Introduction
Cancer is the leading cause of mortality worldwide;
1,762,450 new cancer cases and 606,880 cancer mortalities were
estimated in 2019 (1). Furthermore,
27,510 new cases of gastric cancer leading to 11,140 deaths are
expected in the United States in 2019 (1). The incidence of GC is particularly high
in eastern Asian countries, such as China, Japan and South Korea
(2). In China, GC is the second
leading cause of cancer-related morbidity and mortality (3). Despite the developments in multimodal
therapy strategies, such as surgery, chemotherapy, radiotherapy,
target therapy and immunotherapy, the prognosis of advanced GC
remains poor (4). Thus, identifying
novel molecular biomarkers for GC is crucial to the improvement of
the therapeutic effects and of the survival of patients with
GC.
The mixed lineage kinase domain-like protein (MLKL)
is a component of the receptor-interacting kinase 1
(RIP1)/receptor-interacting kinase 3 (RIP3)/MLKL pathway, which is
involved in cell necroptosis (5,6).
Necroptosis, which is a form of programmed necrotic death, is
characterized by increased cell volume, cell rounding, nuclear
membrane dilation, chromatin condensation, cytoplasmic membrane
disruption, organelle swelling, and inadequate caspase activation
(6–11). The interaction between RIP3 and RIP1
recruits the downstream effector MLKL protein to form necrosomes,
where MLKL is phosphorylated (5,12). MLKL
depletion in cancer cells induces spontaneous phosphorylation of
H2A histone family member X, an early marker for DNA damage, which
suggests that MLKL may serve a crucial role in response to DNA
damage (13). Previous studies have
demonstrated that MLKL serves as a potential prognostic biomarker
for patients with multiple carcinomas (14–17).
These data suggest that MLKL may serve a distinctive role in
certain cancers, including GC. However, systematic studies on MLKL
expression and its prognostic value in human cancers are still
insufficient.
In the present study, the expression of MLKL in GC
samples and normal tissues was evaluated, the association between
clinicopathologic and prognostic parameters in patients with GC was
assessed in a large database, and a potential underlying mechanism
was investigated.
Materials and methods
Oncomine database analysis
Oncomine database (http://www.oncomine.org), a publicly accessible online
cancer microarray database, was used to analyze MLKL mRNA
expression levels in tumor and normal tissues. The thresholds were
set as follows: the fold change was defined as 2 and P-value was
set to 0.01. Cancer types, genes, datasets, sample sizes, fold
changes, t-test results and P-values were obtained from studies
that showed statistically significant differences.
Cancer Cell Line Encyclopedia (CCLE)
analysis
The CCLE database (https://portals.broadinstitute.org/ccle) is an online
compilation of gene expression, chromosomal copy number and
parallel sequencing data from 947 human cancer cell lines. The CCLE
database was used to analyze the mRNA expression levels of MLKL in
a series of cancers. Information about cancer types, genes,
datasets, sample sizes, fold changes, t-test results and P-values
were collected.
Kaplan-Meier (KM) Plotter database
analysis
The KM Plotter (http://kmplot.com/analysis) was used to evaluate the
prognostic values of MLKL and fatty acid 2-hydroxylase (FA2H) in
GC. The data from 882 patients with GC were obtained from the KM
database (tumor-node-metastasis (TNM) stages: T1, n=14; T2, n=253;
T3, n=208; T4, n=39; N0, n=76; N1+2+3, n=437; M0, n=459; M1, n=58;
surgical treatment only, n=393). Using the selected parameters, the
analysis was performed on the data from 631 patients with GC for
overall survival (OS) analysis and from 522 patients in first
progression (FP) analysis. The patients were split into high and
low expression groups according to the median values of mRNA
expression, and Kaplan-Meier survival plots were obtained. Logrank
test P<0.05 was considered to indicate a statistically
significant difference. Survival outcome, hazard ratios (HR), 95%
CI and P-values were summarized from the KM plotter webpage.
Patient samples
A total of 25 patients with histopathologically
confirmed GC and complete follow-up data were recruited between
March and September 2017 from the Xiangcheng People's Hospital. The
inclusion criteria were as follows: i) Histopathological diagnosis
of adenocarcinoma; ii) patients who had not received anti-tumor
treatment prior to the study; and iii) computed tomography of the
chest, abdomen and pelvis did not show evidence of distant
metastasis. The exclusion criteria were as follows: i) Patients who
cannot tolerate surgical treatment; and ii) patients who did not
allow specimen provision. There were 17 men and 8 women, aged
between 38 and 71 years (median age, 53 years). Written informed
consent was obtained from all patients and the protocol of the
study was approved by the Institutional Review Board of Suzhou
University (Suzhou, China; approval no. 2016958021; 5 March 2016).
