Introduction
Gastric carcinoma (GC) is one of the most common
types of malignancy and is the second leading cause of
cancer-related mortality worldwide (1). Despite advances in surgical techniques,
adjuvant chemotherapy, radiotherapy and immune therapy, the
morbidity and mortality rates remain high (2). Therefore, new strategies to improve the
diagnosis and prognosis of GC are urgently required.
Tumour metastasis is a complex process that serves a
vital role in the progression and outcome of the cancer (3). Therefore, novel effective strategies
that target metastasis are required. Previous studies have
suggested that adrenergic receptor antagonists could be used in
novel therapeutic approaches in the treatment of various types of
cancer, including prostate and breast carcinoma (4,5). Tumour
metastasis involves the migration of cancer cells from the primary
tumour via lymphoid/hematopoietic pathways and is regulated by
exogenous signalling molecules, including G protein coupled
receptor (GPCR) ligands, chemokines and neurotransmitters (6,7). A
previous study has reported that norepinephrine, a
stress-associated neurotransmitter, is a potent inducer of
migration in cancer cell lines (8).
Adrenergic receptor antagonists include two major
groups; α receptor and β receptor specific antagonists (9,10).
Adrenergic receptor α1 (ADRA1) receptor is classified into three
different subtypes ADRA1A, ADRA1B and ADRA1D, that differ in their
tissue distribution, cell signalling, pharmacology and
physiological roles (11). ADRA1
receptors are ubiquitous in the majority of human tissues; ADRA1B
is predominantly expressed on the cell surface, ADRA1A is mainly
expressed on the cell surface and intracellularly, and ADRA1D is
primarily localized perinuclear (12).
It is well-known that ADRA1 is a member of the GPCR
family, which interacts with a heterotrimeric G protein containing
the Gαq/11/14/16 subunits (13). The
Gαq subunit is the main activator of phospholipase CE, which
promotes the cleavage of phosphatidylinositol-4,5-diphosphate into
diacylglycerol and inositol-1,4,5-triphosphate. This subsequently
promotes the release of Ca2+ from intracellular stores
to activate protein kinase C (PKC) (14).
Previous studies have suggested that adrenergic
signals in cells can promote the development of cancer (15,16).
However, little is understood regarding the associations between
the expression levels of ARDA1A, ARDA1B and ARDA1D and the risk of
GC. Therefore, the aim of the present study was to identify the
associations between the expression levels of individual ADRA1
subtypes and GC prognosis. The current study may provide insights
into the potential functional roles of ADRA1 subtypes in GC.
Materials and methods
Patient and disease
characteristics
Metabolic gEne Rapid Visualizer (http://merav.wi.mit.edu/) was used to generate
boxplots of the expression levels of ADRA1 subtypes in normal
gastric tissues and primary GC tumours. Subsequently, clinical data
were obtained for 379 patients with GC, including sex, grade
(17), age, Tumour-Node-Metastasis
(TNM) stage (18), targeted therapy,
events (metastases or deaths), survival time, mortality status and
mRNA expression levels of ADRA1A, ADRA1B and ADRA1D, from OncoLnc
(http://www.oncolnc.org/) and Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520)
databases (19,20). The data presented here are based on
studies published on The Cancer Genome Atlas (TCGA) (21,22). To
analyse the difference in the expression levels in cancer compared
with the adjacent tissues, three genes (ADRA1A, ADRA1B and ADRA1D0
were searched in the GEPIA (http://gepia.cancer-pku.cn/) database separately.
Correlation and functional enrichment
analysis of the ADRA1 subfamily
The online Database for Annotation, Visualization,
and Integrated Discovery (DAVID; v.6.8; http://david.ncifcrf.gov/tools.jsp) (23) was used to perform functional
enrichment. This included gene ontology (GO) functional analysis
and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway
analysis. In the present study, GO functional analysis consisted of
molecular function, cellular component, and biological process
terms. KEGG pathway analysis was performed on the ADRA1 gene
subfamily. The gene function prediction website GeneMANIA
(http://genemania.org/) was used to construct
gene-gene interaction networks (24).
Survival analysis
Using the TCGA data, 379 patients with GC were
divided into high- and low-expression groups according to the 50%
cut-off values (median values). Overall survival (OS) and median
survival time (MST) were applied to estimate patient prognosis.
Kaplan-Meier analysis with a log-rank test was used to identify
associations between ADRA1A, ADRA1B and ADRA1D mRNA expression
levels and patient survival. Cox regression analysis was then used
to evaluate statistically significant factors, including age and
TNM stage.
Joint-effects analysis
In the TCGA database, the expression levels of
ADRA1A, ADRA1B and ADRA1D were significantly different between
tumour and non-tumour tissues. Therefore, joint-effects analysis
was performed with the following combinations: i) ADRA1A and
ADRA1B; i) ADRA1A and ADRA1D; iii) ADRA1B and ADRA1D; and iv)
ADRA1A, ADRA1B and ADRA1D (Table I).
A Cox regression model was adjusted for TNM stage, age and sex in
keeping with the aforementioned combinations.
 | Table I.Grouping according to the expression
levels of two or three selected genes. |
Table I.
