Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies and is emerging as the second leading cause of
cancer-associated mortality worldwide (1). Numerous factors, including inherited
genetic factors, chronic viral infection, non-alcoholic fatty liver
disease, tobacco consumption and chronic alcohol abuse, contribute
to the development and progression of HCC (2,3).
However, to the best of our knowledge, the precise molecular
mechanisms that induce hepatocarcinogenesis remain unknown. Despite
improvements in therapeutic strategies, including surgical
resection, radiotherapy, chemotherapy, adjunctive therapy and liver
transplantation, the survival rate of HCC remains low due to high
rates of recurrence and metastasis (4–6).
Therefore, there is an urgent requirement to understand the
detailed molecular mechanisms that underlie hepatocarcinogenesis
and identify novel targets for the development of HCC
treatments.
MicroRNAs (miRNAs/miRs) are a group of endogenous,
small, non-coding and single-stranded RNAs, consisting of ~22
nucleotides, which have emerged as novel regulators of gene
expression (7). miRNAs modulate gene
expression by binding to complementary sequences within the
3′-untranslated region (3′-UTR) of mRNAs, which leads to mRNA
degradation or translational repression (8). miRNAs are involved in the development
of disease by regulating cell proliferation, apoptosis and
differentiation (9). Altered
expression levels of miRNAs have been identified in numerous types
of cancer and these dysregulated miRNAs can contribute to
carcinogenesis by functioning as oncogenes or tumor suppressors
(10,11). A number of studies have suggested
that various miRNAs are involved in the progression of HCC and are
potential therapeutic targets and prognostic biomarkers (12–15).
However, to the best of our knowledge, the precise role of miRNAs
in HCC remains largely unknown. Therefore, miRNA-mediated molecular
mechanisms that affect HCC development and progression should be
further investigated.
Cell-cycle-related and expression-elevated protein
in tumor (CREPT), also termed the regulation of nuclear pre-mRNA
domain containing 1B gene, has previously been identified as a
potential oncogene in various types of cancer (16). The CREPT gene is located on human
chromosome 20, which is a highly amplified region in numerous types
of cancer (17,18). CREPT encodes a protein of 326 amino
acids, which contains a regulation of nuclear pre-mRNA domain and
is highly conserved across species (16). CREPT mRNA and protein have been
revealed to be highly expressed in numerous types of clinical
cancer tissues and cancer cell lines, including lung, liver,
breast, prostate, stomach, colon, uterus endometrium and uterine
cervical cancer (16,19). A high expression level of CREPT is
correlated with tumor stage, metastasis and a poor survival rate
(19–21). Previous studies have demonstrated
that CREPT promotes tumor growth in vivo and in vitro
by accelerating cell growth and cell cycle progression (22,23).
Therefore, CREPT may serve as a potential and promising target for
the development of anticancer treatments.
Previous studies have reported that miR-300 is
aberrantly expressed in multiple types of human cancer and serves
an important role in tumor progression (24–26).
However, there is limited understanding regarding the role of
miR-300 in HCC. The present study aimed to investigate the
expression, biological function and regulatory mechanism of miR-300
in liver cancer. It was demonstrated that miR-300 expression was
significantly decreased in HCC tissues and cell lines. Functional
experiments revealed that miR-300 can regulate the proliferation,
colony formation and cell cycle progression of liver cancer cells
in vitro. Notably, CREPT was identified as a target gene of
miR-300. In addition, it was revealed that miR-300 can regulate
CREPT expression and the Wnt/β-catenin signaling pathway in HCC
cells. Restoration of CREPT expression partially reversed the
antitumor effect of miR-300. In conclusion, the present results
demonstrate that miR-300 inhibits the growth of HCC cells by
targeting CREPT, which may provide a novel miRNA target for HCC
treatment.