Fresh gastric cancer and normal tissues (~2 cm from the tumor) were
collected from the patients, immediately stored in liquid nitrogen,
and lysed with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.). None of the patients had received chemotherapy
or radiotherapy prior to surgery. Pathological variables including
depth of invasion, lymph node metastasis, distant metastasis, TNM
stage and anti-tumor therapy were obtained. The Cancer Staging
Manual of the American Joint Committee on Cancer (version 8)
(18) was used for patient
staging.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed as described previously
(19). Total RNA was isolated from
frozen GC and normal tissues with TRIzol®. Complementary
DNA was prepared using oligo(dT) primers with Primer Script RT Mix
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
protocol. Expression levels of MLKL mRNA were determined with
SYBR-Green PCR kit (Takara Biotechnology Co., Ltd.) in an ABI PRISM
7500 fast Sequence Detection System (Applied Biosystems; Thermo
Fisher Scientific, Inc.). RT-qPCR reactions were performed as
follows: 95°C for 20 sec, 95°C for 10 sec and 60°C for 45 sec for
40 cycles. The relative expressions levels were expressed as
2−ΔΔCq (20) using
β-actin as the reference gene. Real-time PCR primers were as
follows: MLKL forward, 5′-TTCACCCATAAGCCAAGGAG-3′ and reverse,
5′-GGATCTCCTGCATGCATTTT-3′; and β-actin forward,
5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse,
5′-GCTGTCACCTTCACCGTTCC-3′.
Statistical analysis
Statistical analyses were performed with GraphPad
Prism 6 (GraphPad Software, Inc.) and SPSS Statistics 19.0 (IBM
Corp.). Student's paired t-test was used to compare the expression
levels of MLKL between gastric cancer and normal tissues. P<0.05
was considered to indicate a statistically significant difference.
Data are presented as the mean ± standard deviation.
Results
MLKL mRNA expression levels in human
cancers
To assess the MLKL mRNA expression levels in tumor
and normal tissues in multiple cancers, the Oncomine database was
used. The database contained 257 unique analyses and 504 unique
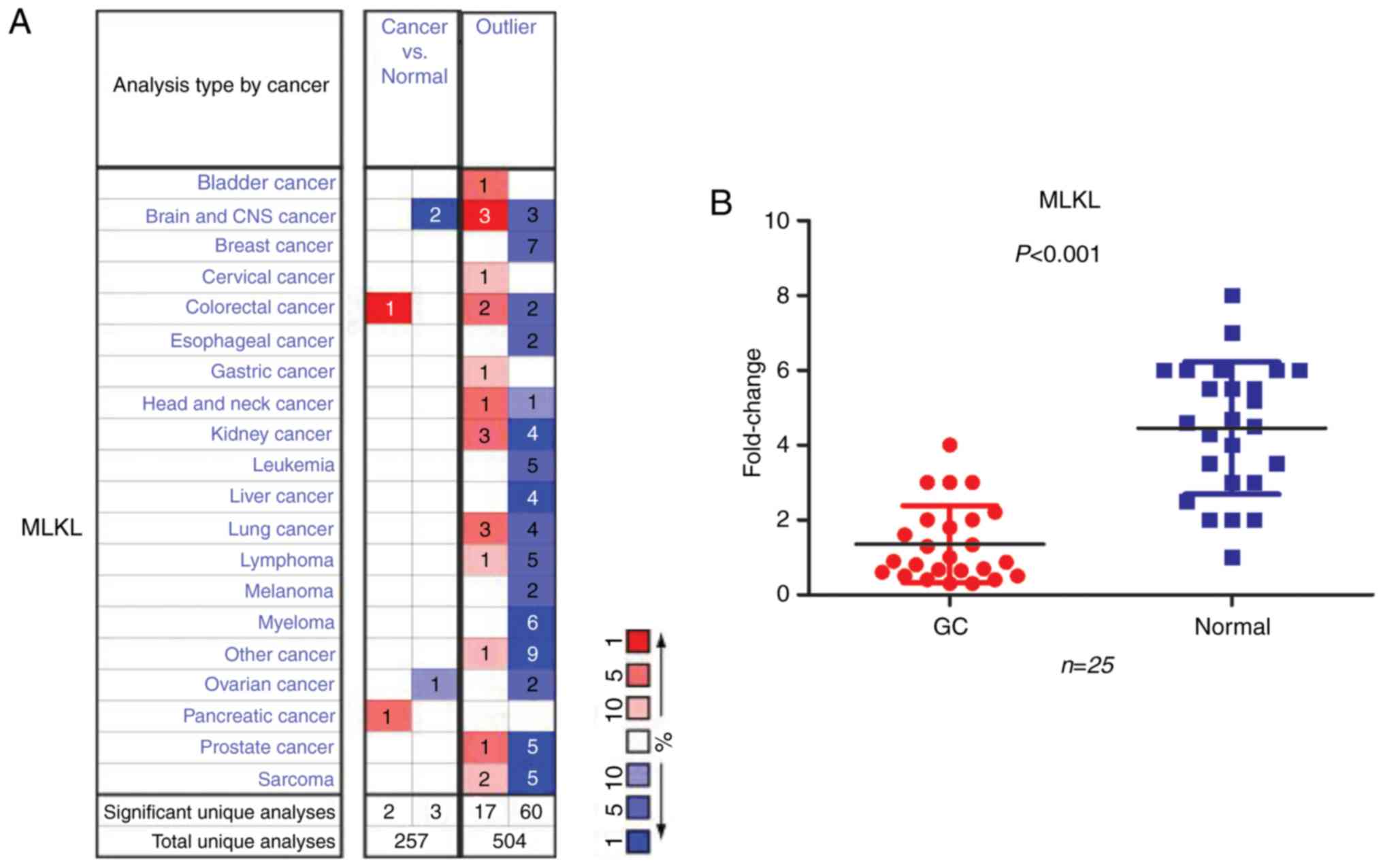
analyses without outliers (Fig. 1A).
