUAD. Moreover, the downregulation of miR-198-5p in
LUAD tissues was associated with shorter survival time and served
as an independent prognostic factor. Unfortunately, the primary
focus was the influence of miR-198-5p on OS, and not all patients
diagnosed with LUAD received further anticancer treatment at the
hospital. Other characteristics, such as weight, height, underlying
diseases and treatment modality, were not fully available, which
further limited the study.
In the present study, 12 online miRNA-mRNA target
prediction tools were applied to predict the target mRNAs of
miR-198-5p, which had not yet been performed for LUAD, to the best
of our knowledge. To further enhance the reliability of the study,
the predicted targets of miR-198-5p in LUAD were cross-referenced
with the DEGs in TCGA. As miR-198-5p was downregulated in LUAD, the
upregulated DEGs were selected from TCGA for further analysis.
Regarding the potential underlying mechanism of
miR-198-5p in LUAD, the bioinformatics analysis showed the p53
signaling pathway as the most enriched pathway, followed by cell
cycle, Parkinson's disease, Alzheimer's disease, oxidative
phosphorylation, progesterone-mediated oocyte maturation, platinum
drug resistance and NAFLD. The p53 signaling pathway, Alzheimer's
disease, oxidative phosphorylation, progesterone-mediated oocyte
maturation, platinum drug resistance and NAFLD have not been
previously reported in the context of miR-198-5p in LUAD. The p53
gene is a well-known tumor suppressor, and the p53 pathway is
involved in several crucial processes of tumorigenesis, such as
cell cycle arrest (53). A previous
study revealed enhanced cell apoptosis and cell cycle arrests in
LUAD cells by miR-198-5p, by targeting serine
hydroxymethyltransferase 1 (54).
This study did not investigate the involvement of the p53 pathway.
A lower miR-198-5p expression was speculated to be one of the
candidate biomarkers for Parkinson's disease (36), which was substantiated by
bioinformatics analysis to some extent. In the GO BP category, the
putative targets participated mainly in mitotic nuclear division.
For CC, the target genes were enriched mainly in the mitochondrial
respiratory chain complex IV. For MF, the target genes associated
with cytochrome c oxidase activity.
The potential targets of miR-198-5p were CCNB2,
AURKB, CCNA2, TOP2A, AURKA, BIRC5, CENPN, RACGAP1, PRC1 and CDKN3,
whose expression was significantly upregulated in LUAD samples
(according to TCGA data). The upregulation of eight of these hub
genes was subsequently validated at the protein level, where data
from THPA showed positive staining in LUAD samples, whereas no
staining was detected in non-tumor lung tissues. The overexpression
of these hub genes could be ascribed to the dysfunction or
downregulation of miR-198-5p. The miRNA-target gene network is
quite complex; one mRNA may be regulated by disparate miRNAs, and a
single miRNA molecule could target multiple genes in different
diseases (55–57). Hence, identifying the exact mechanism
of miR-198-5p and its target genes requires further investigation.
The findings of the present study provide a computational biology
perspective, rather than a focus on specific target genes.
Several limitations of the present study should be
noted. When distinguishing between LUAD and non-tumor tissues, the
expression of miR-198-5p in bodily fluid samples was not clear,
which limits its application in diagnosis. As mentioned, the
treatment-associated information of the patients was incomplete,
which weakened the reliability of the outcomes. Other experimental
methods such as fluorescence in situ hybridization (FISH)
were not used to verify the downregulation of miR-198-5p in LUAD
samples, as RT-qPCR is regarded as the golden standard for miRNA
quantification (58,59) and the level of certain miRNAs in
NSCLC detected by FISH was consistent with that detected by RT-Qpcr
(60). Moreover, the findings of the
present study are based mainly on online databases and in
silico bioinformatics analyses. Based on TCGA and THPA, the hub
genes of miR-198-5p were upregulated in LUAD both at the RNA level
and the protein level; however, their upregulation in LUAD tissues
remains to be further verified by quantitative means, such as
RT-qPCR and western blotting. The role of miR-198-5p and its
mechanism in LUAD should be investigated with further in
vitro and in vivo experiments.
In conclusion, miR-198-5p is downregulated in LUAD
tissues, and it functions as a prognostic factor. Based on the
combined results of the in-house RT-qPCR, data mining and
bioinformatics analysis, miR-198-5p was demonstrated to be involved
in the development of LUAD by targeting various signaling
pathways.
