Introduction
Gastric cancer (GC), a genetic disease involving
multiple changes in the genome, remains a primary cause of male
mortality in developing countries, with its incidence continuously
increasing in recent years (1,2). In
spite of the improvements achieved in the techniques employed for
diagnosis and treatment, the majority of patients are diagnosed
during the advanced stages of disease, and thus suffer a poor
prognosis (3–5). Therefore, improving the early diagnosis
and treatment of GC is a major strategy for decreasing the rate of
mortality.
Over the past few decades, a large number of studies
have focused on the aberrant expression of protein-coding RNAs in
different types of cancer, which have provided promising approaches
for cancer diagnosis and treatment (6,7).
However, along with the advances made in high-resolution
microarrays and genome-wide sequencing technology, an increasing
body of evidence from genomics and transcriptomics studies has
suggested that long non-coding RNAs (lncRNAs) may be effective
biomarkers for cancer detection and molecular targets for cancer
therapy (8–11). The lncRNA actin filament-associated
protein 1 antisense RNA 1 (AFAP1-AS1) has been demonstrated to be
upregulated in ovarian cancer, gallbladder cancer, esophageal
squamous cell carcinoma, pancreatic ductal adenocarcinoma and
colorectal cancer (12–16). However, whether lncRNA AFAP1-AS1 is
involved in the development and tumorigenesis of GC remains
unknown. The aim of the present study was to determine the clinical
significance and biological functions of AFAP1-AS1 in GC.
Materials and methods
Patients and tissue samples
A total of 52 GC samples and the corresponding
adjacent tissues were obtained from patients who underwent surgery
during October 2013 and August 2015 at Nanjing Hospital Affiliated
with Nanjing Medical University (Nanjing, China). The inclusion
criteria for patient recruitment were as follows: i) The first
diagnosis of the patient is GC (age 20–85 years old); ii) the
patients' cardiopulmonary function was confirmed to be normal by
pathological examination prior to surgery; iii) the patients'
blood, liver and kidney function were basically normal; iv) none of
the patients received radiotherapy or chemotherapy prior to
surgery; and v) all patients were diagnosed with clinical TNM stage
(cTNM) I–IV GC (17) and had
detailed clinical data and basic personal information available.
The exclusion criteria for patient recruitment were: i) Patients
who has undergone radiotherapy and chemotherapy prior to surgery;
ii) patients who were in poor physical condition and would not be
able to tolerate the required examination; iii) patients in which
GC was not the primary tumor, but a result of secondary tumor
metastasis; and iv) diagnosis of secondary diseases upon admission,
in particular other types of cancer besides GC. The tumor tissues
and adjacent non-tumor tissue (the distance from tumor surgical
margin ≥5 cm) were collected during surgical resection. All
specimens were immediately stored at −80°C in a freezer until
subsequent use. The present study was approved by the Ethics
Committee of Nanjing First Hospital, Nanjing Medical University
(Jiangsu, China). Written informed consent was obtained from all of
the patients prior to participation.
RNA isolation, reverse transcription
(RT), and RT-quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from the cancerous and
paracancerous tissues using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). cDNA was obtained with the
Prime-Script™ RT-PCR kit (Takara Biotechnology Co., Ltd.).
AFAP1-AS1 expression was measured by RT-qPCR with the following
primer sequences: AFAP1-AS1 forward, 5′-TCGCTCAATGGAGTGACGGCA-3′;
and AFAP1-AS1 reverse, 5′-CGGCTGAGACCGCTGAGAACTT-3′. The results
were normalized to GAPDH using the following primers: GAPDH
forward, 5′-GTCAACGGATTTGGTCTGTATT-3′; and GAPDH reverse,
5′-AGTCTTCTGGGTGGCAGTGAT-3′. RT-qPCR reactions were performed with
ABI7500 System and SYBR Green PCR Master Mix (Thermo Fisher
Scientific, Inc.). The PCR was performed using the following
conditions: Initial denaturation at 95°C for 30 sec; 40 cycles of
95°C for 5 sec and 60°C for 30 sec with final annealing and
extension at 60°C for 60 sec and 95°C for 15 sec. The expression
level of AFAP1-AS1 in GC tissues and corresponding paracancerous
tissues was analyzed using the 2ΔΔCq method (18), and the fold change of target genes
was analyzed.
Cell culture
The human GC MKN-45, MGC-803 and AGS cell lines were
purchased from Shanghai Institute of Cell Biology. Cells were
cultured in RPMI-1640 medium supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), penicillin (100 U/ml), and
streptomycin (100 U/ml) at 37°C in a humidified incubator with 5%
CO2.
Small interfering RNA (siRNA)
preparation and transfection
For gene knockdown, cells were seeded for 10–12 h
and transfected with either 10 nM siRNA or scramble control siRNA
(Invitrogen; Thermo Fisher Scientific, Inc.) using Lipofectamine™
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in Opti-MEM
(Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of the
AFAP1-AS1 targeting siRNAs are presented in Table I.
 | Table I.Sequences of the AFAP1-AS1-targeting
siRNAs. |
Table I.