Grouping according to the expression
levels of two or three selected genes.
| Group | Composition |
|---|
| I | Low ARDA1A + low
ARDA1B |
| II | Low ARDA1A + high
ARDA1B |
|
| High ARDA1A + low
ARDA1B |
| III | High ARDA1A + high
ARDA1B |
| IV | Low ARDA1A + low
ARDA1D |
| V | Low ARDA1A + high
ARDA1D |
|
| High ARDA1A + low
ARDA1D |
| VI | High ARDA1A + high
ARDA1D |
| VII | Low ARDA1B + low
ARDA1D |
| VIII | Low ARDA1B + high
ARDA1D |
|
| High ARDA1B + low
ARDA1D |
| IX | High ARDA1B + high
ARDA1D |
| X | Low ARDA1A + low
ARDA1B + low ARDA1D |
| XI | High ARDA1A + low
ARDA1B + high ARDA1D |
|
| Low ARDA1A+ high
ARDA1B + high ARDA1D |
|
| Low ARDA1A + low
ARDA1B + low ARDA1D |
|
| High ARDA1A + high
ARDA1B + high ARDA1D |
|
| High ARDA1A + low
ARDA1B + low ARDA1D |
|
| Low ARDA1A + high
ARDA1B + low ARDA1D |
| XII | High ARDA1A + high
ARDA1B + high ARDA1D |
Statistical analysis
OS was evaluated by Kaplan-Meier survival analysis
followed by a log-rank test. Cox proportional hazards regression
model was used for univariate and multivariate survival analysis.
GraphPad Prism v.7.0 (GraphPad Prism, Inc.) was used to construct
vertical scatter plots and survival curves. All statistical
analyses were performed using SPSS v.22.0 software and compared
using an independent-samples t-test (IBM, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinicopathological features of the
patient cohort
Detailed characteristics of the 379 included
patients obtained from the TCGA database are presented in Table II. It was identified that TNM stage
and age were significantly associated with MST (P<0.001 and
P=0.007, respectively); however, no significant associations were
revealed for grade, sex or targeted therapy (all P>0.05;
Table II).
 | Table II.Clinical data for the 379 patients
with gastric carcinoma. |
Table II.
Clinical data for the 379 patients
with gastric carcinoma.
| Variable | Patients
(n=379) | No. of events
(%) | MST, days | HR (95% CI) | Log-rank
P-value |
|---|
| Grade |
| 1 +
2 | 146 | 52 (35.6) | 1,294 | Ref. | 0.07 |
| 3 | 233 | 99 (42.5) | 801 | 1.333