In 77 of the 504 unique analyses without outliers, 67 revealed that
MLKL mRNA expression levels were lower in tumors compared with
normal tissues, whereas 20 analyses indicated an opposite result.
Only one study revealed an increased MLKL mRNA expression level in
GC. In addition, MLKL mRNA expression levels in GC and normal
tissues were determined in a cohort of 25 patients by RT-qPCR; the
results indicated that MLKL mRNA levels were significantly lower in
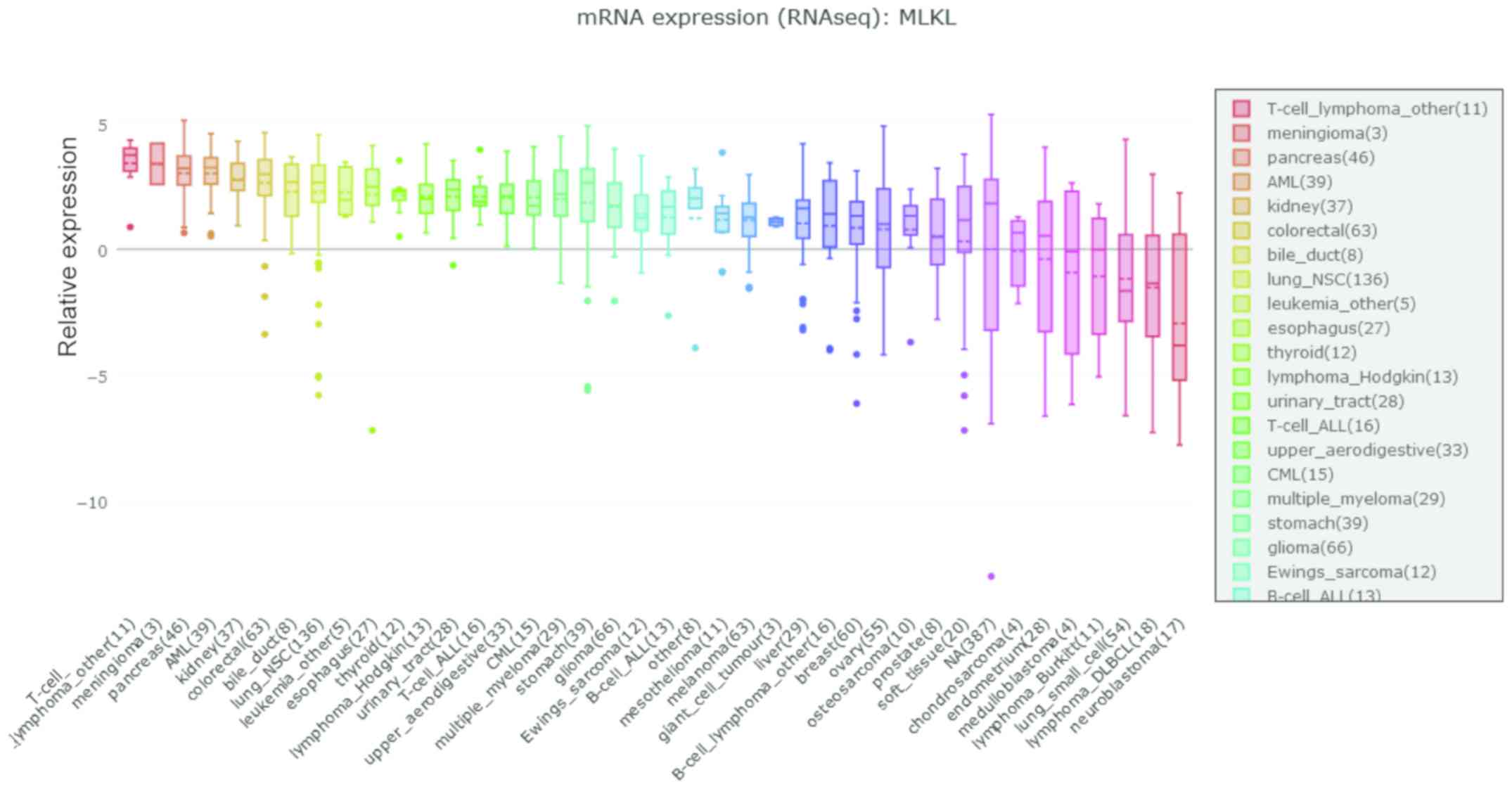
tumors compared with those in normal tissues (P<0.001; Fig. 1B). Additionally, CCLE analysis
demonstrated that MLKL was not upregulated in GC cell lines
(Fig. 2).