Sequences of the AFAP1-AS1-targeting
siRNAs.
| siRNAs | Forward | Reverse |
|---|
| Negative control |
5′-GCGACGAUCUGCCUAAGA-3′ |
5′-AUCUUAGGCAGAUCGUCG-3′ |
| siRNA1 |
5′-GUCCCAGCUUACACUUGUATT-3′ |
5′-UACAAGUGUAAGCUGGGACTT-3′ |
| siRNA2 |
5′-GGGCUUCAAUUUACAAGCATT-3′ |
5′-UGCUUGUAAAUUGAAGCCCTT-3′ |
| siRNA3 |
5′-CCUAUCUGGUCAACACGUATT-3′ |
5′-UACGUGUUGACCAGAUAGGTT-3′ |
Transfection experiments were performed at 10–12 h
after cell growth. Cell proliferation was in the logarithmic growth
phase, and the cell area size occupied 50–60% of the culture dish
area. Initial detection may be performed at multiple concentration
gradients, to detect the interference efficiency of different
concentrations of siRNA. Using a 10 nmol (1.2 µl) interference
concentration as an example, 100 µl Opti-MEM, 1.2 µl negative
control (NC)/interference reagents (siRNA), and 12 µl HiPerFect
(Qiagen GmbH) were added to each tube. After 48 h, the cells were
harvested and RT-qPCR was performed as aforementioned to detect the
interference efficiency.
MTT assays
Cells were seeded onto 96-well culture plates at a
density of 1×103 cells per well and incubated at 37°C overnight.
Then, the cells were incubated with different concentration (1–10
µm) of curcumin for 72 h. After 24, 48, 72 or 96 h, 20 µl MTT
dissolved in 100 µl RPMI-1640 was added to each well and incubated
for an additional 4 h. Then, 150 µl dimethyl sulfoxide was added to
dissolve formazan crystals. Optical density was detected at a
wavelength of 490 nm using an enzyme-labeled analyzer.
Flow cytometry analysis
The GC MGC-803 cell line was harvested following
treatment and washed with PBS. Then, cells were resuspended and
stained with fluorescein isothiocyanate-Annexin V and propidium
iodide Apoptosis Detection Kit (cat. no. 556547; BD Biosciences)
for apoptosis analysis. A flow cytometer (CYTOMICS FC 500; Beckman
Coulter, Inc.) was used to analyze the number of apoptotic MGC-803
cells, and the cell cycle distribution. The fluorescence signal was
detected at an emission wavelength of 530 nm. The percentage of
cells in the G0-G1, S, and G2-M phases was counted and compared.
All data were analyzed using Flowjo v10 software (Tree Star,
Inc.).
In vitro wound healing assay
Wounds were created in adherent cells using a 20 µl
sterile pipette tip after 48 h of transfection with si-AFAP1-AS1 or
si-NC. Cells were then washed 3 times with PBS to remove any
free-floating cells and debris. Medium without serum was added, and
the cells were incubated at 37°C in a humidified atmosphere
containing 5% CO2. Wound healing was observed after 0, 12, or 24 h,
respectively, and images of the cells were captured using digital
light microscope at ×200 magnification.
Statistical analysis
All experiments were performed at least 3 times, and
the data are presented as the mean ± standard deviation. The
differential expression levels of AFAP1-AS1 between the cancerous
and adjacent tissues were analyzed with paired Student's t-tests.
Categorical data in Table II were
analyzed using the χ2 test. A one-way analysis of variance followed
by the Bonferroni's post-hoc test was used to evaluate numerical
differences for multiple comparisons. For GC tissues and the
corresponding non-cancerous adjacent tissues, the fold change of
the target gene was calculated using the 2-∆∆Cq method (15). All statistical analyses were
performed with SPSS 23 software (IBM Corp.) and GraphPad Prism 7.0
(GraphPad Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
 | Table II.Associations between AFAP1-AS1 and
clinicopathological characteristic of 52 primary gastric cancer
samples. |
Table II.
Associations between AFAP1-AS1 and
clinicopathological characteristic of 52 primary gastric cancer
samples.
