Introduction
As a type of cancer that originates from the
prostate, a gland of the male reproductive system, prostate
carcinoma is one of the most frequently diagnosed and leading
causes of cancer associated-mortality in males (1). The early stages of prostate carcinoma
development lack classic cancer symptoms, leading to a high
prevalence of tumor metastasis by the time of diagnosis (2). However, patients in the advanced stages
of disease exhibit difficulty in urinating, pain in the pelvis and
back when urinating, in addition to blood in the urine (3). In spite of efforts to improve treatment
of the disease, the mortality rate of prostate carcinoma remains
high (4,5).
Due to the abnormally accelerated proliferation of
cancer cells, which requires increased energy input, glucose
metabolism is considered to be a promising therapeutic target for
cancer therapy (6,7). In mammalian cells, glucose transporter
1 (GLUT-1) mediates the transport of glucose across the plasma
membrane (8). GLUT-1 is reportedly
overexpressed in the majority of, if not all human cancers
(including prostate carcinoma) and the iinhibition of GLUT-1
resulted in the inhibited growth of cancer cells (9). A growing body of literature has
revealed that GLUT-1 may participate in the development and
progression of cancer by interacting with long non-coding RNAs
(lncRNAs) (10), a group of
non-coding RNA transcripts >200 nucleotides, associated with
cancer biology (11). Despite a
recent report of the reduced expression of lncRNA MORT in 16 types
of cancer (12), the functionality
of lncRNA MORT in cancer remains unknown. The present study aimed
to investigate the involvement of lncRNA MORT in prostate
carcinoma, and its potential interaction with glucose uptake
pathways.
Materials and methods
Human specimens and cell lines
Tumor and adjacent healthy tissue specimens were
obtained from 60 male patients with prostate carcinoma who were
admitted to the People's Hospital of Xinjiang Uyghur Autonomous
Region (Urumqi, China) between February 2016 and March 2018. The
inclusion criteria were as follows: i) Patients were diagnosed by
pathological biopsy; ii) patients had complete medical records; and
iii) patients were willing to cooperate with researchers, and gave
written informed consent. Exclusion criteria: i) Patients with
other diseases; ii) patients who could not understand the
experimental protocol; and iii) patients had received treatment
within 3 months of admission. The patient age range was between
34–70 years (mean, 49.3±6.1 years). There were 10 cases of stage I,
12 cases of stage II, 18 cases of stage III and 20 cases of stage
IV cancer. Tumor sizes were determined using magnetic resonance
imaging (GE Signa HDxt 1.5T, GE, USA). The present study was
approved by the Ethics committee of the People's Hospital of
Xinjiang Uyghur Autonomous Region, and all participants gave
written informed consent to participate.
Cells of the human prostate carcinoma cell line
22Rv1 were purchased from the American Type Culture Collection
(ATCC, Manassas, VA, USA), and cultured in Dulbecco's Modified
Eagle's medium supplemented with 10% fetal bovine serum (cat. no.
30–2020; ATCC) at 37°C, 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was used to extract total RNA
from the tissue specimens and cells. Tissues retrieved from liquid
nitrogen were ground up prior to the addition of TRIzol reagent,
whilst 22Rv1 cells were directly mixed with the reagent to extract
total RNA. Following reverse transcription using the High-Capacity
cDNA Reverse Transcription kit (Applied Biosystems™; Thermo Fisher
Scientific, Inc.), PCR reactions were prepared using the
SYBR® Green Quantitative RT-qPCR kit (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany). CFX96 Touch Real-Time PCR
Detection system (Bio-Rad Laboratories, Inc.) was used to perform
all qPCR reactions. Three replicates were set for each reaction. RT
and PCR steps were conducted according to the manufacturer's
protocol. Primers for lncRNA MORT and β-actin were designed and
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The
primer sequences were as follows: lncRNA MORT forward,
5′-GTTGTAGCATGCATCAGAAC-3′, and reverse,
5′-CAGGACACCCAACAGCCCAA-3′; GLUT-1 forward,
5′-CAGTTCGGCTATAACACTGGT-3′, and reverse, 5′-GCCCCCGACAGAGAAGAT-3′;
β-actin forward, 5′-GACCTCTATGCCAACACAGT-3′, and reverse,
5′-AGTACTTGCGCTCAGGAGGA-3′. Thermocycling conditions were as
follows: 95°C for 1 min, and then 40 cycles of 95°C for 10 sec,
55°C for 30 sec and 72°C for 30 sec. The expression level of lncRNA
MORT was normalized to that of the endogenous β-actin control using
the 2−ΔΔCq method (13).