(0.977–1.818) |
|
| Sex |
|
Male | 247 | 104 (42.1) | 2,030 | Ref. | 0.229 |
|
Female | 132 | 47 (35.6) | 874 | 1.236
(0.875–1.745) |
|
| Age, years |
|
≥60 | 124 | 38 (30.6) | 475 | Ref. | 0.007 |
|
<60 | 255 | 113 (44.3) | 792 | 0.604
(0.418–0.874) |
|
| TNM stage |
| I | 50 | 12 (24.0) | 2,197 | 0.270
(0.133–0.550) | <0.001 |
| II | 119 | 33 (27.7) | 1,686 | 0.372
(0.214–0.645) |
|
|
III | 164 | 78 (47.6) | 766 | 0.649
(0.400–1.053) |
|
| IV | 34 | 21 (61.8) | 476 | Ref. |
|
| NA | 12 |
|
|
|
|
| Targeted
therapy |
| No | 188 | 79 (42.0) | 805 | Ref. | 0.063 |
|
Yes | 162 | 59 (38.4) | 1,294 | 0.726
(0.518–1.018) |
|
| NA | 29 |
Analysis of ADRA1 subfamily gene
expression in GC
Expression levels of ADRA1A, ADRA1B and ADRA1D in
primary GC tissues and normal tissues were analysed using the
online tool (GEPIA; http://gepia.cancer-pku.cn/) and illustrated by
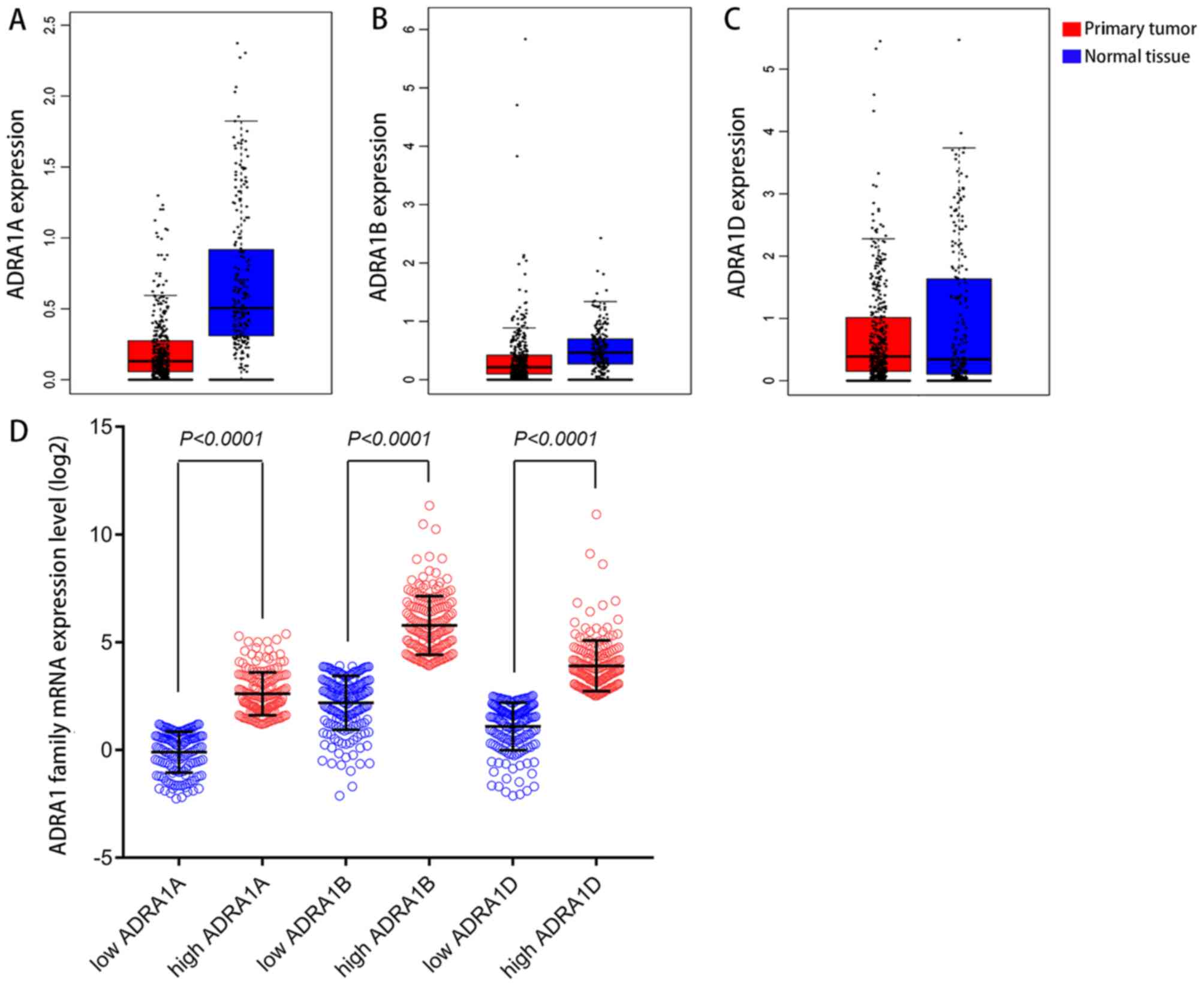
boxplots (Fig. 1A-C). The expression
levels of ADRA1A were higher in normal gastric tissues compared
with primary gastric tumours. However, no marked differences in the
expression levels of ADRA1B and ADRA1D were revealed between
primary GC tumours and normal tissues. The low expression groups
and high expression groups for these genes obtained from the GEO
and TCGA databases were analysed by Scatter diagrams and compared
using an independent-samples t-test (Fig. 1D).
GO and KEGG pathway analysis of the
ADRA1 subfamily
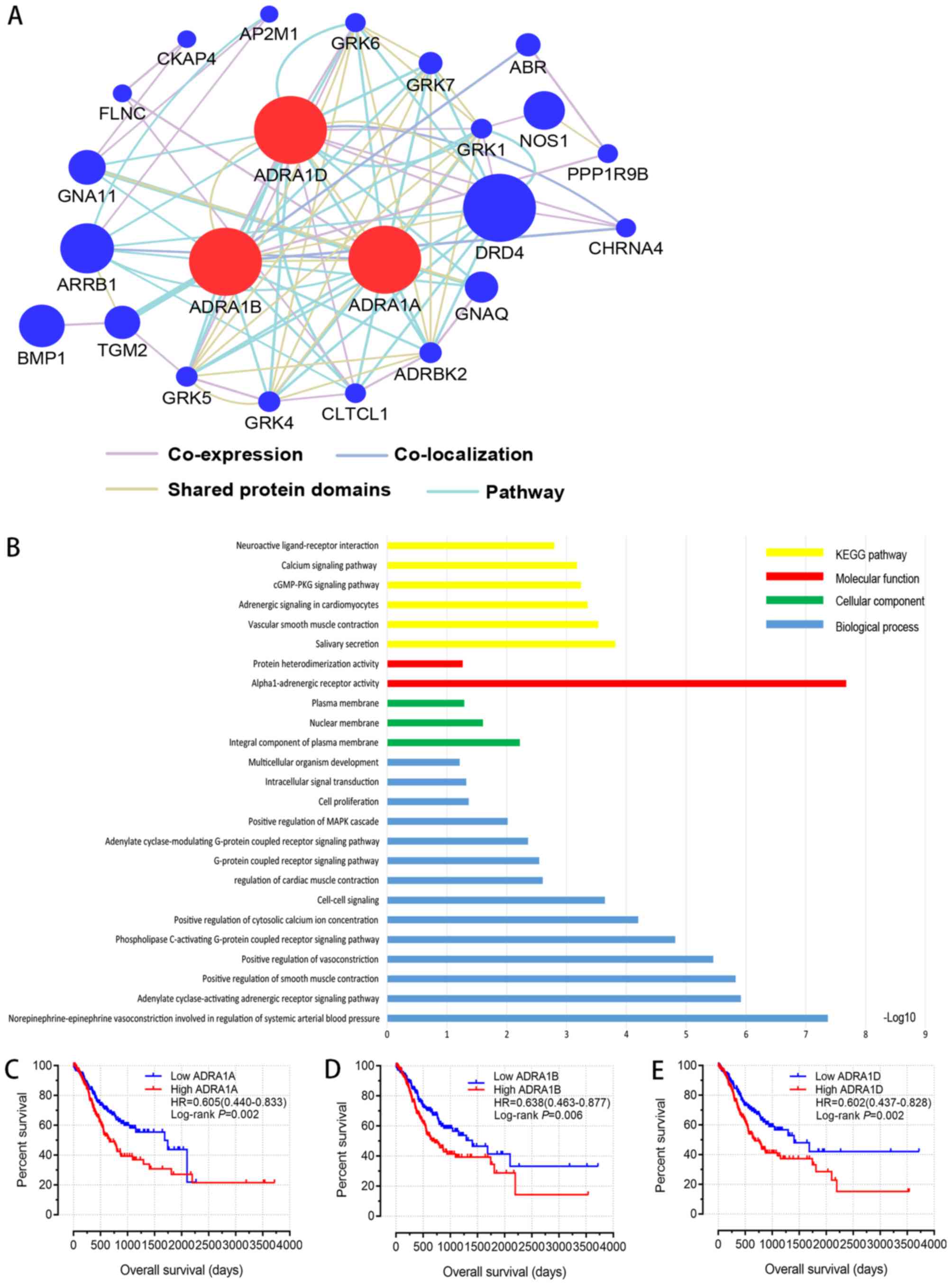
The results of GO analysis of ADRA1A, ADRA1B and
ADRA1D including molecular function, cellular component and
biological process terms, are presented in Fig. 2B. KEGG enrichment analysis identified
certain signalling pathways associated with ADRA1 subfamily
members, such as ‘Calcium signalling pathway’, ‘cGMP-PKG signalling
pathway’ and mitogen-activated protein kinase (MAPK) signalling
(Fig. 2B). Furthermore, the
GeneMANIA website (http://genemania.org/) was used to analyse interaction
networks of ADRA1A, ADRA1B and ADRA1D. The interaction network
between the ADRA1 subfamily and other genes presented in Fig. 2A.