| Variables | Over expression of
AFAP1-AS1, n (%) | Normal expression of
AFAP1-AS1, n (%) | χ2
value | P-value |
|---|
| Age, y |
|
| 0.084 | 0.772 |
| ≥60 | 20 (52.6) | 6 (42.9) |
|
|
|
<60 | 18 (47.4) | 8 (57.1) |
|
|
| Sex |
|
| 2.608 | 0.106 |
| Male | 28 (73.7) | 7 (50) |
|
|
|
Female | 10 (26.3) | 7 (50) |
|
|
| Diameter, cm |
|
| 3.791 | 0.052 |
| ≥5 | 13 (34.2) | 9 (64.3) |
|
|
|
<5 | 25 (65.8) | 5 (35.7) |
|
|
| Sites |
|
| 0.259 | 0.611 |
|
Upper | 16 (47.1) | 7 (50) |
|
|
|
Lower | 22 (52.9) | 7 (50) |
|
|
| Differentiation |
|
| 1.481 | 0.224 |
|
Well/Mid | 18 (47.4) | 4 (28.6) |
|
|
|
Poorly | 20 (52.6) | 10 (71.4) |
|
|
| Lymphatic
metastasis |
|
| 6.656 | 0.010 |
|
Yes | 12 (31.6) | 10 (71.4) |
|
|
| No | 26 (68.4) | 4 (28.6) |
|
|
| TNM stage |
|
| 7.137 | 0.008 |
|
I+II | 14 (36.8) | 11 (78.6) |
|
|
|
III+IV | 24 (63.2) | 3 (21.4) |
|
|
Results
Expression of LncRNA AFAP1-AS1 in GC
tissues
RT-qPCR was used to examine AFAP1-AS1 mRNA
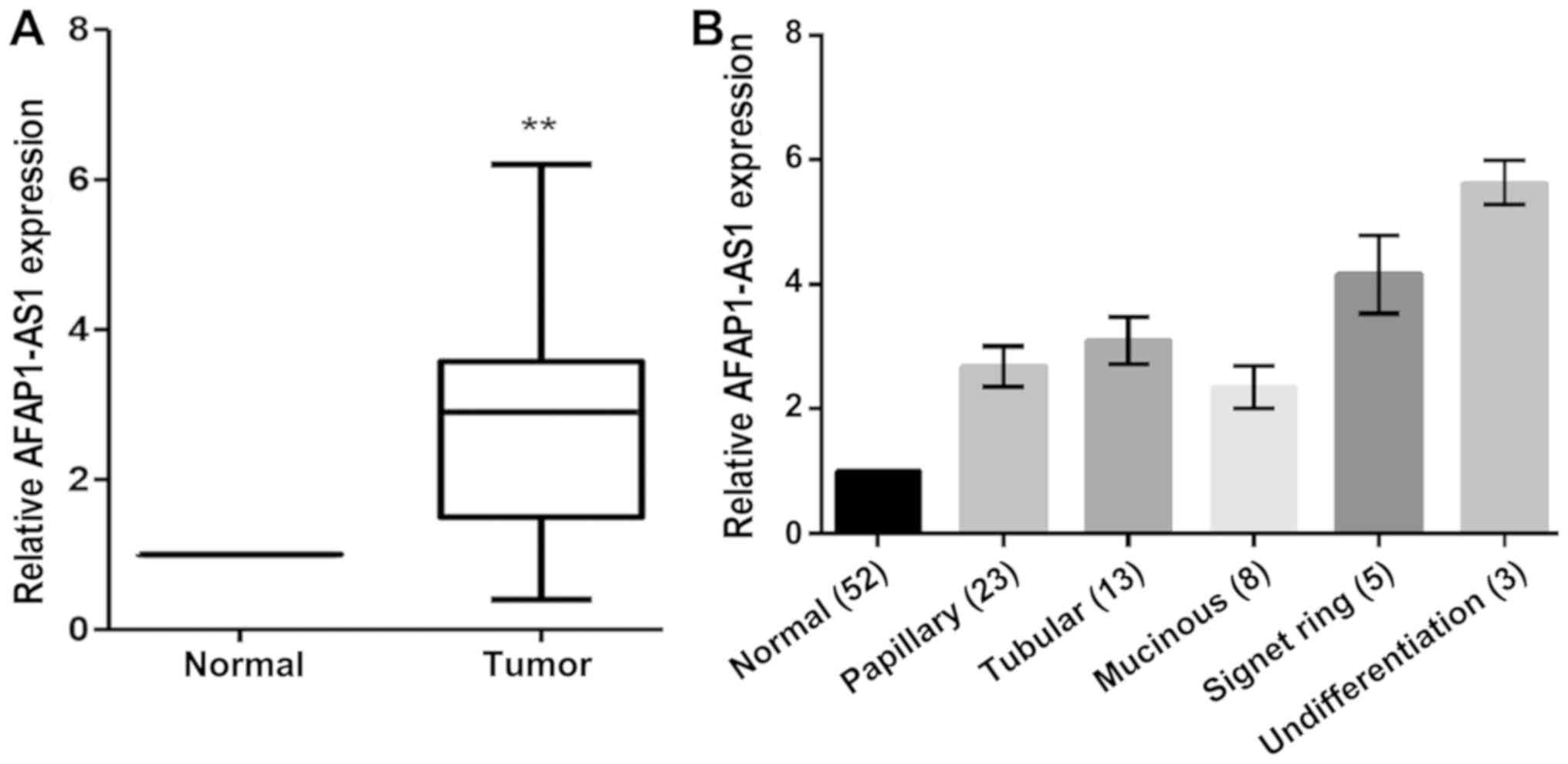
expression in 52 cases of GC. The results demonstrated that lncRNA
AFAP1-AS1 expression levels were significantly upregulated in GC
tissues when compared with the corresponding non-cancerous tissues
(n=52; P<0.05; Fig. 1).