Transfection
The pSF-MinCMV-KrYFP vector expressing lncRNA MORT
or GLUT-1, and the empty control vector were designed and
synthesized by Sangon Biotech Co., Ltd. The 22Rv1 cell line was
cultivated overnight to reach 70–80% confluence. Lipofectamine
2000® (cat. no. 11668-019, Invitrogen; Thermo Fisher
Scientific, Inc.) was used to transfect 10 nM vector into
5×105 cells/sample. Negative control cells were
transfected with empty vector, and control cells were treated with
Lipofectamine 2000 only. In cases of co-transfection, 10 nM MORT
and 10 nM GLUT-1 vectors were transfected at the same time.
Following experiments were performed at 24 h post-transfection.
Glucose uptake assay
Cell glucose uptake was assessed only in cases in
which the overexpression rate of lncRNA MORT reached 200%
(determined using RT-qPCR). Briefly, to initiate glucose uptake,
22Rv1 cells (3×105 cells harvested at 24 h
post-transfection) were washed with PBS, and incubated with 10 ml
Krebs-Ringer-HEPES (KRH) buffer (120 mM NaCl, 1.2 mM
MgSO4, 25 mM HEPES pH 7.4, 1.3 mM CaCl2, 1.3
mM KH2PO4 and 5 mM KCl) containing 1 µCi of
[3H]-2-deoxyglucose at 37°C for 30 min. Cells were
subsequently washed with ice-cold KRH buffer to terminate glucose
uptake, and lysed (10 mM Tris-HCl, pH 8.0, 0.2% SDS) prior to the
measurement of radioactivity using liquid scintillation
spectrometry using 1220 QUANTULUS Ultra Low Level Liquid
Scintillation Spectrometer (PerkinElmer, Inc.). Glucose uptake was
presented as disintegrations per minute (DPM).
Cell proliferation assay
Cell proliferation was detected only in cases in
which the overexpression rate of lncRNA MORT reached 200%, 24 h
post-transfection. Briefly, cells were collected 24 h
post-transfection, and single 0.1 ml cell suspensions per well
(4×104 cells/ml) were transferred to a 96-well plate.
Cells were cultured at 37°C and 10 µl Cell Counting Kit-8 solution
(Sigma-Aldrich; Merck KGaA) was added at 24, 48, 72 and 96 h. Cells
were cultured for an additional 4 h at 37°C. Dimethyl sulfoxide (10
µl) was added to each well and optical density values were obtained
at 450 nm using CLARIOstar Plus Plate Reader (BMG Labtech,
Ltd.).
Western blot analysis
Following a 24-h transfection period,
radioimmunoassay buffer (Thermo Fisher Scientific, Inc.) was used
to extract the total protein from in vitro cultured cells.
Protein concentrations were determined using a bicinchoninic acid
assay. Proteins were denatured and separated using 12% SDS-PAGE
with 30 µg/lane. Following protein transfer to PVDF membranes, the
membranes were blocked using 5% non-fat milk for 1 h at room
temperature, followed by incubation with the following primary
antibodies: Rabbit anti-human GLUT-1 (1:1,200; cat. no. ab32551,
Abcam, Cambridge, UK) and GAPDH (1:1,000; ab8245, Abcam) at 4°C for
18 h. The membranes were washed and subsequently incubated with
goat anti-rabbit immunoglobulin G-horseradish peroxidase-conjugated
secondary antibody (1:1,000, MBS435036, MyBioSource, Inc., San
Diego, CA, USA) at 24°C for 2 h. Signals were developed using an
enhanced chemiluminescence detection reagent (Sigma-Aldrich; Merck
KGaA), and the data were normalized using ImageJ v1.8.0 software
(National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All experiments were performed in triplicate, and
the data are presented as the mean ± standard deviation. All
statistical analyses were performed using SPSS software (version
19.0; IBM Corp., Armonk, NY, USA). For lncRNA MORT and GLUT-1
expression, data were normalized to the sample with the lowest
value, which was set to 1. For cell proliferation, the data were
normalized to the control group, which was set to 100. Comparisons
between lncRNA MORT expression levels in tumor tissues and adjacent
healthy tissues were determined using a paired Student's t-test.