Association of ADRA1 subfamily members
with survival
Cox proportional hazards regression analysis of the
ADRA1 members revealed that the mRNA expression levels of all three
genes were significantly associated with MST in patients with GC
(Table III). As aforementioned in
Table I, TNM stage and age were also
identified as prognostic factors in TCGA GC cohort. Therefore, a
multivariate Cox regression model was employed next, in order to
explore whether the expression levels of the ADRA1 subfamily genes
were independent prognostic factors following adjustment for age
and TNM stage. The results revealed that the lower expression
levels of ADRA1A [hazard ratio (HR), 0.595; 95% CI, 0.426–0.831;
adjusted P=0.002], ADRA1B (HR, 0.576; 95% CI, 0.412–0.805; adjusted
P=0.001) and ADRA1D (HR, 0.559; 95% CI, 0.398–0.787; adjusted
P=0.001) were significantly associated with favourable MST in
patients with GC, following adjustment for age and TNM stage
(Table III). The survival curves
of ADRA1A, ADRA1B and ADRA1D are presented in Fig. 2C-E; these revealed that low
expression levels of these genes were significantly associated with
a favourable OS in patients with GC (P=0.002, P=0.006 and P=0.002,
respectively).
 | Table III.Survival analysis according to the
expression levels of ADRA1 subfamily genes. |
Table III.
Survival analysis according to the
expression levels of ADRA1 subfamily genes.
| Gene | Patients
(n=379) | No. of events
(%) | MST, days | Crude HR (95%
CI) | Crude P-value | Adjusted HR (95%
CI)a | Adjusted
P-valuea |
|---|
| ADRA1A |
|
Low | 189 | 61 (32.1) | 1,686 | Ref. | 0.001 | Ref. | 0.002 |
|
High | 189 | 90 (47.6) | 669 | 0.601
(0.434–0.834) |
| 0.595
(0.426–0.831) |
|
| NA | 1 |
|
|
|
|
|
|
| ADRA1B |
|
Low | 189 | 62 (32.9) | 1,407 | Ref. | 0.006 | Ref. | 0.001 |
|
High | 189 | 89 (47.1) | 712 | 0.637
(0.460–0.881) |
| 0.0576
(0.412–0.805) |
|
| NA | 1 |
|
|
|
|
|
|
| ADRA1D |
|
Low | 189 | 58 (30.7) | 1,407 | Ref. | 0.002 | Ref. | 0.001 |
|
High | 189 | 93 (49.2) | 675 | 0.601
(0.433–0.835) |
| 0.559
(0.398–0.787) |
|
| NA | 1 |
|
|
|
|
|
|
Joint-effects analysis of the ADRA1
subfamily
Based on the influence of ADRA1A, ADRA1B and ADRA1D
on patient survival identified by multivariate analysis, a
joint-effects model was employed to evaluate the combined effects
of these genes on the OS of patients with GC. According to the
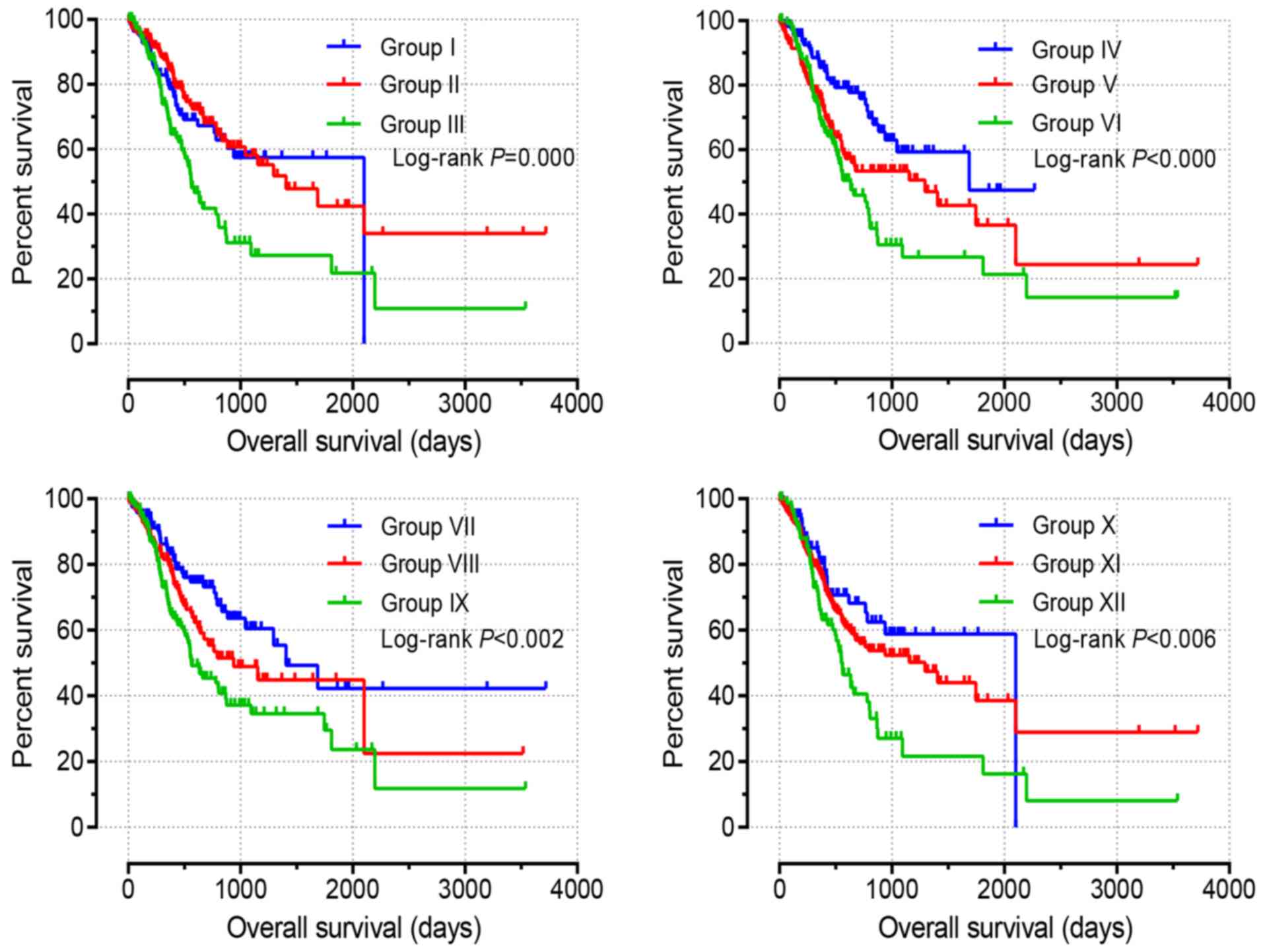
different expression levels of ADRA1A, ADRA1B and ADRA1D, 12 groups