Evaluation of the differences in the clinicopathological
characteristics between the patients with primary GC with and
without AFAP1-AS1 overexpression revealed that the overexpression
of AFAP1-AS1 was positively associated with clinical stage and
lymph node metastasis, although they were not significantly
associated with sex, age, tumor size, tumor location or degree of
differentiation in patients with GC (Table II).
Expression of LncRNA AFAP1-AS1 in GC
cells
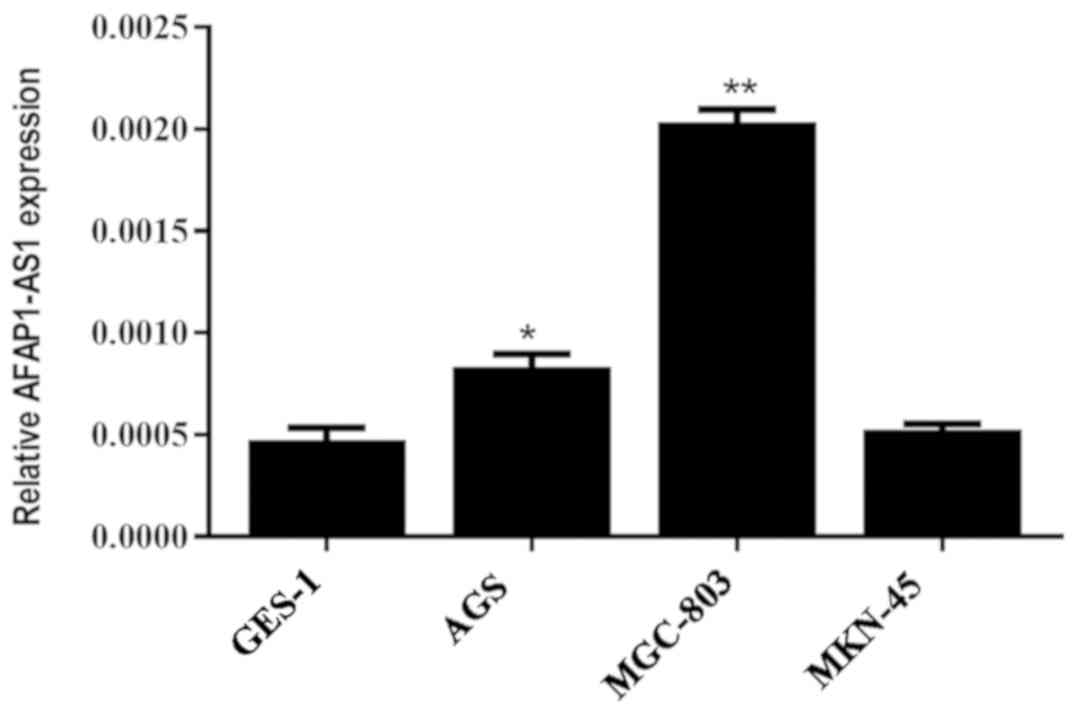
The expression levels of AFAP1-AS1 in the GC MGC-803
and AGS cell lines were significantly increased compared with that
observed in the normal gastric GES-1 cell line (P<0.05); the
expression level in MGC-803 cells was the highest. However, the
expression of AFAP1-AS1 in the GC cell line MKN45 was not
significantly different when compared with the normal gastric cells
(Fig. 2).
Effects of AFAP1-AS1 knockdown on the
proliferation of the GC cell line MGC-803
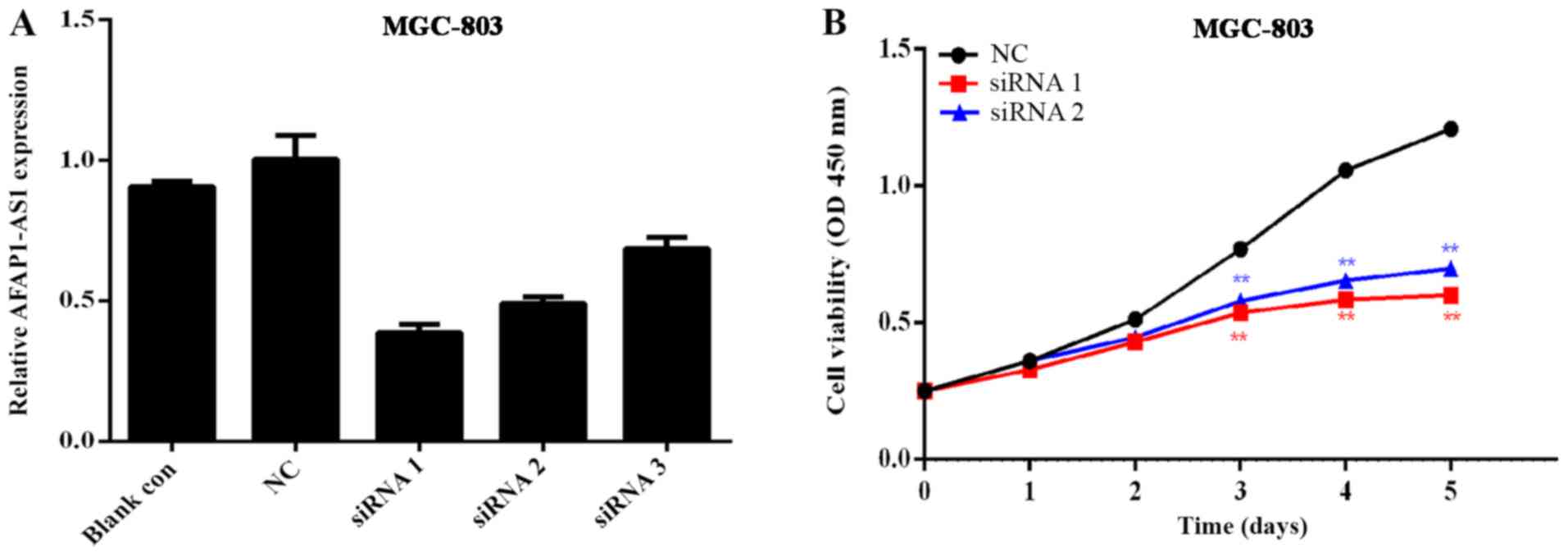
The GC cell line MGC-803, which exhibited the
highest level of AFAP1-AS1 expression, was selected for subsequent
