Introduction
Ovarian cancer has the highest mortality rate of all
gynecological malignancies, and epithelial ovarian cancer (EOC) is
the most frequently encountered (1).
The disease course of ovarian cancer is complex, and symptoms are
non-specific (2). Therefore, at the
point of diagnosis the majority of patients have reached the
advanced stages of disease, when radical treatment is less
successful (3). Current issues in
the diagnosis and treatment of ovarian cancer include a lack of
early diagnosis, limited effective treatment options, and
propensity to chemoresistance and metastatic dissemination
(4,5). Therefore, the discovery of novel
diagnostic markers and therapeutic targets is pivotal to improving
the prognosis for patients with EOC.
Long non-coding RNAs (lncRNAs) are a type of
non-coding RNA >200 nucleotides in length, known as the ‘dark
matter’ of the genome (6). Although
they are not protein-coding, lncRNAs possess complex biological
functions (7,8). Yan et al (9) analyzed the expression of lncRNAs in
5,037 human tissue samples from The Cancer Genome Atlas (TCGA)
database, across 13 types of cancer. It was revealed that, in
different types of cancer tissue, non-coding genes had a specific
expression profile compared with their protein-coding counterparts.
Therefore, the study of lncRNAs may hold promise towards improving
cancer diagnosis and treatment programs (10–12).
However, to the best of our knowledge, there is currently limited
research into the expression and function of lncRNAs in EOC.
Therefore, the present study aimed to improve the understanding of
the functions and molecular mechanisms of lncRNAs in EOC, and how
they may contribute to the early diagnosis and treatment of ovarian
cancer (13,14).
In previous years, the development of chip
technology has greatly enhanced the efficiency of biological
research (15). Bioinformatics is an
interdisciplinary field of biology and computer science, and with
the advent of ‘big data’ in health care, bioinformatics research
has greatly improved the efficiency and progress of health
care-associated research (16).
Materials and methods
Data processing
Ovarian cancer expression files GSE18520 (17), GSE19829 (18), GSE26193 (19), GSE63885 (20), GSE14001 (21), GSE38666 (22) and GSE40595 (23) were retrieved from the Gene Expression
Omnibus (GEO) database (platform GPL570). Datasets involving
patients with EOC were selected; the differential expression levels
of lncRNAs between normal ovarian and ovarian tumor tissues were
analyzed using the R (3.3.3) package ‘limma’ (3.40.2)′ (http://bioconductor.org/packages/release/bioc/html/limma.html),
and disease prognosis was analyzed using the R package ‘survival’.
The low grades included grade 1 and 2 and the high grade included
grade 3. According to the International Federation of Gynaecology
and Obstetrics (FIGO) staging systems, stages III and IV included
advanced stages and stages I and II included early stage. The
median expression of LINC01627 was used as cut-off value for
survival analysis. Where P≥0.1 and I2≤50% indicated no
heterogeneity between each study, meta-analysis was performed using
the fixed effect model. Where P<0.1 and I2>50%
suggested heterogeneity between each study, the random effects
model was used for meta-analysis, and sub-analysis was conducted to
determine the source of heterogeneity.
Tissues sample collection and total
RNA extraction
A total of 20 EOC tissue samples and 10 normal
ovarian tissue samples were collected from patients who underwent
surgery between June 2015 and October 2018 at the Second Affiliated
Hospital of Soochow University (Suzhou, China). The present study
was approved by the Ethics Committee of the Second Affiliated
Hospital of Soochow University (Suzhou, China), and all patients
provided written informed consent. None of the patients had
received chemotherapy or radiotherapy prior to surgery and all
patients were treated with systemic platinum-based chemotherapy
following surgery. Tumor stage was determined according to the FIGO
staging system. Following resection, the tissues were immediately
frozen at −80°C. TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract the total RNA from
ovarian cancer cells and tissue samples.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was reverse transcribed into cDNA using
the PrimeScript™ RT Master Mix (cat. no. RR036A; Clontech
Laboratories, Inc.), and qPCR was performed using the
SYBR® Green Master Mix (Tiangen Biotech Co., Ltd.),
according the manufacturer's protocol. The reaction volume total
was 10 µl, and the thermocycling conditions were: 95°C
pre-denaturation for 3 sec, followed by 40 cycles of 95°C
denaturation for 10 min, 60°C annealing for 40 sec, followed by
72°C extension for 30 sec. The 2−ΔΔCq method was used to
calculate relative expression levels (24). Primers were as follows: LINC01627,
forward 5′-ACTGATACCCACvACAGAAGAAG-3, and reverse
5′-CCCGTCAAGTCGAGTAATATCC-3′; and GAPDH, forward
5′-GCACCGTCAAGGCTGAGAAC-3′, and reverse
5′-GGATCTCGCTCCTGGAAGATG-3′.
Cell culture and transfection
Ovarian cancer cell lines HO8910 and HEY were
purchased from the American Type Culture Collection. The ovarian
cancer cell lines HO8910 and HEY were cultured in RPMI 1640 (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; cat. no. 10099-141; Gibco; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2. Cells were seeded in 6-well plates and
once reached 70% confluency, cells were transfected with 50nM
negative control (NC) or LINC01627 small interfering (si)RNA using
Lipofectamine™ RNAi MAX reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The LINC01627 siRNA
sequence was 5′-GGGTGTACACAGAGTATAA-3′. The negative control
sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. After 24 h of
transfection, subsequent experiments were performed.
Cell migration assay
Following transfection, cells were resuspended in
RPMI medium without FBS at a density of 2×104
cells/well. A total of 100 µl resuspended cells were seeded into
the upper chamber of a Transwell insert. RPMI medium supplemented
with 10% FBS was added to the lower chamber. The cells were
cultured at 37°C with 5% CO2 for 25 h. Migratory cells
were fixed in 70% paraformaldehyde for 30 min and stained with 0.1%
crystal violet for a further 30 min at room temperature. Following
washing with PBS, cells were directly observed using an inverted
microscope and was counted in five randomly selected fields to get
the average using ImageJ (version 1.48; National Institutes of
Health).