were generated, as presented in Table
I. Group XII, with high expression levels of ARDA1A, ARDA1B and
ARDA1D, exhibited the shortest MST of 552 days (adjusted P=0.001).
Group X, with low expression levels of ARDA1A, ARDA1B and ARDA1D,
exhibited the highest MST of 2,100 days (adjusted P=0.002). The
detailed joint-effects analysis results are presented in Table IV.
 | Table IV.Joint-effects analysis of the
prognostic value of combinations of ADRA1A, ADRA1B and ADRA1D
expression levels in gastric carcinoma. |
Table IV.
Joint-effects analysis of the
prognostic value of combinations of ADRA1A, ADRA1B and ADRA1D
expression levels in gastric carcinoma.
| Group | Patients
(n=379) | MST, days | Crude P-value | Crude HR (95%
CI) | Adjusted
P-valuea | Adjusted HR (95%
CI)a |
|---|
| I | 148 | 1,407 | <0.001 | 0.480
(0.327–0.705) | <0.001 | 0.444
(0.297–0.664) |
| II | 114 | 2,100 | 0.005 | 0.552
(0.364–0.838) | 0.004 | 0.531
(0.347–0.813) |
| III | 113 | 560 | <0.001 | Ref. | <0.001 | Ref. |
| IV | 115 | 1,686 | <0.001 | 0.420
(0.271–0.651) | <0.001 | 0.406
(0.260–0.633) |
| V | 147 | 1,294 | 0.083 | 0.729
(0.510–1.043) | 0.031 | 0.665
(0.458–0.963) |
| VI | 114 | 633 | 0.001 | Ref. | <0.001 | Ref. |
| VII | 121 | 1,407 | <0.001 | 0.481
(0.319–0.725) | <0.001 | 0.433
(0.285–0.659) |
| VIII | 134 | 940 | 0.066 | 0.708
(0.490–1.023) | 0.003 | 0.563
(0.385–0.825) |
| IX | 121 | 562 | 0.002 | Ref. | <0.001 | Ref. |
| X | 83 | 2,100 | 0.005 | 0.490
(0.300–0.802) | 0.002 | 0.457
(0.279–0.749) |
| XI | 215 | 1,294 | 0.010 | 0.625
(0.438–0.893) | 0.001 | 0.541
(0.376–0.780) |
| XII | 78 | 552 | 0.006 | Ref. | 0.001 | Ref. |
Associated survival curves, presented in Fig. 3, were evaluated by Kaplan-Meier
analysis with a log-rank test. Low expression levels of ADRA1A,
ADRA1B and ADRA1D (group X) were identified to be significantly
associated with favourable OS. By contrast, high expression levels
of ADRA1A, ADRA1B and ADRA1D (group XII) were revealed to be
significantly associated with poor OS (all P<0.05; Fig. 3).
Discussion
Cancer metastasis can be influenced by numerous
factors, including intracellular signalling molecules and
extracellular components, including cytokines, the extracellular
matrix and neurotransmitters (25).
It has previously been suggested that the nervous system serves an
important role in the progression of cancer. It has been reported
that the sympathetic and parasympathetic nervous systems
participate in the development and distribution of cancer (26).