Prognostic effects of MLKL mRNA
expression in patients with GC
Patients with GC with high MLKL mRNA expression
levels exhibited improved OS (HR=0.80; P=0.045; Fig. 3A) and FP (HR=0.70, p=0.0036; Fig. 3B). In addition, the prognosis of
patients with high and low MLKL expression levels in different T
(depth of invasion), N (lymph node metastasis) and M (distant
metastasis) stages and in patients receiving surgical treatment was
evaluated. In T2 (OS, HR=0.56, P=0.0066; FP, HR=0.56, P=0.0066), N0
(OS, HR=0.36, P=0.011; FP, HR=0.37, P=0.013), N1+2+3 (OS, HR=0.7,
P=0.012; FP, HR=0.7, P=0.0055), M0 (OS, HR=0.7, P=0.011; FP,
HR=0.69, P=0.0055), M1 (OS, HR=0.42, P=0.016; FP, HR=0.43, P=0.026)
and patients receiving surgical treatment alone (OS, HR=0.69,
P=0.01; FP, HR=0.64, P=0.0013), high MLKL expression was
significantly associated with longer OS and longer FP compared with
patients with low MLKL expression. However, in T3, no significant
association between MLKL and the prognosis of patients with GC was
observed (OS, HR=0.77, P=0.14; FP, HR=0.79, P=0.16).