Comparisons among three groups were performed using one-way
analysis of variance followed by a Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
lncRNA MORT is downregulated in tumor
tissues, compared with the adjacent healthy tissues of patients
with prostate carcinoma
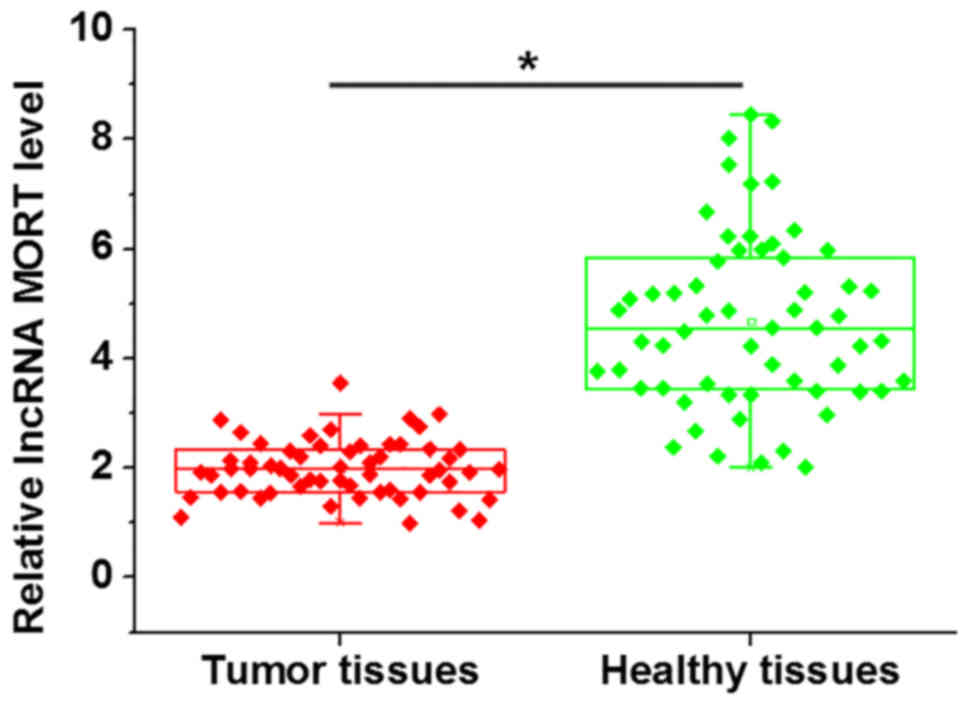
The expression levels of lncRNA MORT in the tumor
tissues of patients with prostate carcinoma were detected using
RT-qPCR. Compared with adjacent healthy tissues, expression levels
of lncRNA MORT were significantly downregulated in tumor tissues
(Fig. 1). In addition, no
significant differences in lncRNA MORT expression were observed
between patients at different cancer stages (data not shown).
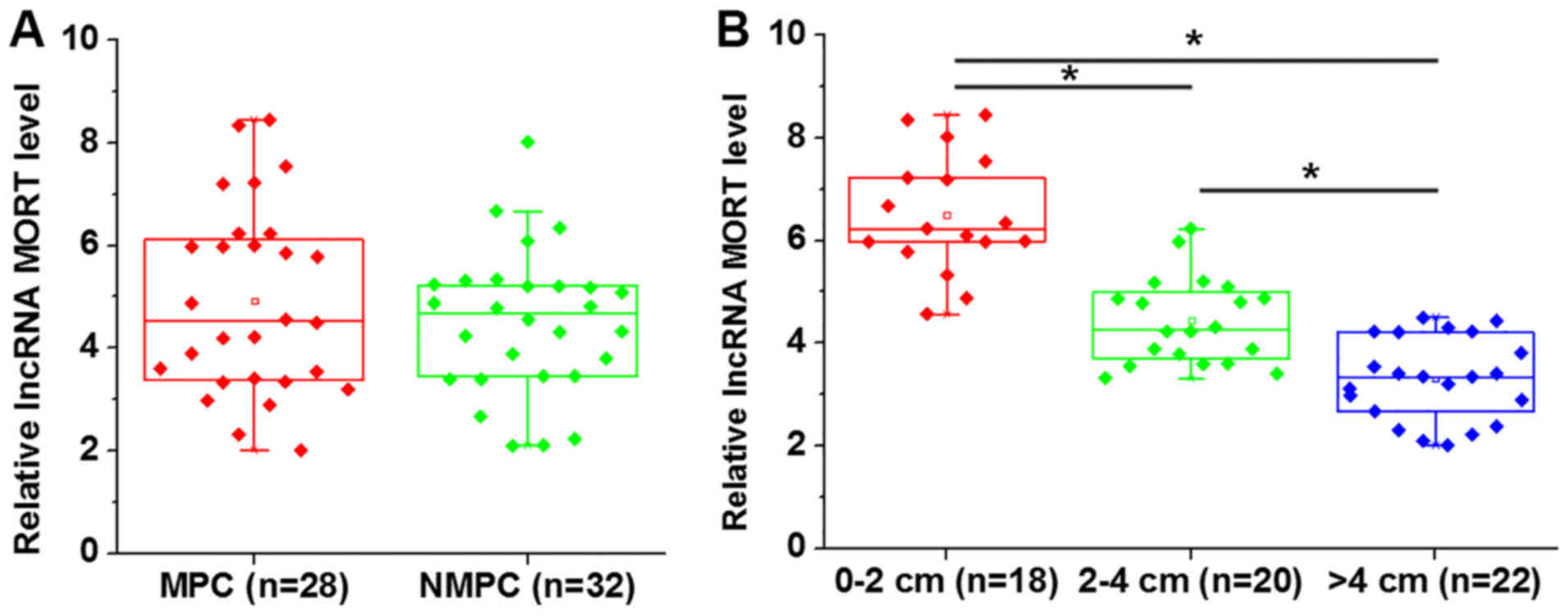
Expression of lncRNA MORT in tumor
tissues is influenced by tumor size, but not tumor metastasis