Materials and methods
Collection of clinical specimens
Hepatocellular carcinoma (HCC) tissue samples (n=20)
and adjacent non-tumor tissue samples (n=20) were obtained from The
Affiliated Hospital of Changchun University of Chinese Medicine
(Changchun, China). All HCC samples were obtained from patients
with HCC who underwent radical surgical resection without
preoperative chemotherapy or radiotherapy between May 2015 and Dec,
2017 Collected tissue samples were immediately frozen in liquid
nitrogen and stored at −80°C until needed. Patient characteristics
are listed in Table I. Written
informed consent was obtained from all patients. The present study
was approved by the Institutional Human Experiment and Ethics
Committee of Changchun University of Chinese Medicine (Changchun,
China) and performed in accordance with the Declaration of
Helsinki.
 | Table I.Associations between miR-300
expression and the clinicopathological variables of hepatocellular
carcinoma. |
Table I.
Associations between miR-300
expression and the clinicopathological variables of hepatocellular
carcinoma.
| Parameter | n | Relative miR-300
expression | P-value |
|---|
| Age, years |
|
| 0.221 |
|
>50 | 11 | 0.25±0.08 |
|
|
≤50 | 9 | 0.20±0.06 |
|
| Gender |
|
| 0.331 |
|
Male | 10 | 0.21±0.06 |
|
|
Female | 10 | 0.23±0.07 |
|
| Histological
differentiation |
|
| 0.404 |
|
Low/no | 10 | 0.19±0.06 |
|
|
Moderate/high | 10 | 0.25±0.07 |
|
| Lymph node
metastasis |
|
| 0.471 |
|
Yes | 10 | 0.24±0.07 |
|
| No | 10 | 0.21±0.06 |
|
| TNM stage |
|
| 0.673 |
| I/II
stage | 10 | 0.21±0.07 |
|
| III/IV
stage | 10 | 0.23±0.07 |
|
Culture of cell lines
The human liver cancer cell lines HepG2, Hep3B and
Huh-7, and the 293T cell line were provided by the Chinese Academy
of Sciences. The normal liver cell line HL-7702 was purchased from
the Bena Culture Collection. Cells were routinely cultured,
according to the manufacturers' protocols. Briefly, HepG2, Hep3B,
Huh-7 and 293T cells were cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS; Sigma-Aldrich; Merck KGaA) and 1%
penicillin/streptomycin mix (Sigma-Aldrich; Merck KGaA). HL-7702
cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS and 1% penicillin/streptomycin
mix (Sigma-Aldrich; Merck KGaA). Cells were maintained in a
humidified incubator (Thermo Fisher Scientific, Inc.) with 5%
CO2 and a temperature of 37°C. Cell line authentication
was performed using STR profiling.
Cell transfection
The miR-300 mimic and miR-300 inhibitor were
purchased from Thermo Fisher Scientific, Inc. The sequences of
miR-300 were as follows: Sense, 5′-UAUACAAGGGCAGACUCUCUCU-3′;
anti-sense, 5′-AGAGAGAGUCUGCCCUUGUAUA-3′. The sequence of the
miR-300 inhibitor was as follows: 5′-GAGAGAGUCUGCCCUUGUAU-3′. The
open reading frame fragments of CREPT were inserted into a pcDNA3.1
vector (Thermo Fisher Scientific, Inc.) to generate the CREPT
expression vector. A total of 2×105 cells were plated in
triplicate overnight in antibiotic-free complete medium in 6-well
plates. The cells were grown overnight and then transfected with
200 µl mature miRNA (100 nM) and RNAiMAX reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol for 48 h. The transfection efficacy was confirmed by
RT-qPCR or western blot analysis. Each experiment was repeated at
least 3 times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (human HCC tissue samples and adjacent
non-tumor tissue samples; the liver cancer cell lines HepG2, Hep3B,
Huh-7 and 293T) was extracted using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. For detection of miR-300 expression, complementary DNA
was generated using the TaqMan MicroRNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) at the
following conditions: 16°C for 30 min, 42°C for 30 min and 85°C for
5 min). PCR amplifications were performed using the TaqMan Small
RNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.). For
detection of mRNA expression, complementary DNA (cDNA) was
synthesized using Moloney Murine Leukemia Virus reverse
transcriptase (Takara Biotechnology Co., Ltd., Dalian, China) at
the following conditions: 37°C for 15 min, followed by 72°C for 10
min) and qPCR amplifications were performed using Power SYBR Green
PCR Master mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.). PCR amplifications were performed using the Applied
Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following thermal
parameters: 95°C for 10 min followed by 40 cycles of 95°C for 15
sec and 60°C for 60 sec. U6 small nuclear RNA and GAPDH were used
as internal controls for normalizing the relative expression levels
of miR-300 and mRNA, respectively. The primer sequences were:
miR-300 forward, 5′-TATACAAGGGCAGACTCTCTCT-3′; and U6 reverse,
5′-CGCAAGGATGACACGCAAATTCGT-3′; CREPT forward,
5′-CACGCGGGACCCATCGTCTC-3′; CREPT reverse,
5′-AGCCTTCATCTGCCTCTCTGGCA-3′: cyclin D1 forward,
5′-CTGGCCATGAACTACCTGGA-3′; cyclin D1 reverse,
5′-GTCACACTTGATCACTCTCC-3′; GAPDH forward,
5′-CATGAGAAGTATGACAACAGCCT-3′; and GAPDH reverse,
5′-AGTCCTTCCACGATACCAAAGT-3′. All RT-qPCR assays were run in
triplicate. All results are presented as the mean ± standard
deviation of three independent experiments. Relative gene
expression analysis was performed using the comparative
2−ΔΔCq method (27).