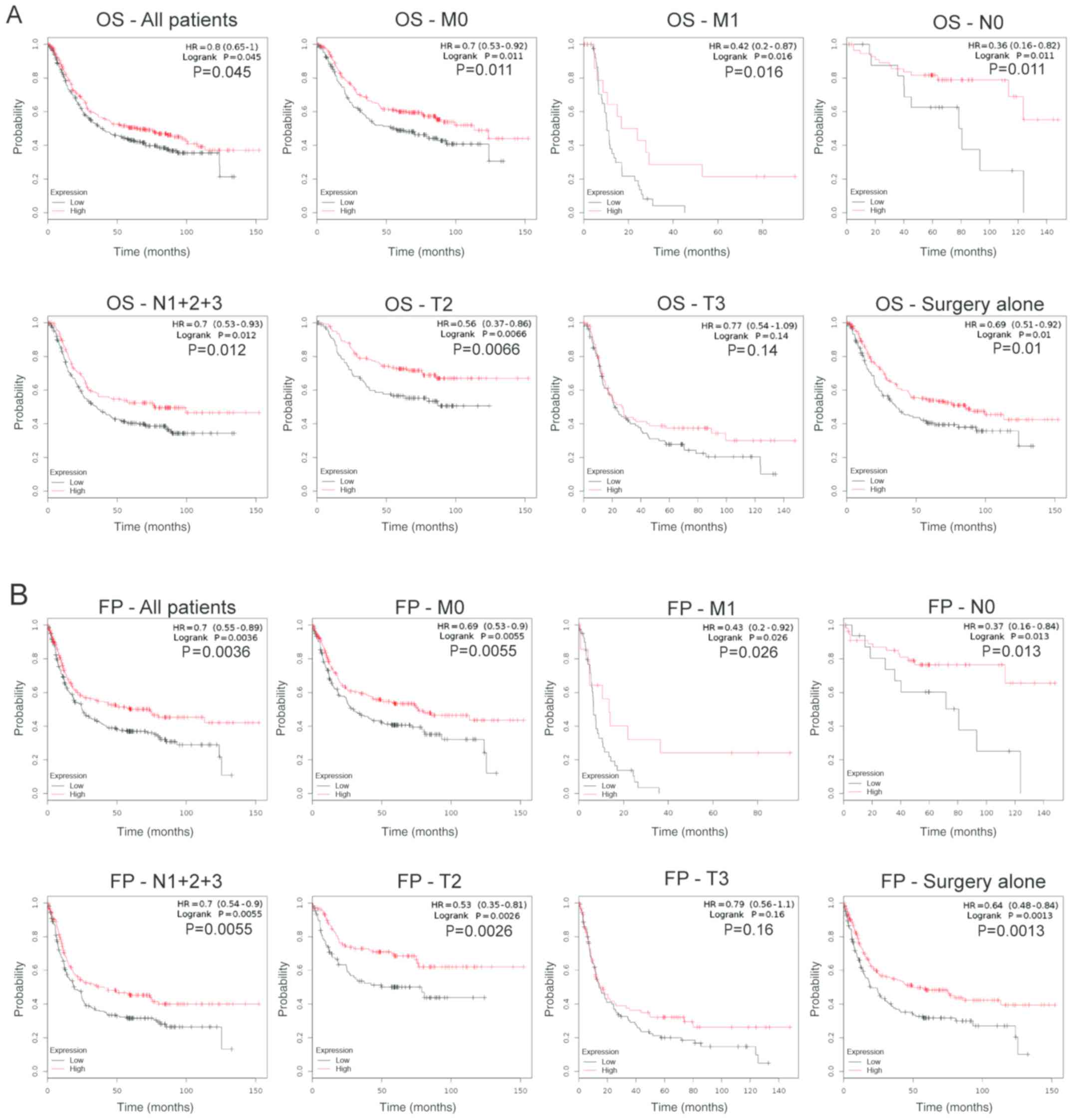 | Figure 3.Prognostic values of MLKL in patients
with GC. Decreased MLKL was related with poor OS (HR=0.80, P=0.045)
and FP (HR=0.70, P=0.0036) in patients with GC. MLKL, mixed lineage
kinase domain-like protein; GC, gastric cancer; OS, overall
survival; FP, first progression; M0, no distant metastasis; M1,
distant metastasis; N0, no lymph node metastasis; N1+2+3, lymph
node metastasis; T, depth of invasion. |
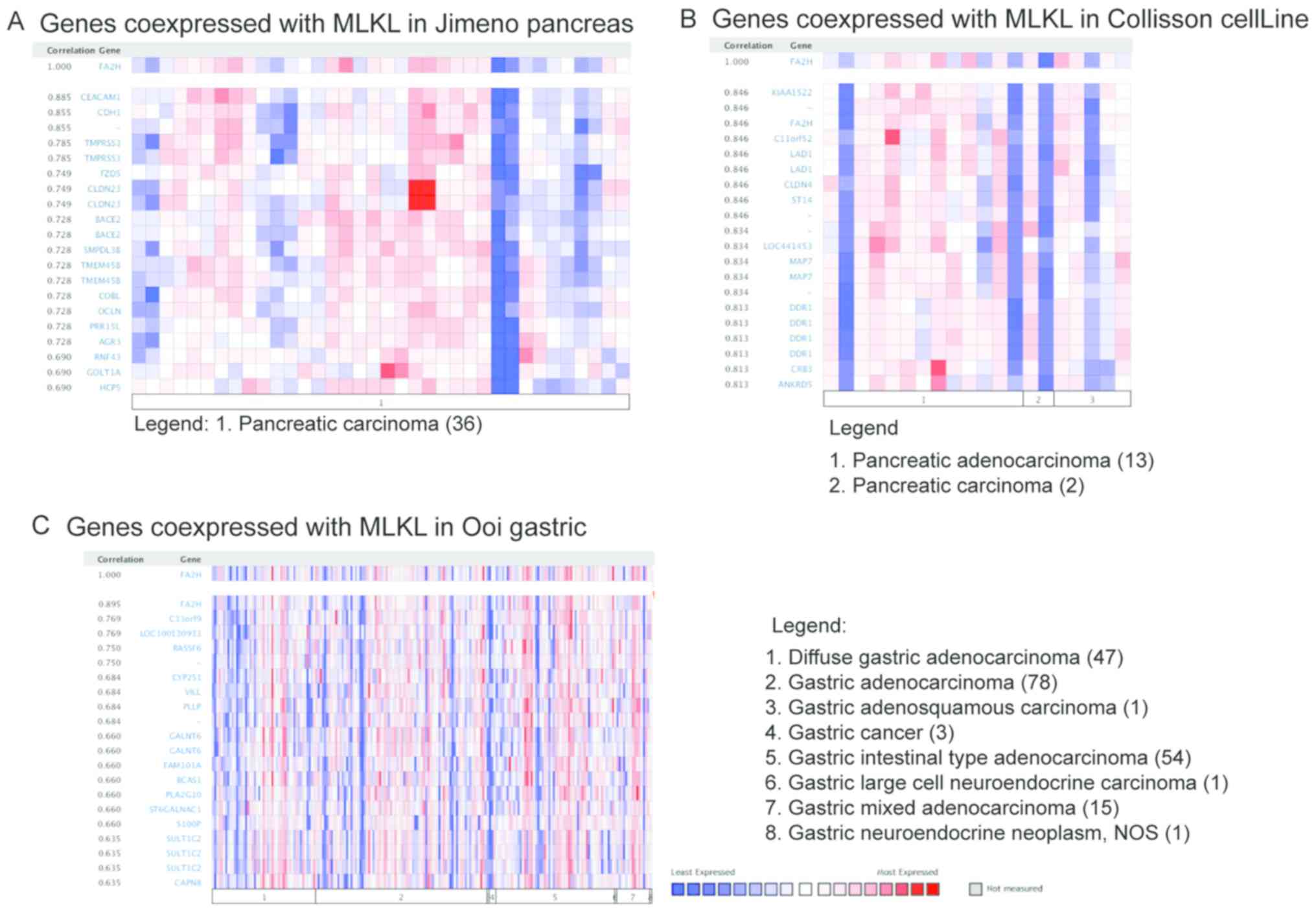
Co-expression analysis of MLKL and
FA2H in tumors
The co-expression of MLKL and other genes were
investigated by using Jimeno Pancreas, Ooi gastric and Collisson
CellLine from the Oncomine database. MLKL expression significantly
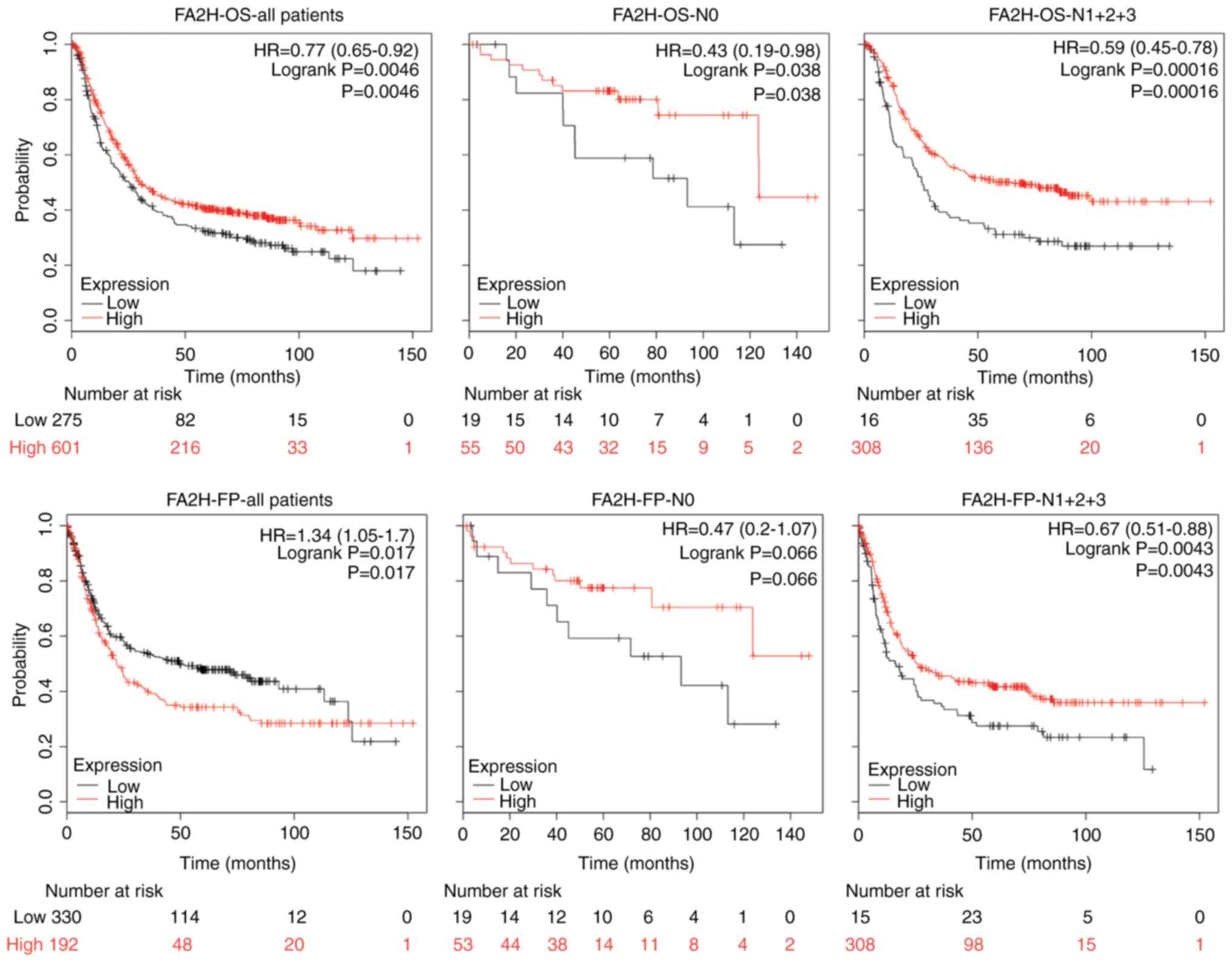
correlated with FA2H in a number of tumors (Fig. 4). In the survival analysis, high FA2H
expression was associated with good OS and poor FP of patients with
GC (OS, HR=0.77, P=0.0046; FP, HR=1.34, P= 0.017; Fig. 5). In patients with lymph node
metastasis, higher FA2H expression was associated with longer OS
and FP (OS, HR=0.59, P=0.00016; FP, HR=0.67, P=0.0043; Fig. 5). By contrast, FA2H had positive
effects on OS of patients with GC without lymph node metastasis
(HR=0.43, P=0.038; Fig. 5).
Therefore, FA2H and MLKL exhibited similar association with the
survival of patients with GC, which suggested that FA2H may be a
downstream molecule of MLKL.
Discussion
MLKL serves important roles in certain malignant
tumors, such as pancreatic cancer, ovarian cancer, colon cancer and
cervical squamous cell carcinoma (14–17). In
patients with cervical squamous cell carcinoma, MLKL expression was
increased in cancer tissue compared with normal cervical tissues
(P=0.004) and was negatively correlated with histological grade and
lymph node metastasis; low MLKL expression was also associated with
poor prognosis (14). Similar
conclusions were obtained from studies on early-stage resected
pancreatic adenocarcinoma, colon cancer and ovarian cancer
(15–17). The results of these studies indicated
that MLKL may be a potential prognostic biomarker for patients with
carcinoma.