experiments to determine the transfection efficiency. As indicated
in Fig. 3A, the knockout effects of
siRNA1 and siRNA2 were more efficient compared with siRNA3, with
knockdown efficiencies of ~65 and ~50%, respectively. Therefore,
these 2 interference sequences were selected for co-transfection in
order to examine the effects on the functions of GC cells.
Using the GC cell line MGC-803, an MTT assay was
performed to generate curves of cell growth over 5 days and the
results determined the proliferation rate of GC MGC-803. Cell
proliferation was significantly inhibited following AFAP1-AS1
downregulation (Fig. 3B). The
results revealed that transfection of MGC-803 cells with siRNA1 and
siRNA2 resulted in a significant decrease in cell proliferation
when compared with the control siRNA-transfected MGC-803 cells
(Fig. 3B).
Effects of AFAP1-AS1 on apoptosis and
the cell cycle in MGC-803 cells
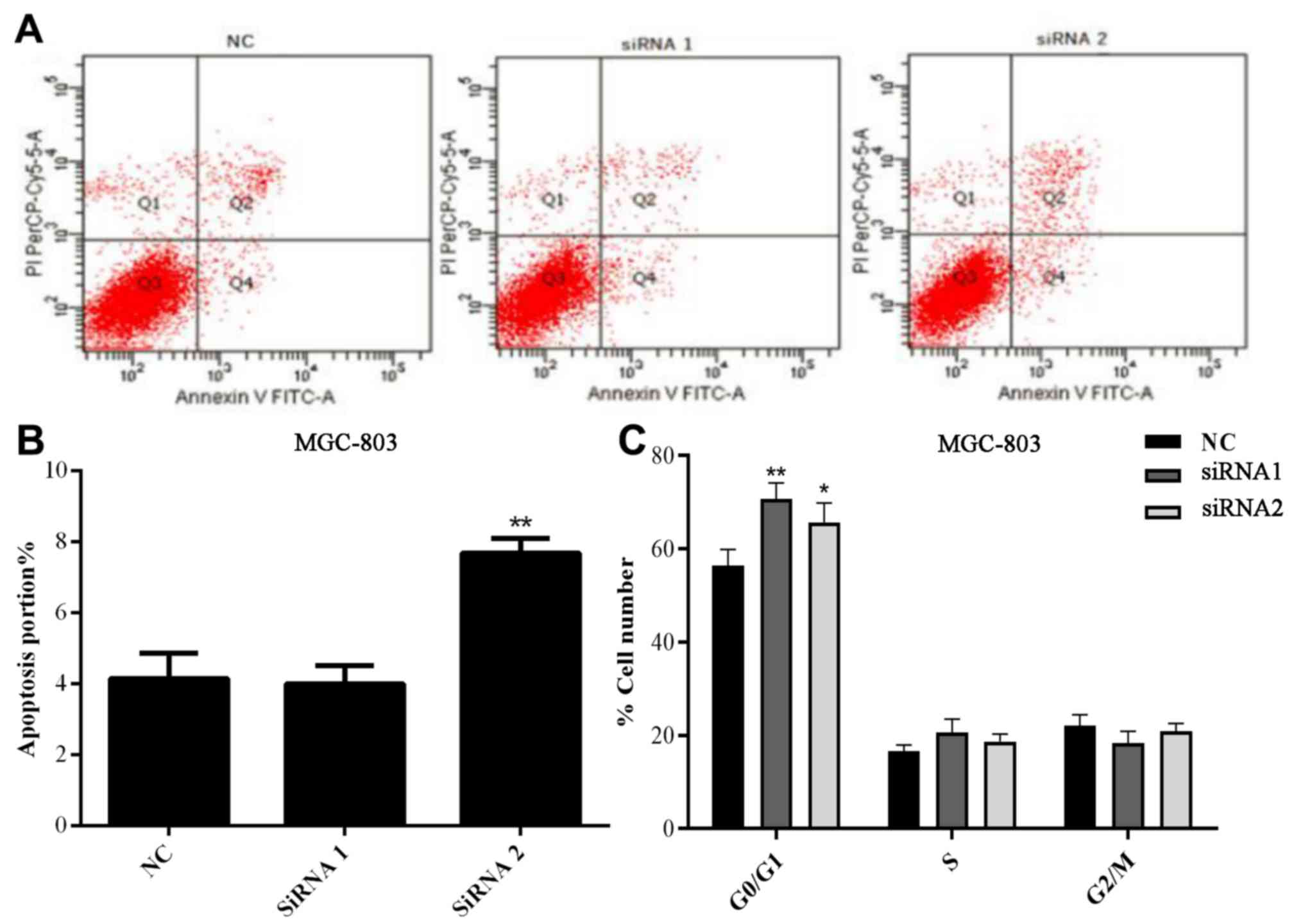
Flow cytometry was conducted to detect the changes
in apoptosis and the cell cycle in MGC-803 cells following
transfection with si-AFAP1-AS1. As demonstrated in Fig. 4A and B, there was no significant