Among the 60 patients with prostate carcinoma, there
were 28 cases of metastatic prostate carcinoma (MPC) and 32 cases
of non-metastatic prostate carcinoma (NMPC). No significant
differences in the expression level of lncRNA MORT in tumor tissues
were revealed between the MPC and NMPC groups (Fig. 2A). There were 18 patients with a
primary tumor of 0–2 cm, 20 cases between 2–4 cm and 22 cases >4
cm. A decrease in the level of lncRNA MORT expression was
associated with an increase in tumor size (Fig. 2B; P<0.05).
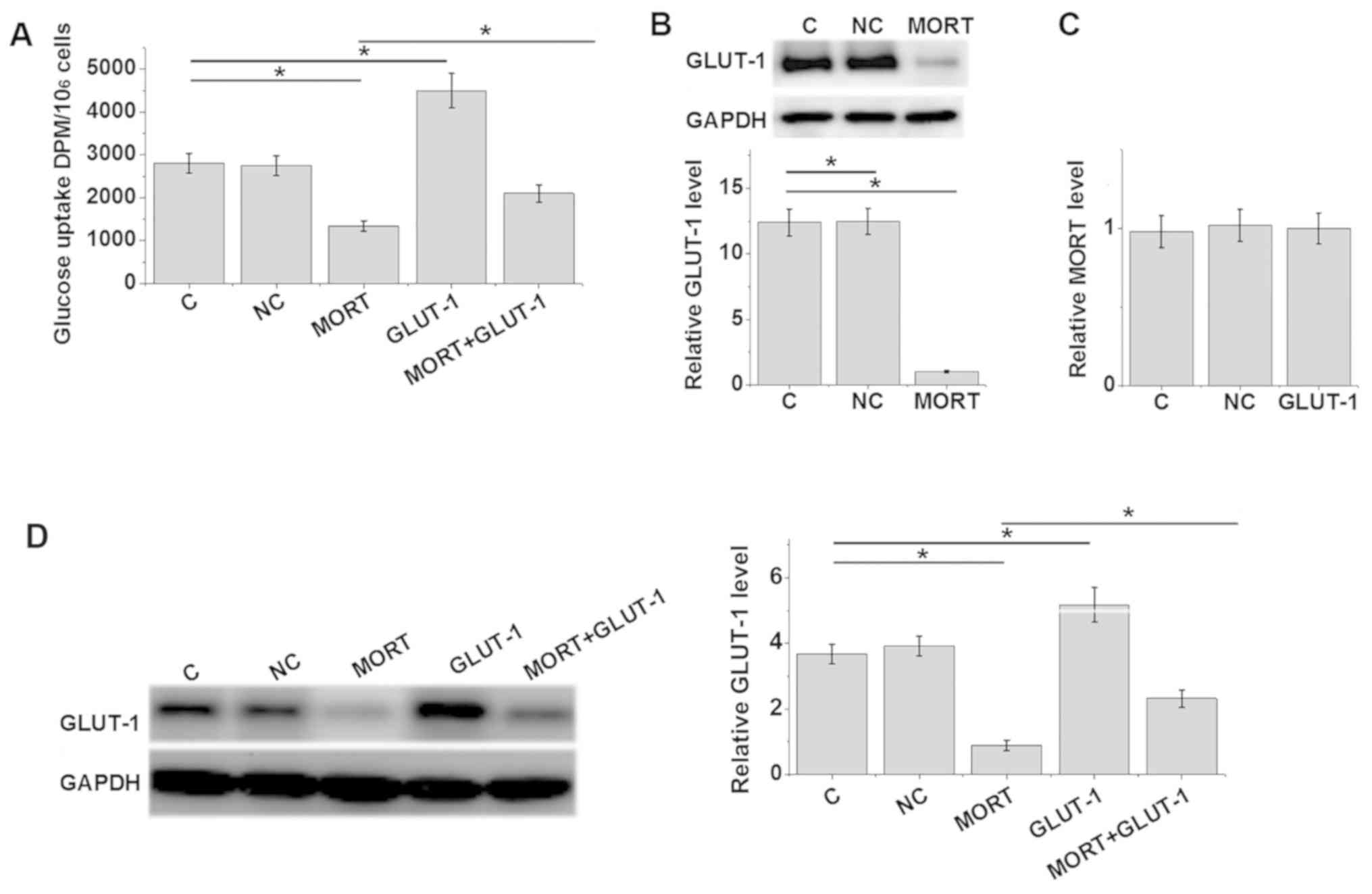
Overexpression of lncRNA MORT inhibits
glucose uptake and GLUT-1 expression in the prostate carcinoma cell
line 22Rv1
The overexpression rate of lncRNA MORT and GLUT-1
reached 200 and 300% 24 h post-transfection, respectively (Fig. S1). Compared with the control and
negative control groups, lncRNA MORT overexpression resulted in