Cell proliferation assay
Cell proliferation was evaluated using a Cell
Counting Kit-8 (CCK-8) assay. HepG2 and Huh-7 cells were seeded
into a 96-well plate (Thermo Fisher Scientific, Inc.) at a density
of 10,000 cells/well. Cells were transfected with 1 pM miR-300
mimic or inhibitor and cultured for 24, 48 and 72 h. Subsequently,
10 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.) was
added to each well. The cells were then cultured for a further 2 h
at 37°C, followed by measurements of the absorbance at 450 nm using
a spectrophotometer (Bio-Rad Laboratories, Inc.). Each condition
was determined in quintuplicates and all experiments were repeated
at least 3 times.
Colony formation assay
After 48 h of transfection, HepG2 and Huh-7 cells
were re-seeded into a six-well plate (Thermo Fisher Scientific,
Inc.) at a density of 1,000 cells/well. And allowed to grow
undisturbed for 7 days. Cells were stained with crystal violet on
the plates at room temperature for 15 min. The number of colonies
(diameter >1 mm) were counted using an inverted light
microscope. Each experiment was repeated at least 3 times.
Cell cycle assay
HepG2 and Huh-7 cells were harvested following the
indicated treatment times and fixed with 75% ice-cold ethanol at
4°C for 24 h. Following washing with ice-cold PBS, cells were
treated with 50 µg/ml RNase and 50 µg/ml propidium iodide (BD
Biosciences; Becton, Dickinson and Company) in 500 µl binding
buffer. Following incubation for 30 min, the cell samples were
subjected to flow cytometry with a flow cytometer (BD Biosciences;
Becton, Dickinson and Company) and data were analyzed using BD
CellFIT software (BD Biosciences; Becton, Dickinson and Company).
Each experiment was repeated at least 3 times.
Bioinformatics analysis and
dual-luciferase reporter assay
Computer-aided algorithms (http://www.targetscan.org/vert_71/) were adopted to
predict the target gene of miR-300. CREPT 3′-UTR fragments
containing miR-300-binding sites or mutant miR-300-binding sites
were cloned into pmirGLO vectors (Promega Cooperation). The
reporter vector was co-transfected with miR-300 mimic into 293T
cells using RNAiMAX reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Following culture for 48 h, relative luciferase activity
(firefly/Renilla) was detected using a Dual-Luciferase Reporter
assay system (Promega Corporation), according to the manufacturer's
protocol. Each experiment was repeated at least 3 times.
Wnt/β-catenin-dependent TOP flash
reporter assay
Wnt/β-catenin signaling was determined by
measurement of TCF-mediated transcriptional activity using a TOP
flash reporter assay. Briefly, HepG2 and Huh-7 cells were
co-transfected with TOP flash vector (2 µg), pRL-TK vector (1 µg)
(Promega Corportation) and 1 nM miR-300 mimic or inhibitor (Thermo
Fisher Scientific, Inc.) for 48 h. Relative luciferase activity
(firefly/Renilla) was detected using a Dual-Luciferase Reporter
assay system (Promega Corporation), according to the manufacturer's
protocol. Each experiment was repeated at least 3 times.