difference in the rate of apoptosis between the NC and siRNA1
groups, but the MGC cells apoptosis rate was significantly
increased in the siRNA2 group (P<0.01).
Following transfection of siRNAs in the MGC803 cell
line, it was demonstrated that there was no marked difference in
the rate of apoptosis between the NC and siRNA1 groups following
APAP1-AS1 silencing (P=0.1064). However, the proportion of cells in
the G0/G1 phase following APAP1-AS1 knockdown was significantly
increased in the siRNA1 and siRNA2 groups compared with that
observed in the NC group (P<0.05; P<0.01 respectively;
Fig. 4C), suggesting that APAP1-AS1
may affect the regulation of the cell cycle in GC.
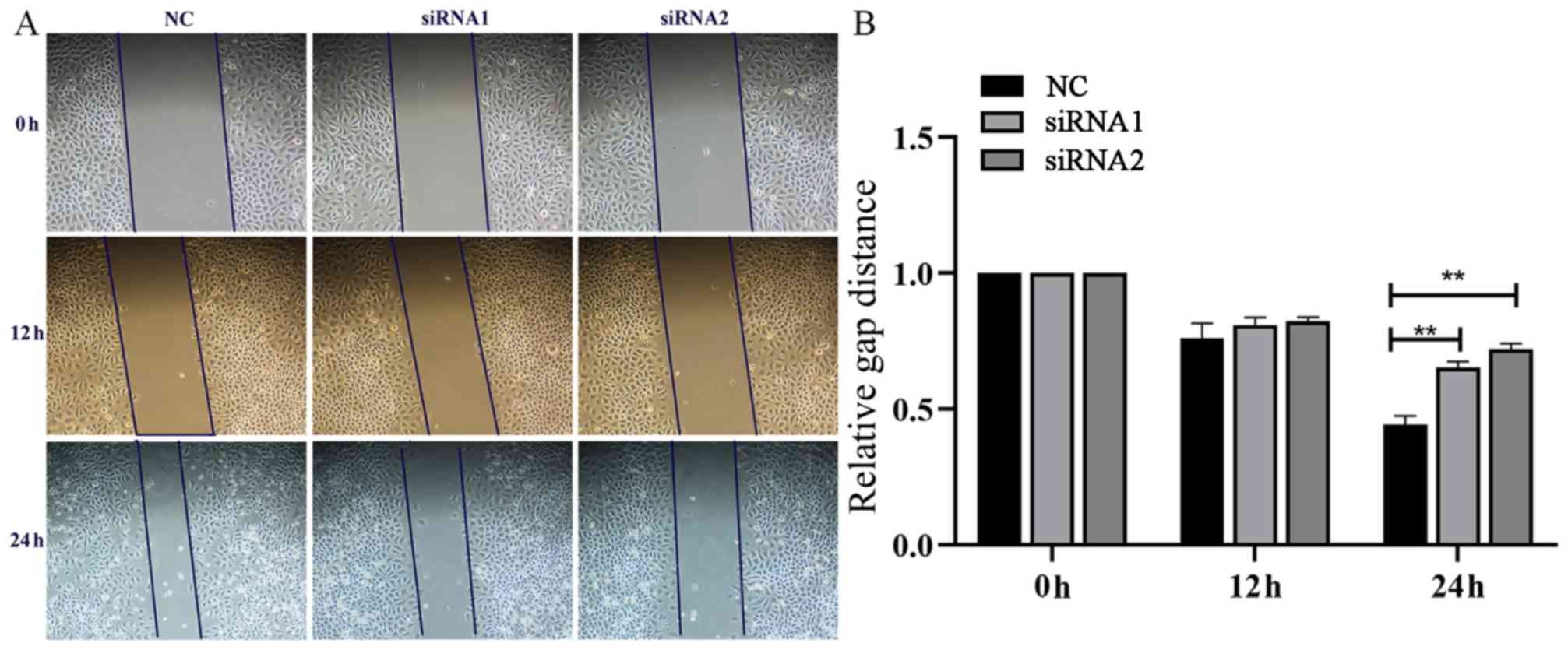
Effects of AFAP1-AS1 on migratory
capabilities in the GC MGC-803 cell line
Using the GC cell line MGC-803, cell scratches were
created at 0, 12, and 24 h after cell interference and images of
the cells were captured. As indicated in Fig. 5, the migratory ability of the GC
MGC-803 cell line was significantly inhibited following knockdown
of AFAP1-AS1.