Cell proliferation assay
Following transfection, HO8910 and HEY cells were
seeded into 96-well plates at a density of 5×103
cells/well. At 24, 48, 72 and 96 h time points, 10 µl Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) solution was
added to each well and the plates were incubated at 37°C for 1 h in
the dark. Absorbance was measured at a wavelength of 450 nm using a
microplate reader.
Statistical analysis
Stata statistical software (StataCorp LP) was used
to perform statistical analyses. The hazard ratio (HR) and 95%
confidence interval (CI) were determined using the log-rank test.
Unpaired t-test was enrolled to compute the difference between two
groups. P<0.05 was considered to indicate a statistically
significant difference. Each experiment was repeated three
times.
Results
LINC01627 is highly expressed in
ovarian cancer and is a prognostic risk factor for EOC
In order to determine potential prognostic factors
for ovarian cancer, a group of differentially expressed genes was
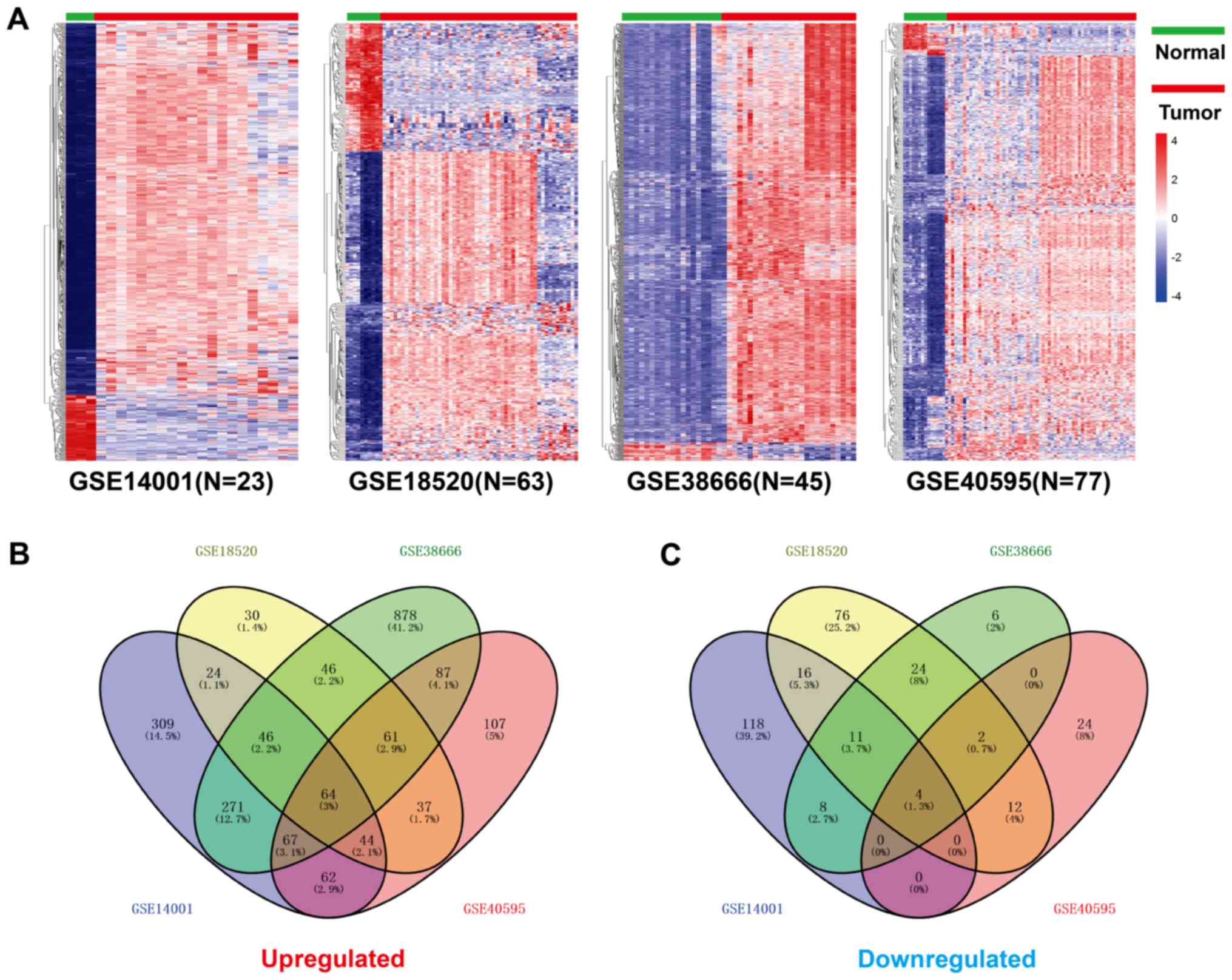
obtained from the GSE14001, GSE18520, GSE38666 and GSE40595
databases using the R package ‘limma’ (Fig. 1A). Using the R package ‘venn’, a
total of 64 lncRNAs were identified as highly expressed in
GSE14001, GSE18520, GSE38666 and GSE4059 (Fig. 1B), and 4 were downregulated (Fig. 1C). Prognostic analysis of the 64
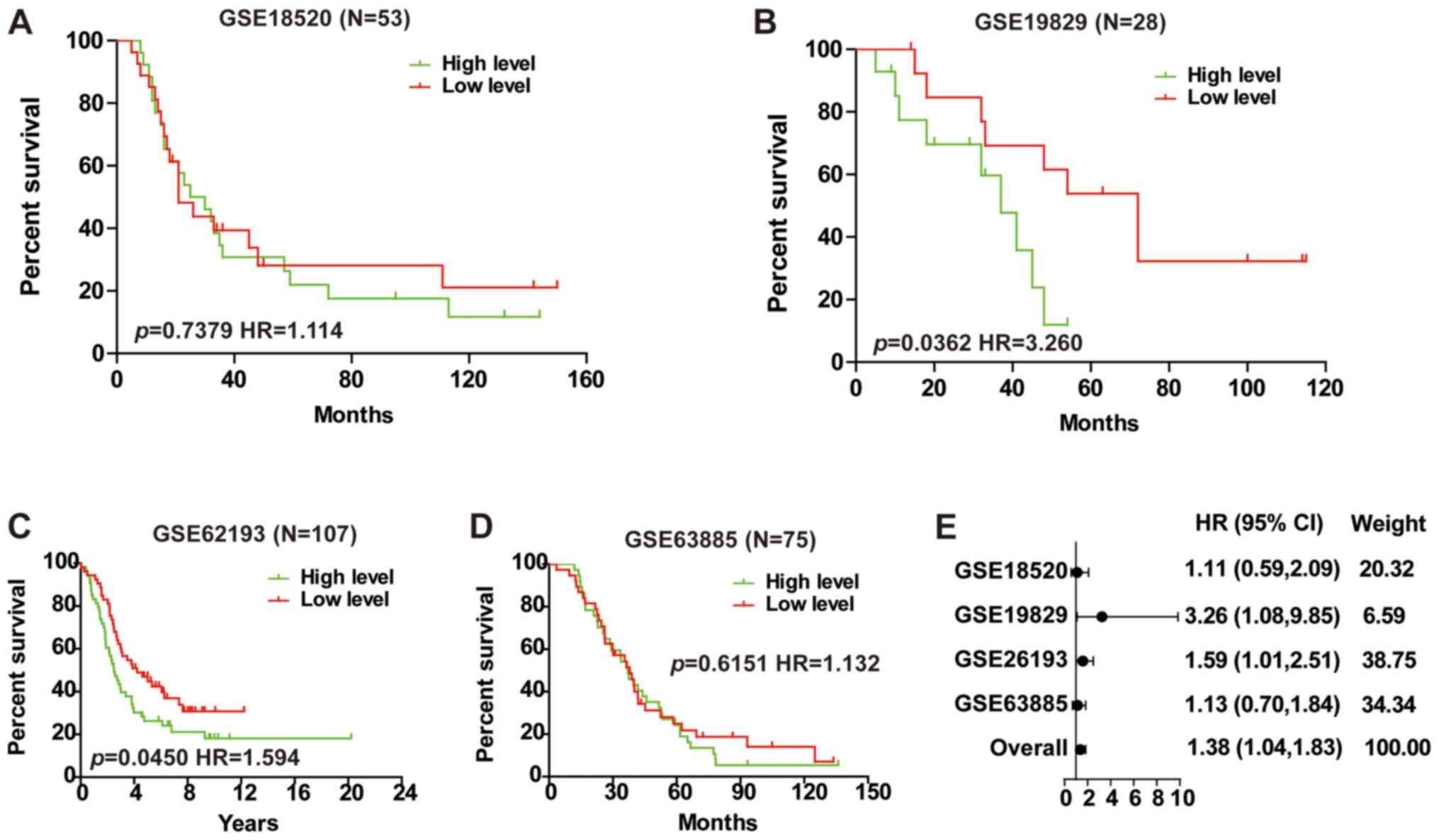
lncRNAs was performed in the GSE18520, GSE19829, GSE62193 and
GSE63885 databases. The results revealed that only LINC01627
expression was negatively correlated with patient outcome in
GSE19829 and GSE62193 (Fig. 2B and
C), whilst no association was observed in GSE18520 or GSE63885
(Fig. 2A and D). To investigate
whether LINC01627 was associated with patient prognosis, a total HR
(1.38) interval of 1.04–1.83 was determined using meta-analysis
(Fig. 2E), suggesting that LINC01627