Western blot analysis
Cell lysates were obtained by lysing cells in cell
lysis buffer (Thermo Fisher Scientific, Inc.) containing protease
inhibitors. Protein concentrations were quantified using the Pierce
Bicinchoninic Acid Protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Proteins (20 µg) were then loaded onto 10%
sodium dodecyl sulfate polyacrylamide gel and separated by
electrophoresis. The separated proteins were transferred to a
polyvinylidene fluoride membrane followed by incubation with 5%
skim milk in TBS and 0.1% Tween-20 (TBST) at 37°C for 1 h. The
membrane was then incubated with appropriate antibodies, including
anti-CREPT (cat. no. GTX119969; 1:2,000; GeneTex, Inc.),
anti-β-catenin (9562; 1:1,000; Cell Signaling Technology, Inc.) and
anti-GAPDH (cat. no. ab9485. 1:2,500; Abcam) at 4°C overnight.
Following washes with TBST, the membrane was incubated with
horseradish peroxidase-labeled goat polyclonal anti-rabbit IgG
secondary antibody (cat. no. ab6721, 1:3,000; Abcam) for 1 h at
room temperature. The immunoblots were visualized using enhanced
chemiluminescent substrate (Thermo Fisher Scientific, Inc.). GAPDH
was used as a loading control protein and was visualized using the
enhanced chemiluminescence system from Pierce (Thermo Fisher
Scientific, Inc.). Gray scale analysis of protein bands was
performed by Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation.
Significant differences were determined using Student's t-test or
one-way analysis of variance followed by Bonferroni's post hoc
test. All statistical analysis was performed with SPSS 19.0
software (IBM Corp.). The correlation between miR-300 and CREPT
expression was determined using Spearman's correlation test.
P<0.05 was considered to indicate a statistically significant
difference. Data are representative of three independent
experiments performed in triplicate.
Results
miR-300 expression level is lower in
HCC tissues and cell lines
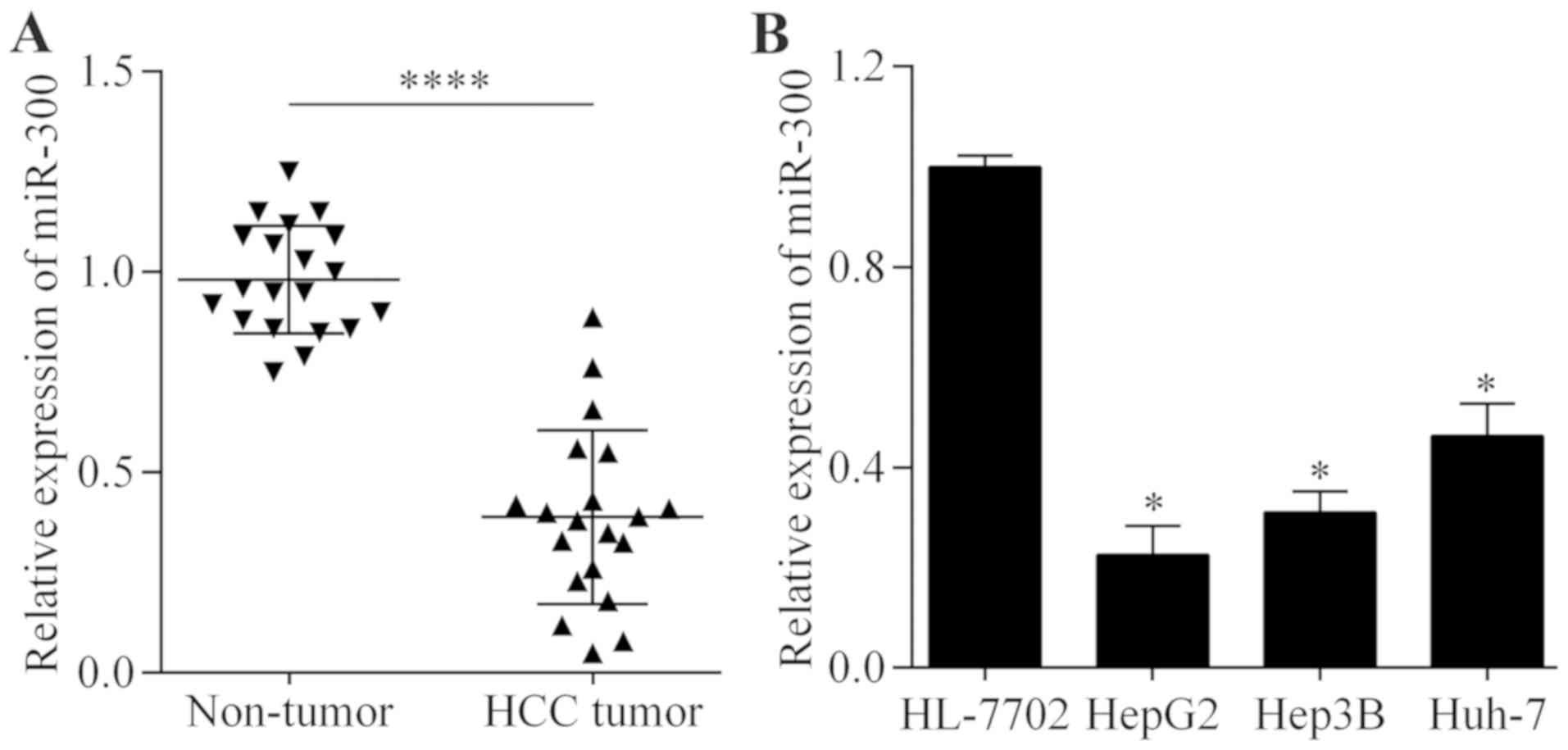