Discussion
An increasing body of evidence has demonstrated that
the upregulation of lncRNAs is involved in the tumorigenesis and
metastasis of GC, a type of malignant neoplasm frequently observed
in East Asia. lncRNAs are a class of nucleic acid sequences
measuring >200 bps in length (19). A number of studies have demonstrated
that lncRNAs participate in a wide range of physiological
processes; they may regulate gene expression at the transcriptional
and post-transcriptional level, and may also regulate cellular
functions through various mechanisms (20). lncRNAs are known to be closely
associated with the development and progression of cancer, and
recently have become a key area of medical research, with potential
in future clinical practice.
GC is one of the most common types of malignant
tumors in humans and is a serious threat to human health. Its
incidence rate has continuously risen over previous years and China
has one of the highest incidence rates of GC in the world. Chen
et al (21) demonstrated that
in China, the GC incidence was the second most common type of tumor
diagnosed. The traditional treatments, including surgery,
radiotherapy and chemotherapy, are not sufficiently effective for
patients with advanced stage GC. Therefore, it is important to
identify valuable biomarkers to improve the percentage of accurate
early diagnoses in patients with GC. According to previous studies,
a number of biomarkers have been identified for the diagnosis or
prognosis of patients with cancer (22,23);
among these, lncRNAs are a key area of interest.
Lately, AFAP1-AS1 has been revealed to function
primarily as an oncogene, regulating gene expression by affecting
mRNA and the corresponding transcriptional protein. A previous
profiling study suggested that the upregulation of lncRNA AFAP1-AS1
affected the proliferation, invasion and survival rates of tongue
squamous cell carcinoma via the Wnt/β-catenin signaling pathway
(24). Yuan et al (25) revealed that the
AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell
stemness and chemoresistance. An additional previous study
indicated that the upregulation of lncRNA AFAP1-AS1 expression was
associated with progression and poor prognosis in nasopharyngeal
carcinoma (26). However, to the
best of our knowledge, its function in carcinogenesis and tumor
progression in GC remains unknown.
In the present study, the results of RT-qPCR
demonstrated that lncRNA AFAP1-AS1 expression in tumor tissues was
significantly increased compared with that of non-cancerous tissues
in 52 matched pairs of patient samples, which is consistent with a
previous study (27). In addition,
the association between AFAP1-AS1 and clinicopathological
characteristics revealed that the overexpression of AFAP1-AS1 was
not aberrantly associated with ex, age, tumor size, tumor location
or degree of differentiation in patients with GC, but was
positively associated with clinical stage and lymph node
metastasis. Higher AFAP1-AS1 mRNA expression levels were also
associated with the occurrence of lymph node metastasis.
To further understand the biological function of
AFAP1-AS1 in GC cells, in vitro experiments were conducted.
The GC MGC-803 cell line exhibited the highest AFAP1-AS1 expression
when compared with the gastric normal cell line GES-1, within a
panel of GC cell lines. Therefore, the MGC-803 cell line was
selected for further analysis. The results demonstrated that the
proliferation rate of MGC-803 cells was significantly inhibited
following knockdown of AFAP1-AS1 expression.
In addition, the proportion of cells in the G0/G1
phase following knockdown was significantly increased compared with
that observed in the NC group, suggesting that AFAP1-AS1 may have
an effect on the regulation of the GC cell cycle. However, the
effect of APAP1-AS1 on the rate of apoptosis was not as marked as
expected. In addition, the migratory ability of the GC MGC-803 cell
line was significantly inhibited following knockdown of AFAP1-AS1
expression.
The present study confirmed that AFAP1-AS1 is highly
expressed in human GC tissues and cell lines, and that it
significantly affected tumor cell invasion and proliferation.
However, its specific mechanism has not been fully elucidated and
requires additional investigation.