Adrenergic neurotransmitters, including
norepinephrine (NE) and epinephrine (E), have been reported to
promote cell migration and invasion in various types of cancer
(6,27). Previous studies have demonstrated
that NE/E may promote tumour progression by promoting angiogenesis
(27). Furthermore, the tumour
stromal cells in the microenvironment of cancer may be affected by
the nervous system (28). A number
of neurotransmitter receptor-associated genes have been identified
to be closely associated with the proliferation and survival of
tumour cells by a large-scale complementary DNA transfection
screening (29).
It is understood that NE/E exert their physiological
functions predominantly by the α and β adrenergic receptors
(30). The ability of NE/E to induce
invasion and anoikis resistance of cancer cells can be mediated by
both ADRA1A and ADRB2 receptors (31). ADRA1 mediates the role of endogenous
adrenaline and NE in multiple target cells. NE induces proton
release and calcium flux, and activates phospholipase C, PKC and
extracellular signal-regulated kinase 1/2 pathways to promote cell
proliferation (32). In addition,
ADRs transactivate epidermal growth factor receptor (EGFR)
signalling via specific matrix metalloproteinases (MMPs) or a
disintegrin and metalloproteinase hydrolysis of latent ligands,
including heparin-binding EGF-like growth factor (HB-EGF) (33). For the first time, Li et al
(30) demonstrated that the ADRA1A
and ADRB2 can inhibit EGFR signalling in cancer. EGFR is
overexpressed in the majority of adenocarcinoma and squamous cell
carcinoma cases and can be selectively targeted by pharmacological
inhibitors currently used in the clinic (34).
It has been reported that ADRA1 can promote the
metastasis of cancer, and continuously activated ADRA induces cell
apoptosis via p53 (35). The tumour
suppressor protein p53 serves an important role in cellular
regulation and acts as an important mediator of apoptotic cell
death (36). Evidence suggests that
p53 not only induces transcription of pro-apoptotic proteins but
also activates the mitochondrial cell death pathway (37). Previous studies have demonstrated
that ADRA1A is also associated with reactive oxygen species (ROS)
production via the EGFR and the nicotinamide adenine dinucleotide
phosphate oxidase signalling pathways (38).
However, little is understood regarding the
association between ADRA1 mRNA expression and the prognosis of GC.
Using data from TCGA and GEO databases that included mRNA
expression profiles of the ADRA1 genes and the associated clinical
information from patients with GC, the present study investigated
the associations between ADRA1 family members expression and
patient prognosis. In addition, the current study assessed whether
any ADRA1 genes, alone or in combination, could be used as
biomarkers for predicting the prognosis of GC. The results
suggested that the expression levels of ADRA1A in normal tissue
were higher compared with that in primary tumour tissue. In
addition, survival analysis demonstrated that low expression levels
of ADRA1A, ADRA1B or ADRA1D were associated with a favourable OS in
patients with GC. Furthermore, joint-effects analysis demonstrated
that the combination of low levels of all three ADRA1A, ADRA1B and
ADRA1D was significantly associated with a favourable OS. By
contrast, the combination of high expression levels of ADRA1A,
ADRA1B and ADRA1D was associated with a poor OS.
Furthermore, functional analysis and KEGG enrichment
identified specific signalling pathways associated with ADRA1 genes
including ‘Calcium signalling pathway’, ‘cGMP-PKG signalling
pathway’ and mitogen-activated protein kinase (MAPK) signalling.
These pathways serve important roles in cancer. For example,
studies have demonstrated that numerous biological functions are
regulated by MAPK signalling, including cell proliferation,
apoptosis and metastasis (39,40). In
addition, cGMP/PKGIβ regulates breast cancer cell migration and
invasion through the actin/myosin-associated protein, caldesmon
(CaD) (41). Analysis of gene-gene
interactions showed that certain genes were associated with members
of the ADRA1 subfamily [G protein-coupled receptor kinases (GRK)1,
GRK4, GRK5, GRK6, GRK7, ARRB1, DRD4, GNAQ, ADRBK2] and some of
these genes serve important roles in the regulation of tumour
biology. For example, GRKs can modulate GPCR signalling by
interacting with the ligand-activated GPCR and phosphorylating its
intracellular domain (42). It has
been demonstrated that GPCRs affect multiple aspects of cancer
biology, such as vascular remodelling, invasion and migration
(42).
Peng et al (43) reported that ADRA1A was highly
expressed in the peripheral serum of patients with hysterocarcinoma
and associated with tumour stage and lymph node metastasis status.
Powe et al (44) demonstrated
that ADRA1B expression was associated with breast cancer
progression and prognosis. Notably, the expression levels of ADRA1A
were higher in normal gastric tissues compared with primary gastric
tumour tissues in the current study. However, the present study
also revealed that low levels of ADRA1 were favourable for patient
outcome. There may be a number of reasons for these contradictory
results. Firstly, it is understood that certain genes act as tumour
suppressors during the early stage of tumorigenesis, while they
serve as tumour promoters in later stages. For example,
transforming growth factor-β (TGF-β) is a cytokine that exhibits
dual activities in numerous types of cancer (45). TGF-β is an important mediator of
cancer invasion, metastasis and angiogenesis; however, it also
exhibits antitumor functions (46).
Another possible explanation may be that ADRA1 serves different
roles in gastric tumour tissue and in normal tissue. Despite the
low expression of ADRA1 in gastric cancer tumour tissues, it also
serves a role in promoting cancer cell proliferation and metastasis
(47). Therefore, the present
results require further experimental validation in order to fully
elucidate the role of ADRA1 in GC.