significantly reduced glucose uptake (Fig. 3A) and inhibited GLUT-1 expression
(Fig. 3B) (P<0.05). Furthermore,
GLUT-1 overexpression promoted glucose uptake, and attenuated the
effects of lncRNA MORT overexpression on glucose uptake (Fig. 3A; P<0.05). However, GLUT-1
overexpression did not significantly effect lncRNA MORT expression
(Fig. 3C; P>0.05). Glucose uptake
levels were consistent with GLUT-1 protein levels (Fig. 3D).
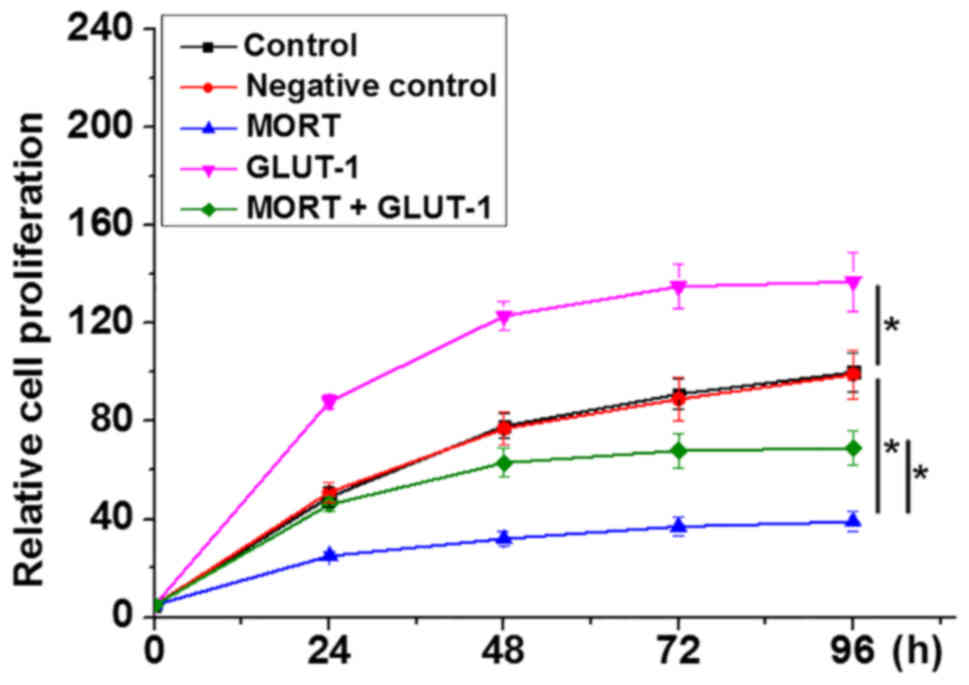
Overexpression of lncRNA MORT inhibits
cell proliferation by suppressing GLUT-1
Compared with the control and negative control
groups, overexpression of lncRNA MORT significantly inhibited,
whilst GLUT-1 overexpression promoted the proliferation of 22Rv1
cells (Fig. 4; P<0.05). In
addition, GLUT-1 overexpression attenuated the effects of lncRNA
MORT on cell proliferation (P<0.05).