To investigate whether miR-300 may serve a role in
HCC, the expression level of miR-300 in HCC tissues was examined by
RT-qPCR. It was identified that the expression level of miR-300 was
significantly decreased in HCC tissue samples compared with
adjacent non-tumor tissue samples (P<0.0001; Fig. 1A). However, no correlation was
identified between miR-300 expression and tumor stage or histologic
grade (Table I). Furthermore, it was
revealed that the miR-300 expression level was significantly
decreased in a number of liver cancer cell lines compared with the
normal liver cancer cell line (P<0.05; Fig. 1B). This altered expression level of
miR-300 indicates a possible role of miR-300 in HCC.
miR-300 inhibits the growth of liver
cancer cells in vitro
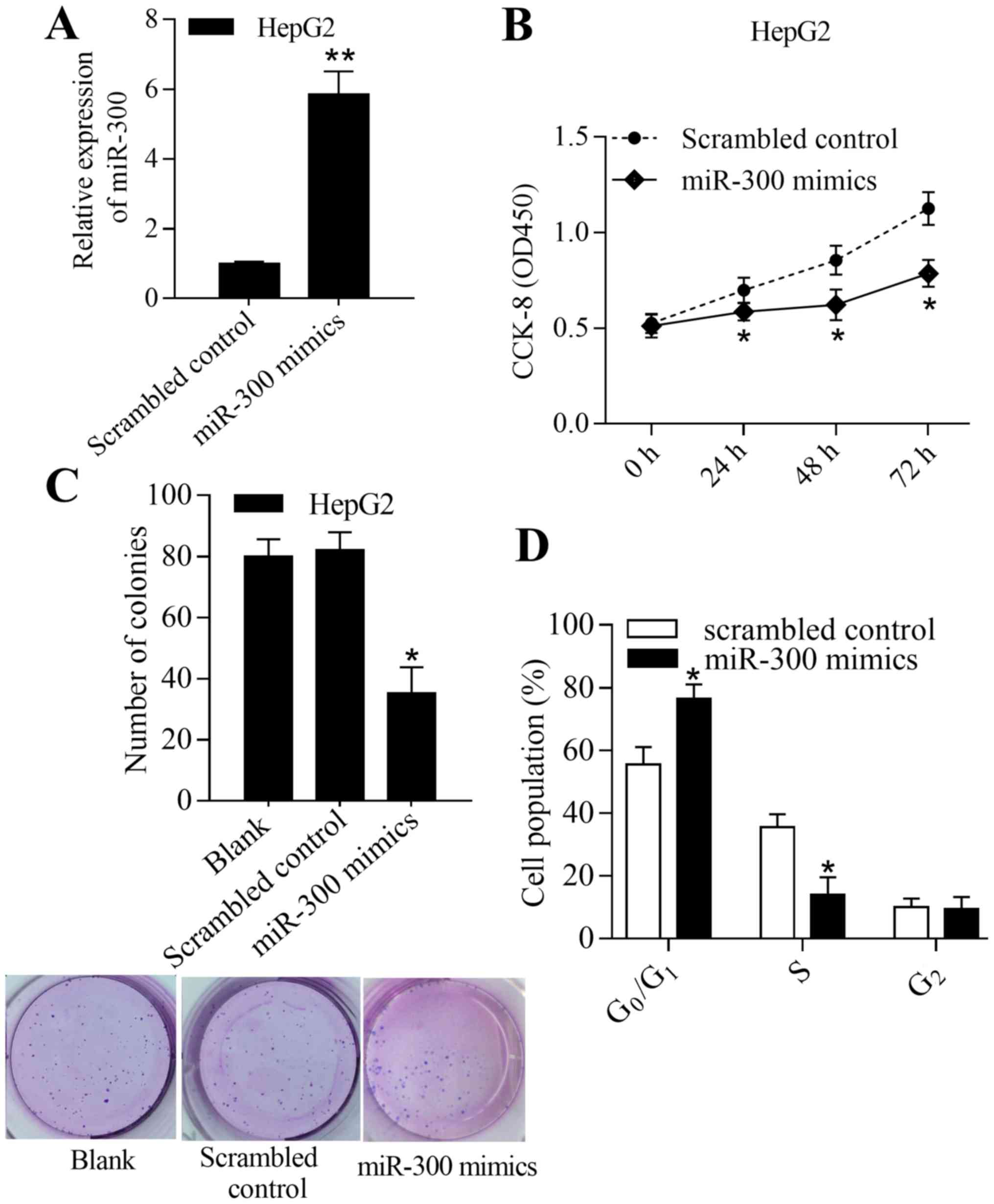
To investigate the biological function of miR-300 in
liver cancer, the effect of miR-300 overexpression or inhibition on
liver cancer cell growth was determined in vitro.