may be a prognostic factor in predicting patient outcome.
LINC01627 is a prognostic risk factor
for high-grade EOC
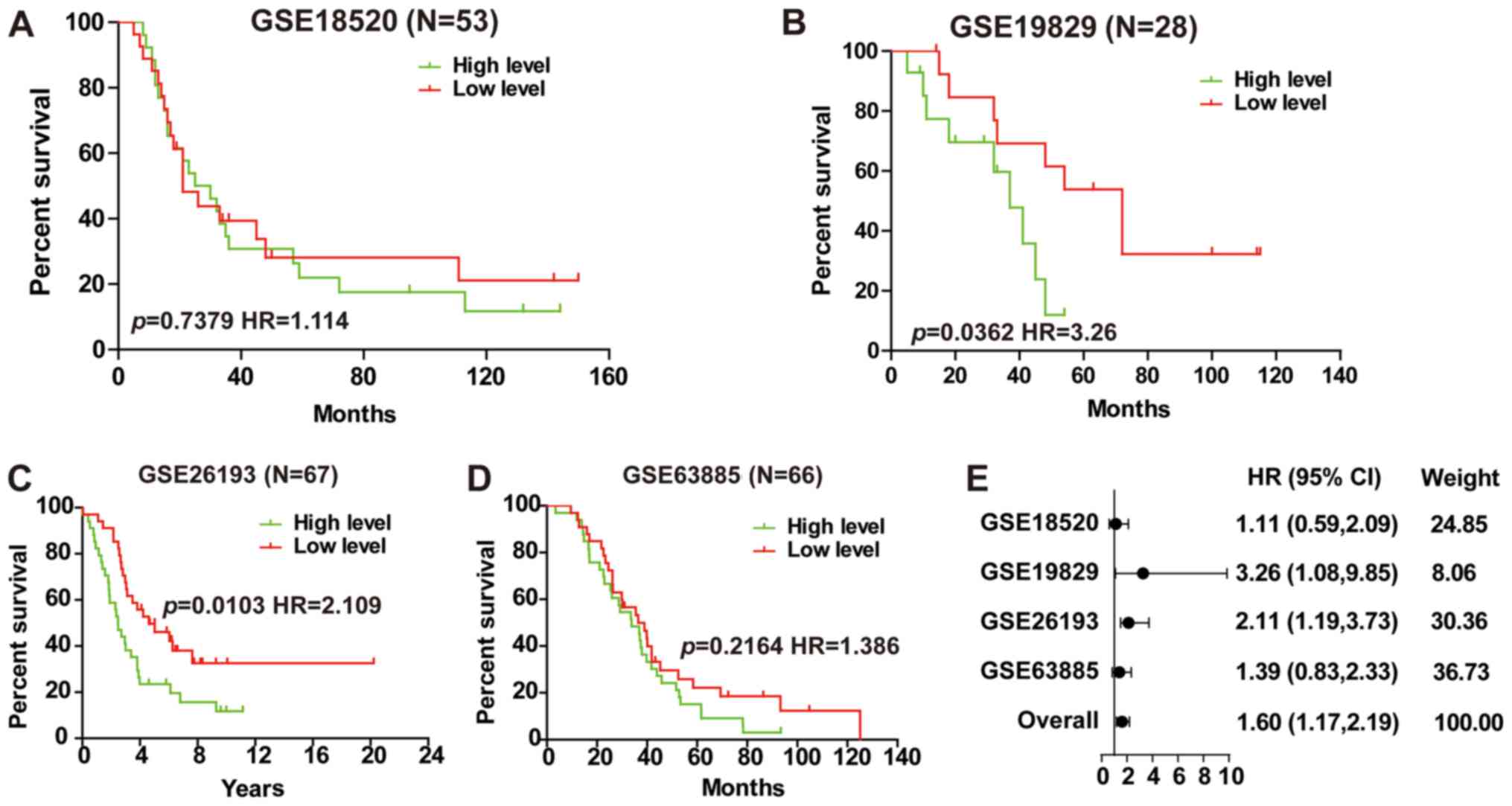
Further analysis of the prognostic implications of
LINC01627 in high-grade EOC illustrated that the expression of
LINC01627 was negatively correlated with patient prognosis in
GSE19829 and GSE26193 (Fig. 3B and
C), whilst no correlation was observed in GSE18520 or GSE63885
(Fig. 3A and D). A total HR (1.60)
interval of 1.17–2.19 was determined using meta-analysis (Fig. 3E), suggesting that LINC01627 may be
regarded as a prognostic factor for high-grade EOC.
LINC01627 is a prognostic risk factor
for advanced EOC
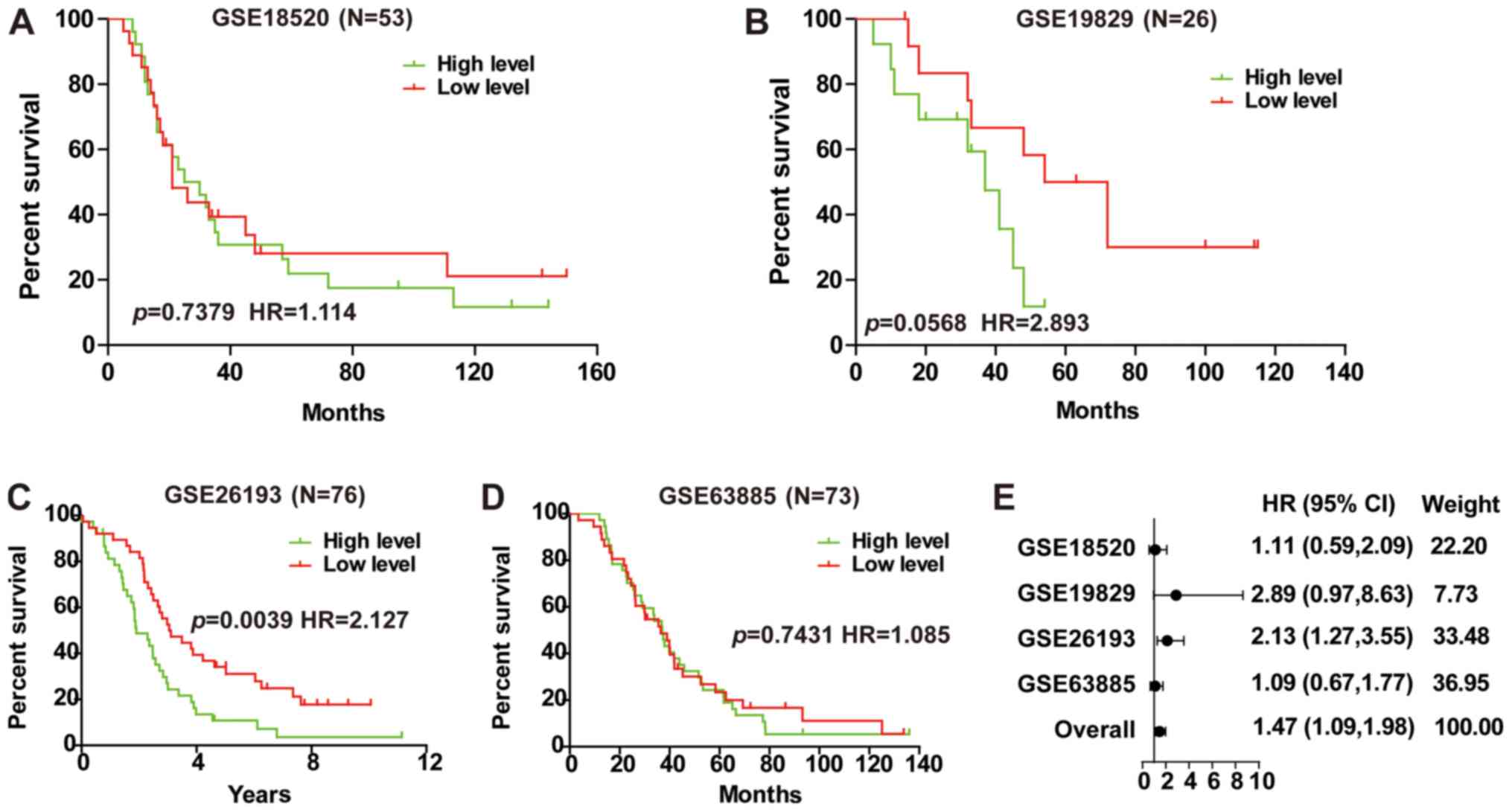
Investigation into the prognostic implications of
LINC01627 in advanced EOC demonstrated that LINC01627 was a
prognostic risk factor within GSE26193 (Fig. 4C), whilst no correlation was revealed
in GSE18520, GSE19829 or GSE63885 (Fig.