In the present study, MLKL mRNA levels were
significantly decreased in GC tissues compared with those in normal
tissues. In addition, decreased MLKL was associated with poorer OS
and FP in patients with GC. These findings were consistent with
those of Ertao et al (21),
which demonstrated that loss of MLKL may be involved in GC
carcinogenesis and tumor progression, and MLKL may serve as a
useful prognosis predictor in patients with GC.
The underlying mechanisms of MLKL in the prognosis
of patients with GC are complicated. To the best of our knowledge,
phosphorylated MLKL promotes necroptosis, which is a
caspase-independent form of regulated cell death (22,23).
Necroptosis is mediated by the kinase activities of RIP1 and RIP3
and MLKL (24–28). Deubiquitylation of RIPK1 to RIPK3
induces RIPK3 oligomerization and activation, and the subsequent
activation of MLKL, which compromises the integrity of the plasma
membrane, leading to the release of intracellular proinflammatory
molecules (24). This process
underlies the immunogenic nature of necroptotic cancer cells and
their ability to induce efficient anti-tumor immunity. When MLKL
expression is decreased, cell necroptosis mediated by MLKL is also
reduced, leading to continuous proliferation of tumor cells
(25–28).
FA2H catalyzes the formation of 2-hydroxy fatty
acids, which are precursors for hFA-sphingolipids (29). FA2H is required for the synthesis of
hFA-galactolipids in myelin and hFA-ceramide and
hFA-glucosylceramide in the epidermis (29). However, the relationship between FA2H
and tumors remains unclear. In human cancer cell lines, including
Hela, COS-7, HepG2 and A549 (30),
FA2H exhibits positive effects on the efficacy of the anti-cancer
agent PM02734 (31). Another study
has demonstrated that Δ9-tetrahydrocannabinol induces FA2H
expression in human breast cancer MDA MB 231 cells through
peroxisome proliferator-activated receptor isoform-selective
agonists and antagonists (32). The
results of the present study demonstrated that MLKL expression is
significantly correlated with FA2H in certain tumors. In addition,
FA2H was associated with prognosis of patients with GC. However, no
previous studies have reported an association between FA2H and GC
or explored the role of FA2H in other tumors.
In conclusion, mRNA expression levels and prognostic
values of MLKL in GC were comprehensively analyzed; the results
demonstrated that MLKL mRNA expression levels were significantly
lower in GC compared with normal tissues, and that patients with GC
with low MLKL expression exhibited poorer prognosis compared with
patients with high MLKL expression. MLKL may be a prognostic factor
for GC; the mechanism of MLKL activity in GC may be through the
RIP1/RIP3/MLKL pathway, which participates in cancer cell necrosis
and other mechanisms that have not yet been clarified. Further
experiments are required to investigate the potential of anti-tumor
treatments that target MLKL. In Oncomine co-expression analysis,
MLKL expression was significantly correlated with FA2H in some
tumors. In addition, FA2H was associated with the overall survival
prognosis in patients with GC; the mechanism of FA2H affecting the
prognosis of patients with GC must be further explored.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the figshare repository (https://figshare.com/account/articles/8397182).
Authors' contributions
LS and TH designed the study and applied for
Research Ethics Board approval. WS recruited patients and collected
data. WY and LS analyzed data and prepared figures. WS wrote the
manuscript. All authors approved the final version of the
manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Review
Board of Suzhou University (register ID number: 2016958021). All
participants included in this study provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moore MA, Eser S, Igisinov N, Igisinov S,
Mohagheghi MA, Mousavi-Jarrahi A, Ozentürk G, Soipova M, Tuncer M
and Sobue T: Cancer epidemiology and control in North-Western and
Central Asia-past, present and future. Asian Pac J Cancer Prev. 11
(Suppl 2):S17–S32. 2010.
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Apicella M, Corso S and Giordano S:
Targeted therapies for gastric cancer: Failures and hopes from
clinical trials. Oncotarget. 8:57654–57669. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun L, Wang H, Wang Z, He S, Chen S, Liao
D, Wang L, Yan J, Liu W, Lei X and Wang X: Mixed lineage kinase
domain-like proteinmediates necrosis signaling downstream of RIP3
kinase. Cell. 148:213–227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q,
Luo J and Liu ZG: Mixed lineage kinase domain-like is a key
receptor interacting protein 3 downstream component of TNF-induced
necrosis. Proc Natl Acad Sci USA. 109:5322–5327. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vanlangenakker N, Vanden Berghe T and
Vandenabeele P: Many stimuli pull the necrotic trigger, an
overview. Cell Death Differ. 19:75–86. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tenev T, Bianchi K, Darding M, Broemer M,
Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K
and Meier P: The ripoptosome, a signaling platform that assembles
in response to genotoxic stress and loss of IAPs. Mol Cell.