Overexpression of miR-300 was achieved by transfection of the
miR-300 mimic into HepG2 cells (Fig.
2A). The results demonstrated that overexpression of miR-300
significantly inhibited proliferation and colony formation
(P<0.05; Fig. 2B and C).
Furthermore, overexpression of miR-300 significantly increased the
number of cells in the G0/G1 phase and
decreased the number of cells in the S phase (P<0.05; Fig. 2D). In addition, inhibition of miR-300
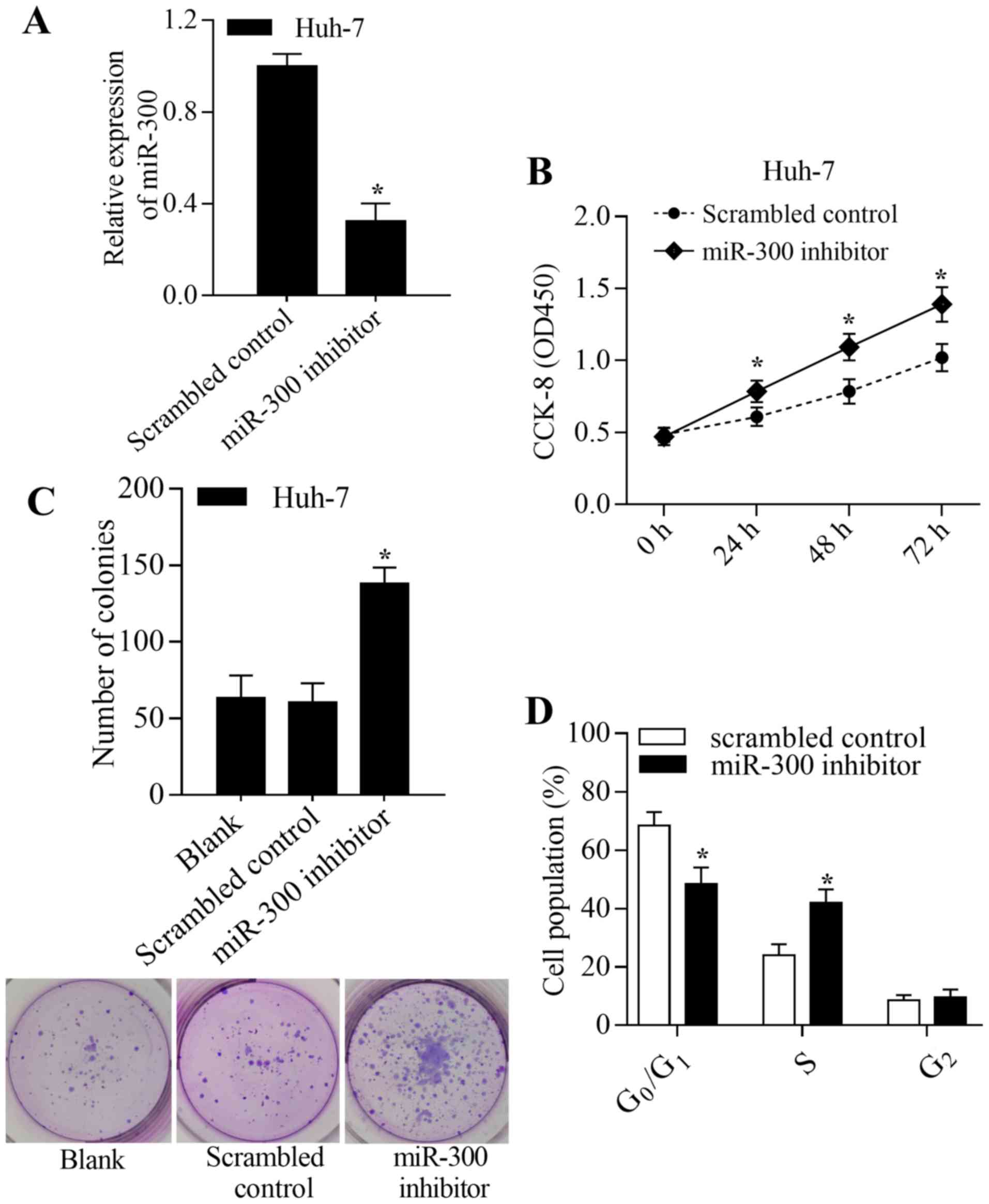
was achieved by transfection of the miR-300 inhibitor into Huh-7
cells (Fig. 3A). Inhibition of
miR-300 significantly promoted proliferation and colony formation
of Huh-7 cells (P<0.05; Fig. 3B and
C). Inhibition of miR-300 significantly decreased the number of
cells in the G0/G1 phase and increased the
number of cells in the S phase compared with the control
(P<0.05; Fig. 3D). In summary,
these results suggest that miR-300 inhibits liver cancer cell
growth by regulating proliferation, colony formation and cell cycle
transition.