Acknowledgements
Not applicable.
Funding
The study was supported by the Youth Science
Foundation of Guangxi Medical University, Nanning, China (grant no.
GXMUYSF2014032), the Central Government Guide Local Science and
Technology Development Project, China (grant no. ZY18076006), Fund
of National Natural Science Foundation of China (grant no.
NSFC81560469), Natural Science Foundation of Guangxi, China (grant
no. 2017GXNSFAA198016), and Guangxi Degree and Postgraduate
Education Reform and Development Research Projects, China (grant
no. JGY2019050)
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YYF, SSW, LJP, and GC conceived and designed the
study. YYF, SSW, JCH, YYL, and YNG collected, extracted and
analyzed the data. YYF, JCH, and YNG drafted the manuscript. LJP
and GC critically revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The Medical Ethics Committee of the First Affiliated
Hospital of Guangxi Medical University (Nanning, China) approved
the present study (approval no. 2015 KY-E-041). Each participant
signed an informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AUC
|
area under the curve
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
CI
|
confidence interval
|
|
DEGs
|
differentially expression genes
|
|
DOR
|
diagnostic odds ratio
|
|
FISH
|
fluorescence in situ
hybridization
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
LUAD
|
lung adenocarcinoma
|
|
LA-MPE
|
LUAD-associated malignant pleural
effusion
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
MF
|
molecular function
|
|
miRNA/miR
|
microRNA
|
|
NAFLD
|
non-alcoholic fatty liver disease
|
|
NLR
|
negative likelihood ratio
|
|
NSCLC
|
non-small cell lung cancer
|
|
PLR
|
positive likelihood ratio
|
|
PPI
|
protein-protein interaction
|
|
ROC
|
receiver operating characteristic
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
SMD
|
standard mean difference
|
|
TCGA
|
The Cancer Genome Atlas
|
|
THPA
|
The Human Protein Atlas
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ettinger DS: Ten years of progress in
non-small cell lung cancer. J Natl Compr Canc Netw. 10:292–295.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noone AM, Cronin KA, Altekruse SF,
Howlader N, Lewis DR, Petkov VI and Penberthy L: Cancer incidence
and survival trends by subtype using data from the surveillance
epidemiology and end results program, 1992–2013. Cancer Epidemiol
Biomarkers Prev. 26:632–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berindan-Neagoe I, Monroig Pdel C,
Pasculli B and Calin GA: MicroRNAome genome: A treasure for cancer
diagnosis and therapy. CA Cancer J Clin. 64:311–336. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin Y, Lv Y, Liang R, Yuan C and Zhang J,
He D, Zheng X and Zhang J: Four-miRNA signature as a prognostic
tool for lung adenocarcinoma. Onco Targets Ther. 11:29–36. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murakami T, Kikuchi H, Ishimatsu H, Iino
I, Hirotsu A, Matsumoto T, Ozaki Y, Kawabata T, Hiramatsu Y, Ohta
M, et al: Tenascin C in colorectal cancer stroma is a predictive
marker for liver metastasis and is a potent target of miR-198 as
identified by microRNA analysis. Br J Cancer. 117:1360–1370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Luo H, Xiao N, Duan J, Wang Z and
Wang S: Long noncoding RNA SChLAP1 accelerates the proliferation
and metastasis of prostate cancer via targeting miR-198 and
promoting the MAPK1 pathway. Onco Res. 26:131–143. 2018. View Article : Google Scholar
|
|
12
|
Hu Y, Tang Z, Jiang B, Chen J and Fu Z:
miR-198 functions as a tumor suppressor in breast cancer by
targeting CUB domain-containing protein 1. Oncol Lett.
13:1753–1760. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Zhao H, Xin Y and Fan L:
MicroRNA-198 inhibits proliferation and induces apoptosis of lung
cancer cells via targeting FGFR1. J Cell Biochem. 115:987–995.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mirsadraee S, Oswal D, Alizadeh Y, Caulo A
and van Beek E Jr: The 7th lung cancer TNM classification and
staging system: Review of the changes and implications. World J
Radiol. 4:128–134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression
profiles-database and tools update. Nucleic Acids Res.