In conclusion, the results of the present study
demonstrated that lncRNA AFAP1-AS1 is highly expressed in GC,
suggesting that AFAP1-AS1 may be a GC-specific lncRNA and serve
play an important role in the development of GC. Knockdown of
lncRNA AFAP1-AS1 inhibited cell proliferation and migration in
MGC-803 cells. These results indicated that lncRNA AFAP1-AS1 may
function as an oncogene in GC and may be a novel marker for the
diagnosis and therapeutic targeting of GC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Jiangsu
Natural Science Foundation (grant no. BK20151087), awarded to
Professor Hongyong Cao.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZXL designed the project, collected patient data and
performed the experiments. ZLD collected the patient data and
performed the experiments. DWR analyzed the data. WWT contributed
to the design of the project, wrote the original draft manuscript
and provided funding. HYC supervised the project throughout,
reviewed and edited the manuscript, provided funding and collected
the patient data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Nanjing First Hospital, Nanjing Medical University
(Jiangsu, China). Written informed consent was obtained from all of
the patients prior to participation.
Patient consent for publication
Written informed consent was obtained from all of
the patients prior to participation.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yagi K, Nozawa Y, Endou S and Nakamura A:
Diagnosis of early gastric cancer by magnifying endoscopy with NBI
from viewpoint of histological imaging: Mucosal patterning in terms
of white zone visibility and its relationship to histology. Diagn
Ther Endosc. 2012:9548092012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yoon SH, Kim YH, Lee YJ, Park J, Kim JW,
Lee HS and Kim B: Tumor heterogeneity in human epidermal growth
factor receptor 2 (HER2)-positive advanced gastric cancer assessed
by CT texture analysis: Association with survival after trastuzumab
treatment. PLoS One. 11:e01612782016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Biffi R, Botteri E, Cenciarelli S, Luca F,
Pozzi S, Valvo M, Sonzogni A, Chiappa A, Leal Ghezzi T, Rotmensz N,
et al: Impact on survival of the number of lymph nodes removed in
patients with node-negative gastric cancer submitted to extended
lymphnode dissection. Eur J Surg Oncol. 37:305–311. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li W, Zheng J, Deng J, You Y, Wu H, Li N,
Lu J and Zhou Y: Increased levels of the long intergenic
non-protein coding RNA POU3F3 promote DNA methylation in esophageal
squamous cell carcinoma cells. Gastroenterology. 146:1714–1726.e5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang SL, Lin RX, Si LH, Cui MH, Zhang XW
and Fan LM: Expression and functional role of long non-coding RNA
AFAP1-AS1 in ovarian cancer. Eur Rev Med Pharmacol Sci.
20:5107–5112. 2016.PubMed/NCBI
|
|
13
|
Ma F, Wang SH, Cai Q, Zhang MD, Yang Y and
Ding J: Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis
and promotes cells proliferation and invasion in gallbladder
cancer. Biomed Pharmacother. 84:1249–1255. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo HL, Huang MD, Guo JN, Fan RH, Xia XT,
He JD and Chen XF: AFAP1-AS1 is upregulated and promotes esophageal
squamous cell carcinoma cell proliferation and inhibits cell
apoptosis. Cancer Med. 5:2879–2885. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han X, Wang L, Ning Y, Li S and Wang Z:
Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes
metastasis in colorectal cancer. Biol Res. 49:362016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW,
Li J, Huo YM, Liu W, Zhang JF, Hong J, et al: Analysis of long
non-coding RNA expression profiles in pancreatic ductal
adenocarcinoma. Sci Rep. 6:335352016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mahul BA, Stephen B, Edge, Frederick L, et
al: AJCC Cancer Staging Manual. https://cancerstaging.org/references-tools/deskreferences/pages/default.aspxSeptember
5–2018
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bergmann JH and Spector DL: Long
non-coding RNAs: Modulators of nuclear structure and function. Curr
Opin Cell Biol. 26:10–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu L and Qu X: Cancer biomarker detection:
Recent achievements and challenges. Chem Soc Rev. 44:2963–2997.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Armitage EG and Barbas C: Metabolomics in
cancer biomarker discovery: Current trends and future perspectives.
J Pharm Biomed Anal. 87:1–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang ZY, Hu M, Dai MH, Xiong J, Zhang S,
Wu HJ, Zhang SS and Gong ZJ: Upregulation of the long non-coding
RNA AFAP1-AS1 affects the proliferation, invasion and survival of
tongue squamous cell carcinoma via the Wnt/β-catenin signaling
pathway. Mol Cancer. 17:32018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan Z, Xiu C, Song K, Pei R, Miao S, Mao
X, Sun J and Jia S: Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ
axis regulates laryngeal carcinoma cell stemness and
chemoresistance. J Cell Mol Med. 22:4253–4262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao
Q, Chen P, Shi L, Lian Y, Jing Y, et al: Upregulated long
non-coding RNA AFAP1-AS1 expression is associated with progression
and poor prognosis of nasopharyngeal carcinoma. Oncotarget.
6:20404–1829. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao
L and Zhang S: Critical role for the long non-coding RNA AFAP1-AS1
in the proliferation and metastasis of hepatocellular carcinoma.
Tumor Biol. 37:9699–9707. 2016. View Article : Google Scholar
|