Discussion
Glucose metabolism is critical for the development
and progression of all types of cancers. The present study
indicated that lncRNA MORT, a recently identified lncRNA which is
silenced in 16 types of cancer (12), is downregulated in prostate
carcinoma. In addition, lncRNA MORT may inhibit the proliferation
of prostate carcinoma cells by inhibiting glucose uptake.
The development and progression of prostate
carcinoma influences the expression of a large set of lncRNAs, a
number of which have reported diagnostic or prognostic value
(14,15). In the present study, downregulated
expression levels of lncRNA MORT were observed in prostate
carcinoma tissues compared with adjacent healthy tissues. This
supported preliminary microarray data revealing that lncRNA MORT
was downregulated in prostate carcinoma tissues (data not shown).
Notably, it was also revealed that the expression level of lncRNA
MORT was effected by tumor size, but not tumor metastasis,
indicating the potential involvement of lncRNA MORT in prostate
tumor growth. This was further confirmed by the in vitro
proliferation assay.
GLUT-1-mediated glucose uptake is associated with
the abnormally accelerated proliferation of cancer cells (16,17).
Consistently, the present study revealed increased glucose uptake
and prostate carcinoma cell proliferation following GLUT-1
overexpression. It is frequently been observed that glucose
metabolism in human cancers is regulated by lncRNAs (18,19). We
reported significantly reduced glucose uptake and inhibited GLUT-1
expression in prostate carcinoma cells following lncRNA MORT
overexpression. However, GLUT-1 overexpression failed to regulate
lncRNA MORT. Therefore, lncRNA MORT may serve as a tumor suppressor
in prostate carcinoma by inhibiting glucose uptake through the
downregulation of GLUT-1. However, GLUT-1 mRNA levels were not
significantly altered following lncRNA MORT overexpression (data
not shown), suggesting that lncRNAs MORT may affect the level of
GLUT-1 by regulating protein accumulation, rather than gene
transcription.
It is worth noting that lncRNA MORT overexpression
has not been reported to significantly affect any of the other
GLUTs. Therefore, lncRNA MORT may specifically regulate GLUT-1 in
prostate carcinoma. Due to limited resources, the present study
only included one prostate cancer cell line. Future studies aim to
include more cell lines to further support the conclusions drawn.
The present study was also limited by a small sample population
size. Future studies with a larger patient cohort are required to
further verify the conclusions. Future studies will also focus on
the diagnostic, prognostic and therapeutic value of lncRNA MORT in
prostate carcinoma, which requires long-term follow-up studies in
addition to an increased sample population.
In conclusion, lncRNA MORT was determined to be
downregulated in prostate carcinoma and may inhibit tumor cell
proliferation by suppressing glucose uptake. Therefore, lncRNA MORT
may serve as a potential therapeutic target for prostate
carcinoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Xinjiang Natural
Science Foundation Funded Project (grant no. 2016D01C126), Xinjiang
‘Tianshan Youth Project’ 2017.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZFS and FG conducted all of the experiments,
analyzed all data and contributed to manuscript writing. DYJ, JXH,
JC and MS also conducted experiments. FBQ was responsible for
clinical studies of prostate cancer and clinical sample collection.
CYL was responsible for experimental design and molecular studies.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the People's
Hospital of Xinjiang Uyghur Autonomous Region, and all participants
gave written informed consent.