4A, B and D). To investigate whether LINC01627 was associated
with the prognosis of patients with advanced EOC, a total HR (1.47)
interval of 1.09–1.98 was determined using meta-analysis (Fig. 4E), suggesting that LINC01627 may be
regarded as a prognostic factor for advanced EOC.
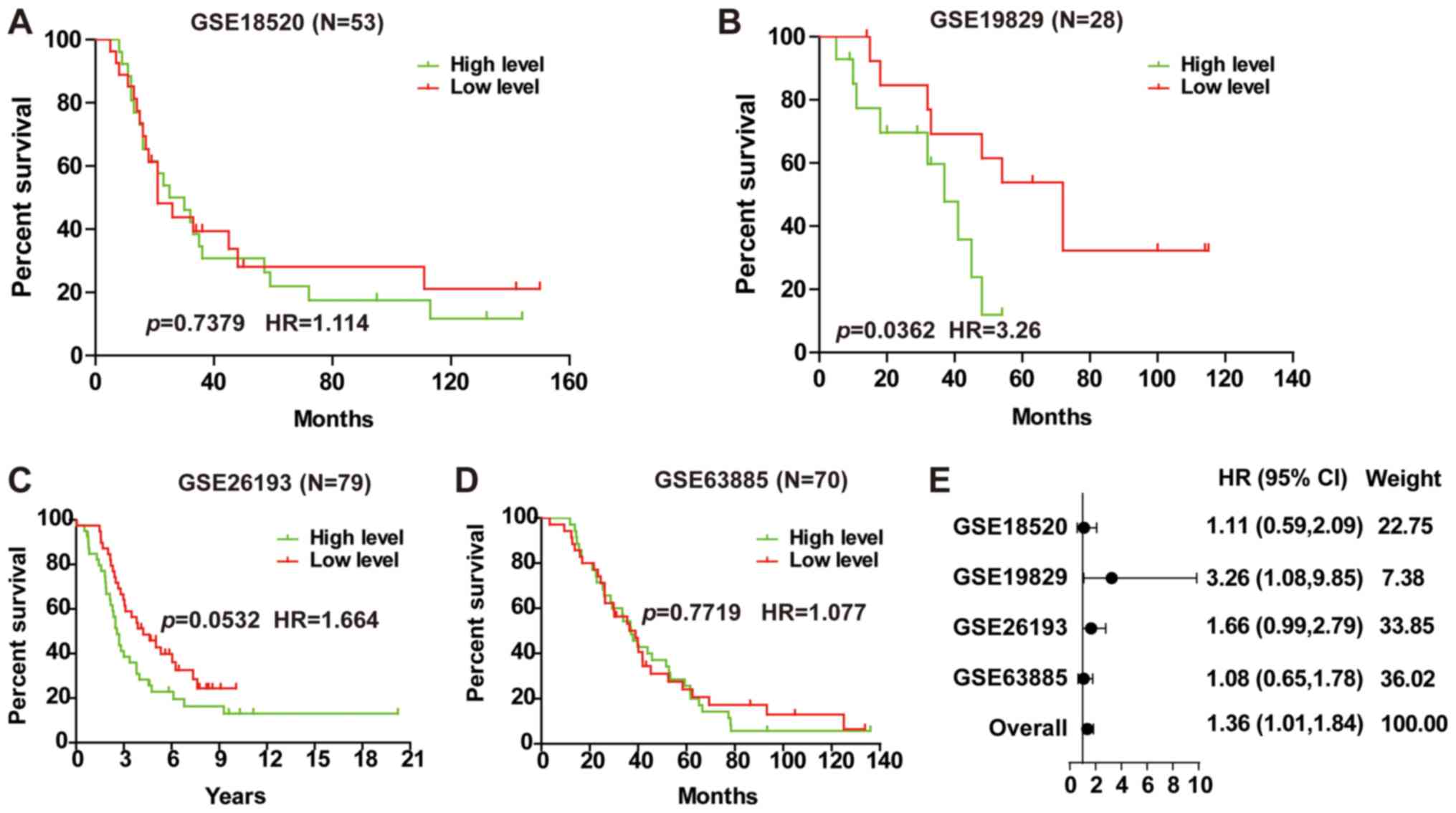
LINC01627 is a prognostic risk factor
for serous EOC
EOC includes serous, mucinous, endometrioid and
other subtypes, of which serous EOC is the most frequently observed
(25). Analysis of the prognostic
implication of LINC01627 in serous EOC illustrated that LINC01627
was a prognostic risk factor within GSE19829 (Fig. 5B), whilst no correlation was observed
in GSE18520, GSE26193 or GSE63885 (Fig.
5A, C and D). To investigate whether LINC01627 is associated
with the prognosis of patients with serous EOC, a total HR (1.36)
interval 1.01–1.84 was determined using meta-analysis (Fig. 5E), suggesting that LINC01627 may be
regarded as a prognostic factor for serous EOC. In addition, it was
indicated that increased expression of LINC01627 was associated
with poor patient prognosis.