35:D760–D765. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Parkinson H, Kapushesky M, Shojatalab M,
Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N,
Lilja P, Lukk M, et al: ArrayExpress-a public database of
microarray experiments and gene expression profiles. Nucleic Acids
Res. 35:D747–D750. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han HS, Yun J, Lim SN, Han JH, Lee KH, Kim
ST, Kang MH, Son SM, Lee YM, Choi SY, et al: Downregulation of
cell-free miR-198 as a diagnostic biomarker for lung
adenocarcinoma-associated malignant pleural effusion. Int J Cancer.
133:645–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seike M, Goto A, Okano T, Bowman ED,
Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et
al: MiR-21 is an EGFR-regulated anti-apoptotic factor in lung
cancer in never-smokers. Proc Natl Acad Sci USA. 106:12085–12090.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nymark P, Guled M, Borze I, Faisal A,
Lahti L, Salmenkivi K, Kettunen E, Anttila S and Knuutila S:
Integrative analysis of microRNA, mRNA and aCGH data reveals
asbestos- and histology-related changes in lung cancer. Genes
Chromosomes Cancer. 50:585–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bjaanaes MM, Halvorsen AR, Solberg S,
Jørgensen L, Dragani TA, Galvan A, Colombo F, Anderlini M,
Pastorino U, Kure E, et al: Unique microRNA-profiles in
EGFR-mutated lung adenocarcinomas. Int J Cancer. 135:1812–1821.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tai MC, Kajino T, Nakatochi M, Arima C,
Shimada Y, Suzuki M, Miyoshi H, Yatabe Y, Yanagisawa K and
Takahashi T: miR-342-3p regulates MYC transcriptional activity via
direct repression of E2F1 in human lung cancer. Carcinogenesis.
36:1464–1473. 2015.PubMed/NCBI
|
|
31
|
Robles AI, Arai E, Mathé EA, Okayama H,
Schetter AJ, Brown D, Petersen D, Bowman ED, Noro R, Welsh JA, et
al: An integrated prognostic classifier for stage I lung
adenocarcinoma based on mRNA, microRNA, and DNA methylation
biomarkers. J Thorac Oncol. 10:1037–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow
KM, Wang G, Li PK and Szeto CC: Glomerular and tubulointerstitial
miR-638, miR-198 and miR-146a expression in lupus nephritis.
Nephrology (Carlton). 17:346–351. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Chang H, Li Y, Zhang T, Zou J,
Zheng X and Wu J: MicroRNAs: Potential regulators involved in human
anencephaly. Int J Biochem Cell Biol. 42:367–374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bakre A, Mitchell P, Coleman JK, Jones LP,
Saavedra G, Teng M, Tompkins SM and Tripp RA: Respiratory syncytial
virus modifies microRNAs regulating host genes that affect virus
replication. J Gen Virol. 93:2346–2356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Choi SY, Yun J, Lee OJ, Han HS, Yeo MK,
Lee MA and Suh KS: MicroRNA expression profiles in placenta with
severe preeclampsia using a PNA-based microarray. Placenta.
34:799–804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cardo LF, Coto E, Ribacoba R, Menéndez M,
Moris G, Suárez E and Alvarez V: MiRNA profile in the substantia
nigra of Parkinson's disease and healthy subjects. J Mol Neurosci.
54:830–836. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qi B, Yao WJ, Zhao BS, Qin XG, Wang Y,
Wang WJ, Wang TY, Liu SG and Li HC: Involvement of microRNA-198
overexpression in the poor prognosis of esophageal cancer. Asian
Pac J Cancer Prev. 14:5073–5076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bi C, Chung TH, Huang G, Zhou J, Yan J,
Ahmann GJ, Fonseca R and Chng WJ: Genome-wide pharmacologic
unmasking identifies tumor suppressive microRNAs in multiple
myeloma. Oncotarget. 6:26508–26518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vychytilova-Faltejskova P, Kiss I, Klusova
S, Hlavsa J, Prochazka V, Kala Z, Mazanec J, Hausnerova J, Kren L,
Hermanova M, et al: MiR-21, miR-34a, miR-198 and miR-217 as
diagnostic and prognostic biomarkers for chronic pancreatitis and
pancreatic ductal adenocarcinoma. Diagn Pathol. 10:382015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao JJ, Yang J, Lin J, Yao N, Zhu Y,
Zheng J, Xu J, Cheng JQ, Lin JY and MA J: Identification of miRNAs
associated with tumorigenesis of retinoblastoma by miRNA microarray
analysis. Childs Nerv Syst. 25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of the tongue. Clin Cancer Res.