In conclusion, the results of the present study
demonstrated that the expression levels of ADRA1A, ADRA1B and
ADRA1D were associated with the prognosis of gastric cancer. High
expression levels of the ADRA1 family were associated with a poor
prognosis and a significant reduction in survival rate. These
results suggested that an increased expression of the ADRA1 family
may somehow promote tumour metastasis. Accordingly, the levels of
ADRA1 genes may serve as potential markers for prognosis
evaluation.
The present study has certain limitations. Firstly,
clinical information and gene expression data used in the study
were obtained from a public database and the data were limited.
Second, the low expression of ADRA1 in gastric tumour tissues and
its role in predicting prognosis is difficult to understand. Due to
insufficient funds, the present study did not perform verification
experiments. Therefore, the specific mechanism of ADRA1 in gastric
cancer tissues requires further experimental validation. Despite
these limitations, to the best of our knowledge, the current study
is the first to systematically investigate the diagnostic and
prognostic values of ADRA1 genes in GC. In summary, ADRA1 genes may
serve as potential novel indicators for the prediction of the
prognosis of patients with GC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81760521).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the author on reasonable request.
Authors' contributions
JC, TW and MH designed the study, YY and ZL
performed the analysis and prepared the manuscript. TW wrote the
manuscript. WW performed the analysis. YQ and HL helped perform the
analysis with constructive discussions. All authors approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ye J, Xu J, Li Y, Huang Q, Huang J, Wang
J, Zhong W and Lin X, Chen W and Lin X: DDAH1 mediates gastric
cancer cell invasion and metastasis via Wnt/β-catenin signaling
pathway. Mol Oncol. 11:1208–1224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harris AM, Warner BW, Wilson JM, Becker A,
Rowland RG, Conner W, Lane M, Kimbler K, Durbin EB, Baron AT and
Kyprianou N: Effect of alpha1-adrenoceptor antagonist exposure on
prostate cancer incidence: An observational cohort study. J Urol.
178:2176–2180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Algazi M, Plu-Bureau G, Flahault A, Dondon
MG and Le MG: Is beta-blocker treatment associated with a decrease
in the risk of cancer. Lett Drug Des Discov. 3:653–661. 2006.
View Article : Google Scholar
|
|
6
|
Entschladen F, Drell TL IV, Lang K, Joseph
J and Zaenker KS: Tumour-cell migration, invasion, and metastasis:
Navigation by neurotransmitters. Lancet Oncol. 5:254–258. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Entschladen F, Drell TL IV, Lang K, Joseph
J and Zaenker KS: Neurotransmitters and chemokines regulate tumor
cell migration: Potential for a new pharmacological approach to
inhibit invasion and metastasis development. Curr Pharm Des.
11:403–411. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lang K, Drell TL IV, Lindecke A, Niggemann
B, Kaltschmidt C, Zaenker KS and Entschladen F: Induction of a
metastatogenic tumor cell type by neurotransmitters and its
pharmacological inhibition by established drugs. Int J Cancer.
112:231–238. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dorn GW II and Liggett SB:
Pharmacogenomics of beta-adrenergic receptors and their accessory
signaling proteins in heart failure. Clin Transl Sci. 1:255–262.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosini M, Bolognesi ML, Giardina D,
Minarini A, Tumiatti V and Melchiorre C: Recent advances in
alpha1-adrenoreceptor antagonists as pharmacological tools and
therapeutic agents. Curr Top Med Chem. 7:147–162. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hein P and Michel MC: Signal transduction
and regulation: Are all alpha1-adrenergic receptor subtypes created
equal? Biochem Pharmacol. 73:1097–1106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Docherty JR: Subtypes of functional
alpha1-adrenoceptor. Cell Mol Life Sci. 67:405–417. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hubbard KB and Hepler JR: Cell signalling
diversity of the Gqalpha family of heterotrimeric G proteins. Cell
Signal. 18:135–150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Theroux TL, Esbenshade TA, Peavy RD and
Minneman KP: Coupling efficiencies of human alpha 1-adrenergic
receptor subtypes: Titration of receptor density and responsiveness
with inducible and repressible expression vectors. Mol Pharmacol.
50:1376–1387. 1996.PubMed/NCBI
|
|
15
|
Fitzgerald PJ: Is norepinephrine an
etiological factor in some types of cancer? Int J Cancer.
124:257–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Adissu HA and Schuller HM: Antagonistic
growth regulation of cell lines derived from human lung
adenocarcinomas of Clara cell and aveolar type II cell lineage:
Implications for chemoprevention. Int J Oncol. 24:1467–1472.
2004.PubMed/NCBI
|
|
17
|
Gobbi PG, Villano L, Pozzoli D, Bergonzi
M, Vanoli A, Tava F, Dionigi P and Corazza GR: Improving the
AJCC/TNM classification for use in early gastric cancer. J
Gastrointest Surg. 15:935–941. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
In H, Solsky I, Palis B, Langdon-Embry M,
Ajani J and Sano T: Validation of the 8th edition of the AJCC TNM
staging system for gastric cancer using the national cancer
database. Ann Surg Oncol. 24:3683–3691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roessler S, Long EL, Budhu A, Chen Y, Zhao
X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al: Integrative
genomic identification of genes on 8p associated with
hepatocellular carcinoma progression and patient survival.
Gastroenterology. 142:957–966.e12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shaul YD, Yuan B, Thiru P, Nutter-Upham A,
McCallum S, Lanzkron C, Bell GW and Sabatini DM: MERAV: A tool for
comparing gene expression across human tissues and cell types.