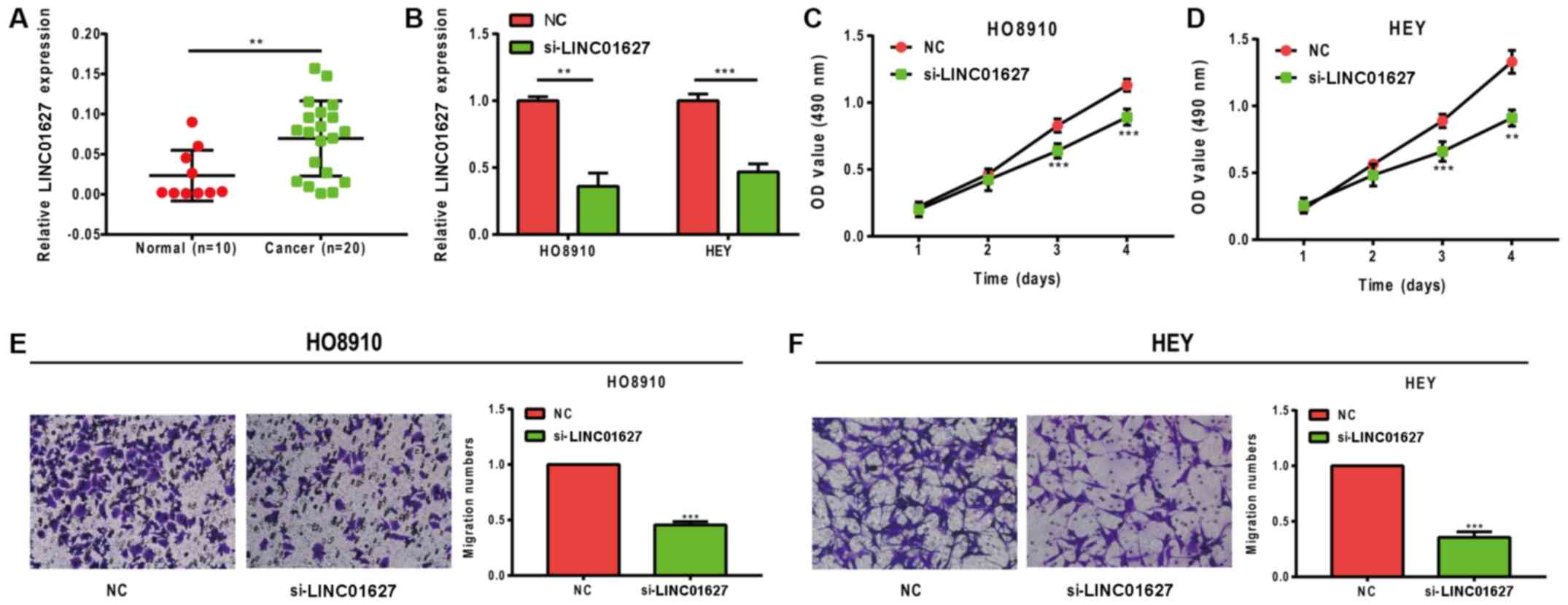
LINC01627 promotes proliferation and
migration in ovarian cancer cells
LINC01627 expression levels were detected in normal
ovarian (n=10) and EOC tissues (n=20). The results revealed that
the expression levels of LINC01627 were significantly increased in
EOC tissues (P=0.0090; Fig. 6A). To
further investigate the functions of LINC01627, siRNA targeting
LINC01627 was established and knockdown assays were subsequently
performed in vitro. Transfection efficiency is illustrated
in Fig. 6B, with the si-LINC01627
group exhibiting significantly decreased levels of LINC01627
compared with the control group, in both cell lines tested. The
results of the CCK-8 assay revealed that cell proliferation was
significantly inhibited following LINC01627-knockdown (Fig. 6C and D). In addition, results from
Transwell assays demonstrated that migration was significantly
abrogated in HO8910 and HEY cells following LINC01627 knockdown
(Fig. 6E and F).
Discussion
With limited early clinical symptoms and the current
limitations of medical treatments, including drug resistance, the
majority of patients with ovarian cancer are in the advanced stages
of disease at the time of diagnosis (26). Although cytoreductive surgery
combined with paclitaxel and cisplatin chemotherapy is able to
effectively relieve symptoms in the majority of patients, the
5-year survival rate of patients with ovarian epithelial tumors is
<40% (27,28). At present, the most frequently used
diagnostic methods for ovarian cancer include ultrasound and the
detection of cancer antigen l25 and human epididymal protein 4
(29), the specificity and
sensitivity of which do not meet the requirements for early
screening. As a result, the early detection and diagnosis of
ovarian cancer has yet to be optimized. To improve the survival
rates of patients with EOC novel tumor biomarkers and therapeutic
targets are required (30).
LncRNAs were discovered in the 1980s and due to
their inability to encode proteins, they were previously considered
to be ‘transcription noise’ (6). In
recent years, an increasing number of studies have indicated that
lncRNAs are able to regulate gene expression at the epigenetic,
transcriptional and post-transcriptional levels (31). In addition, they are likely to serve
a prominent role in the development of cancer (32). It has been reported that there are
significant differences in the expression levels of lncRNAs between
healthy and tumorous tissues, as brain cytoplasmic RNA 1 (BCYRN1)
was highly expressed in breast, lung, tongue and ovarian cancer
(33), and HOX transcript antisense
RNA (HOTAIR) expression levels were significantly higher in tumor
tissues compared with those in normal, healthy tissues (34–36).
Therefore, lncRNAs are potential novel tumor markers and
therapeutic targets for the screening and early diagnosis of cancer
(37–39). The mechanisms of lncRNAs in the
development of ovarian cancer may also be implicated in the
prevention, diagnosis, treatment and prognosis of the disease
(40,41).