43:432–448. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Coupienne I, Fettweis G and Piette J: RIP3
expression induces a death profile change in U2OS osteosarcoma
cells after 5-ALA-PDT. Lasers Surg Med. 43:557–564. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chu WM: Tumor necrosis factor. Cancer
Lett. 328:222–225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zong WX and Thompson CB: Necrotic death as
a cell fate. Genes Dev. 20:1–15. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Z, Han V and Han J: New components of
the necroptotic pathway. Protein Cell. 3:811–817. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paulsen RD, Soni DV, Wollman R, Hahn AT,
Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero
DE, et al: A genome-wide siRNA screen reveals diverse cellular
processes and pathways that mediate genome stability. Mol Cell.
35:228–239. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ruan J, Mei L, Zhu Q, Shi G and Wang H:
Mixed lineage kinase domain-like protein is a prognostic biomarker
for cervical squamous cell cancer. Int J Clin Exp Pathol.
8:150352015.PubMed/NCBI
|
|
15
|
He L, Peng K, Liu Y, Xiong J and Zhu FF:
Low expression of mixed lineage kinase domain-like protein is
associated with poor prognosis in ovarian cancer patients. Onco
Targets Ther. 6:1539–1543. 2013.PubMed/NCBI
|
|
16
|
Colbert LE, Fisher SB, Hardy CW, Hall WA,
Saka B, Shelton JW, Petrova AV, Warren MD, Pantazides BG, Gandhi K,
et al: Pronecrotic mixed lineage kinase domain-like protein
expression is a prognostic biomarker in patients with early-stage
resected pancreatic adenocarcinoma. Cancer. 119:3148–3155. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Guo J, Ding AP, Qi WW, Zhang PH, Lv
J, Qiu WS and Sun ZQ: Association of mixed lineage kinase
domain-like protein expression with prognosis in patients with
colon cancer. Technol Cancer Res Treat. 16:428–434. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ajani JA In H and Sano T: Stomach. AJCC
Cancer Staging Manual. Amin MB. 8th. Springer-Verlag; New York, NY:
2016
|
|
19
|
Xue Y, Ma G, Gu D, Zhu L, Hua Q, Du M, Chu
H, Tong N, Chen J, Zhang Z and Wang M: Genome-wide analysis of long
noncoding RNA signature in human colorectal cancer. Gene.
556:227–234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ertao Z, Jianhui C, Kang W, Zhijun Y, Hui
W, Chuangqi C, Changjiang Q, Sile C, Yulong H and Shirong C:
Prognostic value of mixed lineage kinase domain-like protein
expression in the survival of patients with gastric caner. Tumor
Biol. 37:13679–13685. 2016. View Article : Google Scholar
|
|
22
|
Vandenabeele P, Galluzzi L, Vanden Berghe
T and Kroemer G: Molecular mechanisms of necroptosis: An ordered
cellular explosion. Nat Rev Mol Cell Biol. 11:700–714. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krysko O, Aaes TL, Kagan VE, D'Herde K,
Bachert C, Leybaert L, Vandenabeele P and Krysko DV: Necroptotic
cell death in anti-cancer therapy. Immunol Rev. 280:207–219. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang H, Sun L, Su L, Rizo J, Liu L, Wang
LF, Wang FS and Wang X: Mixed lineage kinase domain-like protein
MLKL causes necrotic membrane disruption upon phosphorylation by
RIP3. Mol Cell. 54:133–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dondelinger Y, Declercq W, Montessuit S,
Roelandt R, Goncalves A, Bruggeman I, Hulpiau P, Weber K, Sehon CA,
Marquis RW, et al: MLKL compromises plasma membrane integrity by
binding to phosphatidylinositol phosphates. Cell Rep. 7:971–981.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu Y, Cui H, Gan H, Xia Y, Wang L, Wang Y
and Sun Y: Necroptosis mediated by receptor interaction protein
kinase 1 and 3 aggravates chronic kidney injury of subtotal
nephrectomised rats. Biochem Biophys Res Commun. 461:575–581. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moriwaki K and Chan FK: RIP3: A molecular
switch for necrosis and inflammation. Genes Dev. 27:1640–1649.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pasparakis M and Vandenabeele P:
Necroptosis and its role in inflammation. Nature. 517:311–320.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alderson NL, Rembiesa BM, Walla MD,
Bielawska A, Bielawski J and Hama H: The human FA2H gene encodes a
fatty acid 2-hydroxylase. J Biol Chem. 279:48562–48568. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alderson NL, Walla MD and Hama H: A novel
method for the measurement of in vitro fatty acid 2-hydroxylase
activity by gaschromatography-mass spectrometry. J Lipid Res.
46:1569–1575. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Herrero AB, Astudillo AM, Balboa MA,
Cuevas C, Balsinde J and Moreno S: Levels of SCS7/FA2H-mediated
fatty acid 2-hydroxylation determine the sensitivity of cells to
antitumor PM02734. Cancer Res. 68:9779–9787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takeda S, Ikeda E, Su S, Harada M, Okazaki
H, Yoshioka Y, Nishimura H, Ishii H, Kakizoe K, Taniguchi A, et al:
Δ(9)-THC modulation of fatty acid 2-hydroxylase (FA2H) gene
expression: Possible involvement of induced levels of PPARα in
MDA-MB-231 breast cancer cells. Toxicology. 326:18–24. 2014.
View Article : Google Scholar : PubMed/NCBI
|