14:2588–2592. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang S, Zhao Y and Wang L: MicroRNA-198
inhibited tumorous behaviors of human osteosarcoma through directly
targeting ROCK1. Biochem Biophys Res Commun. 472:557–565. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nie E, Jin X, Wu W, Yu T, Zhou X, Shi Z,
Zhang J, Liu N and You Y: MiR-198 enhances temozolomide sensitivity
in glioblastoma by targeting MGMT. J Neurooncol. 133:59–68. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang M, Wang J, Kong X, Chen H, Wang Y,
Qin M, Lin Y, Chen H, Xu J, Hong J, et al: MiR-198 represses tumor
growth and metastasis in colorectal cancer by targeting fucosyl
transferase 8. Sci Rep. 4:61452014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cui Z, Zheng X and Kong D: Decreased
miR-198 expression and its prognostic significance in human gastric
cancer. World J Surg Oncol. 14:332016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Marin-Muller C, Li D, Bharadwaj U, Li M,
Chen C, Hodges SE, Fisher WE, Mo Q, Hung MC and Yao Q: A
tumorigenic factor interactome connected through tumor suppressor
microRNA-198 in human pancreatic cancer. Clin Cancer Res.
19:5901–5913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang WT, Wang HL, Yang H, Ren FH, Luo YH,
Huang CQ, Liang YY, Liang HW, Chen G and Dang YW: Lower expressed
miR-198 and its potential targets in hepatocellular carcinoma: A
clinicopathological and in silico study. Onco Targets Ther.
9:5163–5180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gigante M, Pontrelli P, Herr W, Gigante M,
D'Avenia M, Zaza G, Cavalcanti E, Accetturo M, Lucarelli G,
Carrieri G, et al: miR-29b and miR-198 overexpression in
CD8+ T cells of renal cell carcinoma patients
down-modulates JAK3 and MCL-1 leading to immune dysfunction. J
Transl Med. 14:842016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liang YY, Huang JC, Tang RX, Chen WJ, Chen
P, Cen WL, Shi K, Gao L, Gao X, Liu AG, et al: Clinical value of
miR-198-5p in lung squamous cell carcinoma assessed using
microarray and RT-qPCR. World J Surg Oncol. 16:222018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Han HS, Jo YN, Lee JY, Choi SY, Jeong Y,
Yun J and Lee OJ: Identification of suitable reference genes for
the relative quantification of microRNAs in pleural effusion. Oncol
Lett. 8:1889–1895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ,
Dobelbower M, et al: Non-small cell lung cancer, version 5.2017,
NCCN clinical practice guidelines in oncology. J Natl Compr Canc
Netw. 15:504–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zagryazhskaya A and Zhivotovsky B: miRNAs
in lung cancer: A link to aging. Ageing Res Rev. 17:54–67. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ozaki T and Nakagawara A: Role of p53 in
cell death and human cancers. Cancers (Basel). 3:994–1013. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu S, Zhang G, Li P, Chen S, Zhang F, Li
J, Jiang C, Chen X, Wang Y, Du Y, et al: miR-198 targets SHMT1 to
inhibit cell proliferation and enhance cell apoptosis in lung
adenocarcinoma. Tumour Biol. 37:5193–5202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hausser J and Zavolan M: Identification
and consequences of miRNA-target interactions-beyond repression of
gene expression. Nat Rev Genet. 15:599–612. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nakanishi K: Anatomy of RISC: How do small
RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip
Rev RNA. 7:637–660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Duan D, Zheng KX, Shen Y, Cao R, Jiang L,
Lu Z, Yan X and Li J: Label-free high-throughput microRNA
expression profiling from total RNA. Nucleic Acids Res.
39:e1542011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ouyang T, Liu Z, Han Z and Ge Q: MicroRNA
detection specificity: Recent advances and future perspective. Anal
Chem. 91:3179–3186. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li J, Yang H, Li Y, Liu Y, Chen S, Qi C,
Zhang Q, Lan T, He X, Guan XY and Wang L: microRNA-146
up-regulation predicts the prognosis of non-small cell lung cancer
by miRNA in situ hybridization. Exp Mol Pathol. 96:195–199. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ,
Van Schil PE, et al: International association for the study of
lung cancer/americanthoracic society/european respiratory society
internationalmultidisciplinary classification of lung
adenocarcinoma. J Thorac Oncol. 6:244–285. 2011. View Article : Google Scholar : PubMed/NCBI
|