The aim of the present study was to determine the
role of lncRNAs in the development of EOC, to analyse the
expression of LINC01627 in different databases, and to conduct
meta-analysis in order to confirm whether LINC01627 was a
prognostic factor for EOC. The results revealed that LINC01627 was
a prognostic risk factor for EOC. Furthermore, meta-analysis
concluded that LINC01627 was a prognostic risk factor for
high-grade, advanced and serous EOC, as the higher the expression,
the poorer the patient prognosis. It was also concluded that
LINC01627 expression levels may have prognostic significance in
patients with EOC. Upregulation of LINC01627 has been observed in
non-small-cell lung cancer (42);
however, the role of LINC01627 in EOC is yet to be ascertained. In
the present study, to further investigate the oncogenic role of
LINC01627, the expression levels of LINC01627 were determined in
EOC clinical samples using RT-qPCR. The results revealed that
LINC01627 was highly expressed in EOC tissues compared with normal
ovarian tissue samples. Functional cell-based assays further
confirmed that LINC01627 had a role on the proliferation and
migration of HO8910 and HEY cells. To further examine the mechanism
of LINC01627, RNA immunoprecipitation and chromatin
immunoprecipitation assays are going to be performed in future
studies.
With further research, LINC01627 may become a future
therapeutic target for the treatment of ovarian cancer, which may
accelerate the use of lncRNAs from basic research to clinical
application, and provide a novel opportunity for the diagnosis and
treatment of ovarian cancer. These results may subsequently promote
novel ideas for the research, diagnosis and treatment of EOC.
However, since the investigation of lncRNAs is a relatively new
research field, expression profiles in EOC have yet to be fully
established. The understanding of the molecular mechanisms of
lncRNAs is limited and further investigation is required in order
to examine their full role and potential in the development of EOC.
In conclusion, the present study indicated that LINC01627 may be a
prognostic risk factor for EOC, and may predict patient prognosis
in high-grade, advanced and serous disease subtypes.
Acknowledgements
The authors would like to thank Nanjing Qiaoyuan
Biotechnology Co., Ltd for their help with bioinformatics
analysis.
Funding
The present study was funded by The Jiangsu Maternal
and Child Healthcare Research Project (grant no. F201709).
Availability of data and materials
The data used to support the findings of this study
are available from the corresponding author upon request.
Authors' contributions
XS performed most experiments, collected clinical
tissues and analysed data. WZ designed the project and edited the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Soochow University
(Suzhou, China), and all patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Meinsohn MC, Smith OE, Bertolin K and
Murphy BD: The orphan nuclear receptors steroidogenic factor-1 and
liver receptor homolog-1: Structure, regulation, and essential
roles in mammalian reproduction. Physiol Rev. 99:1249–1279. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
George A, Kaye S and Banerjee S:
Delivering widespread BRCA testing and PARP inhibition to patients
with ovarian cancer. Nat Rev Clin Oncol. 14:284–296. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tung N, Domchek SM, Stadler Z, Nathanson
KL, Couch F, Garber JE, Offit K and Robson ME: Counselling
framework for moderate-penetrance cancer-susceptibility mutations.
Nat Rev Clin Oncol. 13:581–588. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ren C, Li X, Wang T, Wang G, Zhao C, Liang
T, Zhu Y, Li M, Yang C, Zhao Y and Zhang GM: Functions and
mechanisms of long noncoding RNAs in ovarian cancer. Int J Gynecol
Cancer. 25:566–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang LQ, Yang SQ, Wang Y, Fang Q, Chen
XJ, Lu HS and Zhao LP: Long noncoding RNA MIR4697HG promotes cell
growth and metastasis in human ovarian cancer. Anal Cell Pathol
(Amst). 2017:82678632017.PubMed/NCBI
|
|
6
|
Esposito R, Bosch N, Lanzos A, Polidori T,
Pulido-Quetglas C and Johnson R: Hacking the cancer genome:
Profiling therapeutically actionable long non-coding RNAs using
CRISPR-Cas9 screening. Cancer Cell. 35:545–557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chu C, Spitale RC and Chang HY:
Technologies to probe functions and mechanisms of long noncoding
RNAs. Nat Struct Mol Biol. 22:29–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li T, Yang XD, Ye CX, Shen ZL, Yang Y,
Wang B, Guo P, Gao ZD, Ye YJ, Jiang KW and Wang S: Long noncoding
RNA HIT000218960 promotes papillary thyroid cancer oncogenesis and
tumor progression by upregulating the expression of high mobility
group AT-hook 2 (HMGA2) gene. Cell Cycle. 16:224–231. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng L, Yuan XQ, Liu ZY, Li WL, Zhang CY,
Zhang YQ, Pan X, Chen J, Li YH and Li GC: High lncRNA H19
expression as prognostic indicator: Data mining in female cancers
and polling analysis in non-female cancers. Oncotarget.
8:1655–1667. 2017.PubMed/NCBI
|
|
13
|
Jiang C, Li X, Zhao H and Liu H: Long
non-coding RNAs: Potential new biomarkers for predicting tumor
invasion and metastasis. Mol Cancer. 15:622016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meryet-Figuière M, Lambert B, Gauduchon P,
Vigneron N, Brotin E, Poulain L and Denoyelle C: An overview of
long non-coding RNAs in ovarian cancers. Oncotarget. 7:44719–44734.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baumgart S, Hölters S, Ohlmann CH, Bohle
R, Stöckle M, Ostenfeld MS, Dyrskjøt L, Junker K and Heinzelmann J:
Exosomes of invasive urothelial carcinoma cells are characterized
by a specific miRNA expression signature. Oncotarget.