Nucleic Acids Res. 44(D1): D560–D566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Anaya J: OncoLnc: Linking TCGA survival
data to mRNAs, miRNAs, and lncRNAs. PeerJ Comp Sci. 2:e672016.
View Article : Google Scholar
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mínguez B, Hoshida Y, Villanueva A,
Toffanin S, Cabellos L, Thung S, Mandeli J, Sia D, April C, Fan JB,
et al: Gene-expression signature of vascular invasion in
hepatocellular carcinoma. J Hepatol. 55:1325–1331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Magnon C, Hall SJ, Lin J, Xue X, Gerber L,
Freedland SJ and Frenette PS: Autonomic nerve development
contributes to prostate cancer progression. Science.
341:12363612013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schuller HM: Neurotransmitter
receptor-mediated signaling pathways as modulators of
carcinogenesis. Prog Exp Tumor Res. 39:45–63. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thaker PH, Han LY, Kamat AA, Arevalo JM,
Takahashi R, Lu C, Jennings NB, Armaiz-Pena G, Bankson JA, Ravoori
M, et al: Chronic stress promotes tumor growth and angiogenesis in
a mouse model of ovarian carcinoma. Nat Med. 12:939–944. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wan D, Gong Y, Qin W, Zhang P, Li J, Wei
L, Zhou X, Li H, Qiu X, Zhong F, et al: Large-scale cDNA
transfection screening for genes related to cancer development and
progression. Proc Natl Acad Sci USA. 101:15724–15729. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Yang XM, Wang YH, Feng MX, Liu XJ,
Zhang YL, Huang S, Wu Z, Xue F, Qin WX, et al: Monoamine oxidase A
suppresses hepatocellular carcinoma metastasis by inhibiting the
adrenergic system and its transactivation of EGFR signaling. J
Hepatol. 60:1225–1234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Perez DM: The adrenergic receptors: In the
21st century. NewYork: Humana Press; 2006
|
|
32
|
Liu W, Wang X, Mei Z, Gong J, Huang L, Gao
X, Zhao Y, Ma J and Qian L: BNIP3L promotes cardiac fibrosis in
cardiac fibroblasts through [Ca2+]i-TGF-β-Smad2/3
pathway. Sci Rep. 7:19062017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oganesian A, Yarov-Yarovoy V, Parks WC and
Schwinn DA: Constitutive coupling of a naturally occurring human
alpha1a- adrenergic receptor genetic variant to EGFR
transactivation pathway. Proc Natl Acad Sci USA. 108:19796–19801.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salazar R and Ciardiello F: Optimizing
anti-EGFR therapy in colorectal cancer. Clin Cancer Res.
21:5415–5416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
García-Cazarín ML, Smith JL, Clair DK and
Piascik MT: The alpha1D-adrenergic receptor induces vascular smooth
muscle apoptosis via a p53-dependent mechanism. Mol Pharmacol.
74:1000–1007. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Okada H and Mak TW: Pathways of apoptotic
and non-apoptotic death in tumour cells. Nat Rev Cancer. 4:592–603.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu B, Chen Y and St Clair DK: Ros and
p53: A versatile partnership. Free Radic Biol Med. 44:1529–1535.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiao L, Pimentel DR, Wang J, Singh K,
Colucci WS and Sawyer DB: Role of reactive oxygen species and
NAD(P)H oxidase in alpha(1)-adrenoceptor signaling in adult rat
cardiac myocytes. Am J Physiol Cell Physiol. 282:C926–C934. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoda Y, Takeshima H, Niwa T, Kim JG, Ando
T, Kushima R, Sugiyama T, Katai H, Noshiro H and Ushijima T:
Integrated analysis of cancer-related pathways affected by genetic
and epigenetic alterations in gastric cancer. Gastric Cancer.
18:65–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Z, Wang W, Xu S, Wang S, Tu Y, Xiong
Y, Mei J and Wang C: The role of MAPK signaling pathway in the
Her-2-positive meningiomas. Oncol Rep. 36:685–695. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwappacher R, Rangaswami H, Su-You J,
Hassad A, Spitler R and Casteel DE: cGMP-dependent protein kinase
Iβ regulates breast cancer cell migration and invasion via
interaction with the actin/myosin-associated protein caldesmon. J
Cell Sci. 126:1626–1636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu S, Sun L, Jiao Y and Lee LTO: The role
of G protein-coupled receptor kinases in cancer. Int J Biol Sci.
14:189–203. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Peng L, Peng W, Hu P and Zhang HF:
Clinical significance of expression levels of serum ADRA1A in
hysterocarcinoma patients. Oncol Lett. 15:9162–9166.
2018.PubMed/NCBI
|
|
44
|
Powe DG, Voss MJ, Habashy HO, Zänker KS,
Green AR, Ellis IO and Entschladen F: Alpha- and beta-adrenergic
receptor (AR) protein expression is associated with poor clinical
outcome in breast cancer: An immunohistochemical study. Breast
Cancer Res Treat. 130:457–463. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang JJ and Blobe GC: Dichotomous roles
of TGF-β in human cancer. Biochem Soc Trans. 44:1441–1454. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Costanza B, Umelo IA, Bellier J,
Castronovo V and Turtoi A: Stromal modulators of TGF-β in cancer. J
Clin Med. 6(pii): E72017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tatsuta M, Iishi H, Baba M, Yano H, Sakai
N, Uehara H, Hirasawa R and Nakaizumi A: Alpha1-adrenoceptor
stimulation enhances experimental gastric carcinogenesis induced by
N-methyl-N′-nitro-N-nitrosoguanidine in Wistar rats. Int J Cancer.
77:467–469. 1998. View Article : Google Scholar : PubMed/NCBI
|