Patient consent for publication
The study followed the tenets of the Declaration of
Helsinki, and informed written consent was obtained from all
patients and controls when the nature and possible consequences of
the study were explained.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gundem G, Van Loo P, Kremeyer B,
Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML,
Högnäs G, Annala M, et al: The evolutionary history of lethal
metastatic prostate cancer. Nature. 520:353–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
US Preventive Services Task Force, ;
Grossman DC, Curry SJ, Owens DK, Bibbins-Domingo K, Caughey AB,
Davidson KW, Doubeni CA, Ebell M, Epling JW Jr, et al: Screening
for prostate cancer: US preventive services task force
recommendation statement. JAMA. 319:1901–1913. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheng L, Zincke H, Blute ML, Bergstralh
EJ, Scherer B and Bostwick DG: Risk of prostate carcinoma death in
patients with lymph node metastasis. Cancer. 91:66–73. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fossa SD, Wiklund F, Klepp O, Angelsen A,
Solberg A, Damber JE, Hoyer M and Widmark A; The Scandinavian
Prostate Cancer Group-7 Investigators, : Ten- and 15-yr prostate
cancer-specific mortality in patients with nonmetastatic locally
advanced or aggressive intermediate prostate cancer, randomized to
lifelong endocrine treatment alone or combined with radiotherapy:
Final results of the scandinavian prostate cancer Group-7. Eur
Urol. 70:684–691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hay N: Reprogramming glucose metabolism in
cancer: Can it be exploited for cancer therapy? Nat Rev Cancer.
16:635–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Semenza GL: Hypoxia-inducible factors:
Coupling glucose metabolism and redox regulation with induction of
the breast cancer stem cell phenotype. EMBO J. 36:252–259. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Olson AL and Pessin JE: Structure,
function, and regulation of the mammalian facilitative glucose
transporter gene family. Annu Rev Nutr. 16:235–256. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Cao Y, Zhang W, Bergmeier S, Qian
Y, Akbar H, Colvin R, Ding J, Tong L, Wu S, et al: A small-molecule
inhibitor of glucose transporter 1 downregulates glycolysis,
induces cell-cycle arrest, and inhibits cancer cell growth in vitro
and in vivo. Mol Cancer Ther. 11:1672–1682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X and Gan B: lncRNA NBR2 modulates
cancer cell sensitivity to phenformin through GLUT1. Cell Cycle.
15:3471–3481. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long Non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vrba L and Futscher BW: Epigenetic
silencing of lncRNA MORT in 16 TCGA cancer types. F1000Res.
7:2112018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee B, Mazar J, Aftab MN, Qi F, Shelley J,
Li JL, Govindarajan S, Valerio F, Rivera I, Thurn T, et al: Long
noncoding RNAs as putative biomarkers for prostate cancer
detection. J Mol Diagn. 16:615–626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wan X, Huang W, Yang S, Zhang Y, Pu H, Fu
F, Huang Y, Wu H, Li T and Li Y: Identification of
androgen-responsive lncRNAs as diagnostic and prognostic markers
for prostate cancer. Oncotarget. 7:60503–60518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sawayama H, Ishimoto T, Watanabe M,
Yoshida N, Baba Y, Sugihara H, Izumi D, Kurashige J and Baba H:
High expression of glucose transporter 1 on primary lesions of
esophageal squamous cell carcinoma is associated with hematogenous
recurrence. Ann Surg Oncol. 21:1756–1762. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gwak H, Haegeman G, Tsang BK and Song YS:
Cancer-specific interruption of glucose metabolism by resveratrol
is mediated through inhibition of Akt/GLUT1 axis in ovarian cancer
cells. Mol Carcinog. 54:1529–1540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng X, Han H, Liu GP, Ma YX, Pan RL,
Sang LJ, Li RH, Yang LJ, Marks JR, Wang W and Lin A: lncRNA wires
up Hippo and Hedgehog signaling to reprogramme glucose metabolism.
EMBO J. 36:3325–3335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun LY, Li XJ, Sun YM, Huang W, Fang K,
Han C, Chen ZH, Luo XQ, Chen YQ and Wang WT: lncRNA ANRIL regulates
AML development through modulating the glucose metabolism pathway
of AdipoR1/AMPK/SIRT1. Mol Cancer. 17:1272018. View Article : Google Scholar : PubMed/NCBI
|