8:58278–58291. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lo CM, Iqbal U and Jack Li YC: Automatic
methods for managements of cancer, medicine, and behavior. Comput
Methods Programs Biomed. 146:A12017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mok SC, Bonome T, Vathipadiekal V, Bell A,
Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, et al: A
gene signature predictive for outcome in advanced ovarian cancer
identifies a survival factor: Microfibril-associated glycoprotein
2. Cancer Cell. 16:521–532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Konstantinopoulos PA, Spentzos D, Karlan
BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA and Cannistra
SA: Gene expression profile of BRCAness that correlates with
responsiveness to chemotherapy and with outcome in patients with
epithelial ovarian cancer. J Clin Oncol. 28:3555–3561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gentric G, Kieffer Y, Mieulet V, Goundiam
O, Bonneau C, Nemati F, Hurbain I, Raposo G, Popova T, Stern MH, et
al: PML-regulated mitochondrial metabolism enhances
chemosensitivity in human ovarian cancers. Cell Metab. 29:156–173
e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lisowska KM, Olbryt M, Dudaladava V,
Pamuła-Piłat J, Kujawa K, Grzybowska E, Jarząb M, Student S,
Rzepecka IK, Jarząb B and Kupryjańczyk J: Gene expression analysis
in ovarian cancer-faults and hints from DNA microarray study. Front
Oncol. 4:62014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tung CS, Mok SC, Tsang YT, Zu Z, Song H,
Liu J, Deavers MT, Malpica A, Wolf JK, Lu KH, et al: PAX2
expression in low malignant potential ovarian tumors and low-grade
ovarian serous carcinomas. Mod Pathol. 22:1243–1250. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lili LN, Matyunina LV, Walker LD, Benigno
BB and McDonald JF: Molecular profiling predicts the existence of
two functionally distinct classes of ovarian cancer stroma. Biomed
Res Int. 2013:8463872013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yeung TL, Leung CS, Wong KK, Samimi G,
Thompson MS, Liu J, Zaid TM, Ghosh S, Birrer MJ and Mok SC: TGF-β
modulates ovarian cancer invasion by upregulating CAF-derived
versican in the tumor microenvironment. Cancer Res. 73:5016–5028.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Erol A, Niemira M and Kretowski AJ: Novel
approaches in ovarian cancer research against heterogeneity, late
diagnosis, drug resistance, and transcoelomic metastases. Int J Mol
Sci. 20(pii): E26492019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Franier B and Thompson M: Early stage
detection and screening of ovarian cancer: A research opportunity
and significant challenge for biosensor technology. Biosens
Bioelectron. 135:71–81. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oberaigner W, Minicozzi P, Bielska-Lasota
M, Allemani C, de Angelis R, Mangone L and Sant M; Eurocare Working
Group, : Survival for ovarian cancer in Europe: The across-country
variation did not shrink in the past decade. Acta Oncol.
51:441–453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shapira I, Oswald M, Lovecchio J, Khalili
H, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, et
al: Circulating biomarkers for detection of ovarian cancer and
predicting cancer outcomes. Br J Cancer. 110:976–983. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dochez V, Caillon H, Vaucel E, Dimet J,
Winer N and Ducarme G: Biomarkers and algorithms for diagnosis of
ovarian cancer: CA125, HE4, RMI and ROMA, a review. J Ovarian Res.
12:282019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li K, Hüsing A, Fortner RT, Tjønneland A,
Hansen L, Dossus L, Chang-Claude J, Bergmann M, Steffen A, Bamia C,
et al: An epidemiologic risk prediction model for ovarian cancer in
Europe: The EPIC study. Br J Cancer. 112:1257–1265. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qin Y, Liu X, Pan L, Zhou R and Zhang X:
Long noncoding RNA MIR155HG facilitates pancreatic cancer
progression through negative regulation of miR-802. J Cell Biochem.
Jun 3–2019.(Epub ahead of print). View Article : Google Scholar
|
|
32
|
Huang Z, Luo Q, Yao F, Qing C, Ye J, Deng
Y and Li J: Identification of differentially expressed long
non-coding RNAs in polarized macrophages. Sci Rep. 6:197052016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu T and Lu YR: BCYRN1, a c-MYC-activated
long non-coding RNA, regulates cell metastasis of non-small-cell
lung cancer. Cancer Cell Int. 15:362015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Martínez-Fernández M, Feber A, Dueñas M,
Segovia C, Rubio C, Fernandez M, Villacampa F, Duarte J,
López-Calderón FF, Gómez-Rodriguez MJ, et al: Analysis of the
Polycomb-related lncRNAs HOTAIR and ANRIL in bladder cancer. Clin
Epigenetics. 7:1092015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang A, Zhao JC, Kim J, Fong KW, Yang YA,
Chakravarti D, Mo YY and Yu J: LncRNA hotair enhances the
androgen-receptor-mediated transcriptional program and drives
castration-resistant prostate cancer. Cell Rep. 13:209–221. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang XD, Xu HT, Xu XH, Ru G, Liu W, Zhu
JJ, Wu YY, Zhao K, Wu Y, Xing CG, et al: Knockdown of long
non-coding RNA HOTAIR inhibits proliferation and invasiveness and
improves radiosensitivity in colorectal cancer. Oncol Rep.
35:479–487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bolha L, Ravnik-Glavač M and Glavač D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo J, Qu J, Wu DK, Lu ZL, Sun YS and Qu
Q: Long non-coding RNAs: A rising biotarget in colorectal cancer.
Oncotarget. 8:22187–22202. 2017.PubMed/NCBI
|
|
39
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3:52015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11:492016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nikpayam E, Tasharrofi B, Sarrafzadeh S
and Ghafouri-Fard S: The role of long non-coding RNAs in ovarian
cancer. Iran Biomed J. 21:3–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang Q, Ni Z, Cheng Z, Xu J, Yu H and Yin
P: Three circulating long non-coding RNAs act as biomarkers for
predicting NSCLC. Cell Physiol Biochem. 37:1002–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|