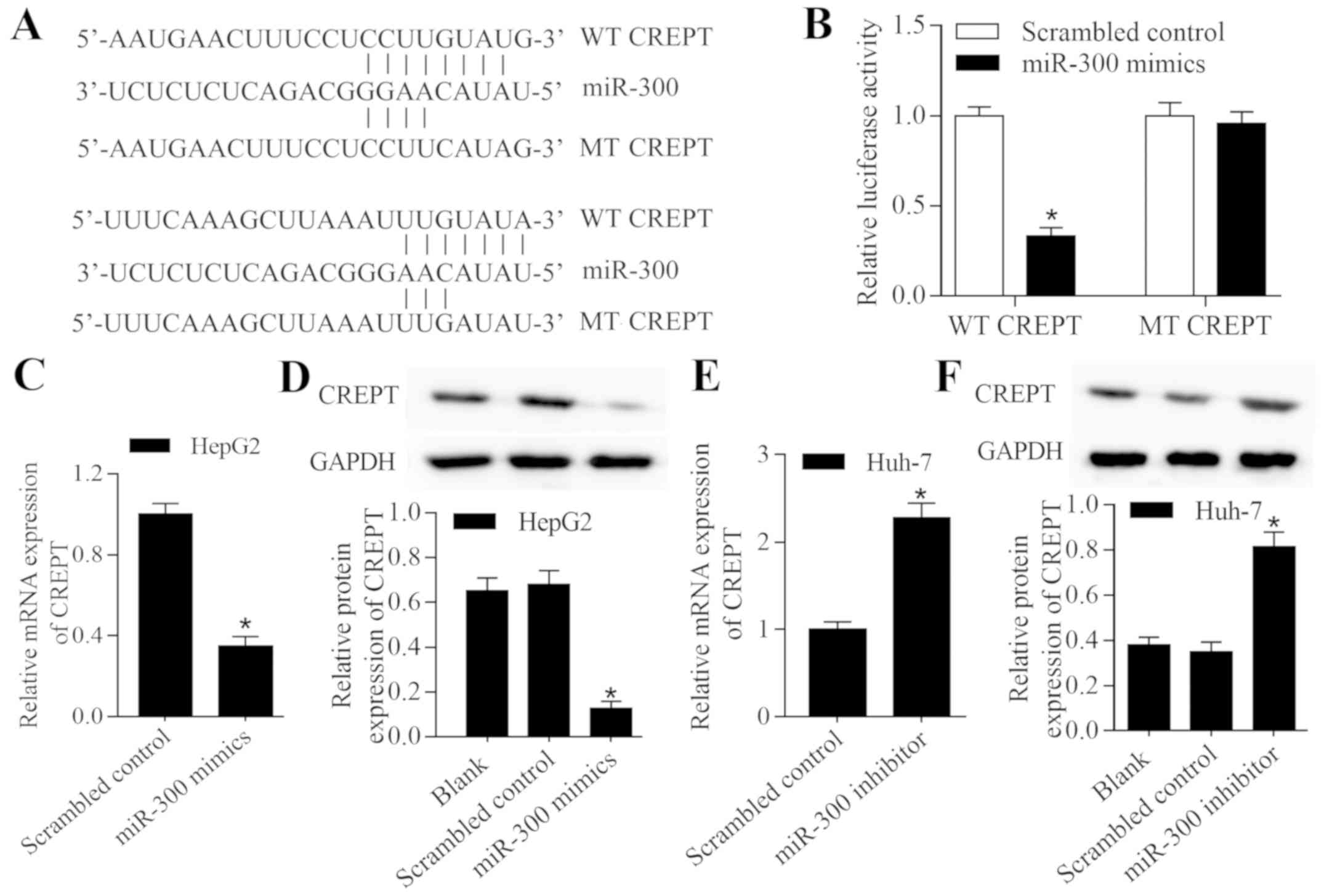
CREPT is a target gene of miR-300 in
liver cancer
It is understood that miRNAs can participate in
tumor progression by repressing target genes (13). Therefore, the present study used
bioinformatics analysis to predict the potential targets of
miR-300. Notably, it was identified that CREPT, an oncogene in
numerous types of cancer (16), is a
putative target gene of miR-300. The 3′-UTR of CREPT contains
putative binding sites for miR-300 (Fig.
4A). A luciferase reporter assay demonstrated that transfection
with miR-300 mimic significantly suppressed the luciferase activity
of a vector containing wild-type CREPT 3′-UTR; however, no effect
on luciferase activity was observed with a vector containing mutant
CREPT 3′-UTR (P<0.05; Fig. 4B).
Subsequently, the regulatory effect of miR-300 on CREPT expression
was examined in liver cancer cells. The results revealed that
overexpression of miR-300 significantly decreased the expression
level of CREPT (P<0.05; Fig. 4C and
D), while inhibition of miR-300 significantly increased the
expression level of CREPT (P<0.05; Fig. 4E and F). In summary, these results
indicate that miR-300 binds to the 3′-UTR of CREPT, which regulates
the expression level.
miR-300 regulates Wnt/β-catenin
signaling in liver cancer cells
CREPT has been reported to be an important regulator
of the Wnt/β-catenin signaling pathway (28,29).
With the understanding that miR-300 regulates CREPT expression, it
was suggested that miR-300 may have a regulatory effect on the
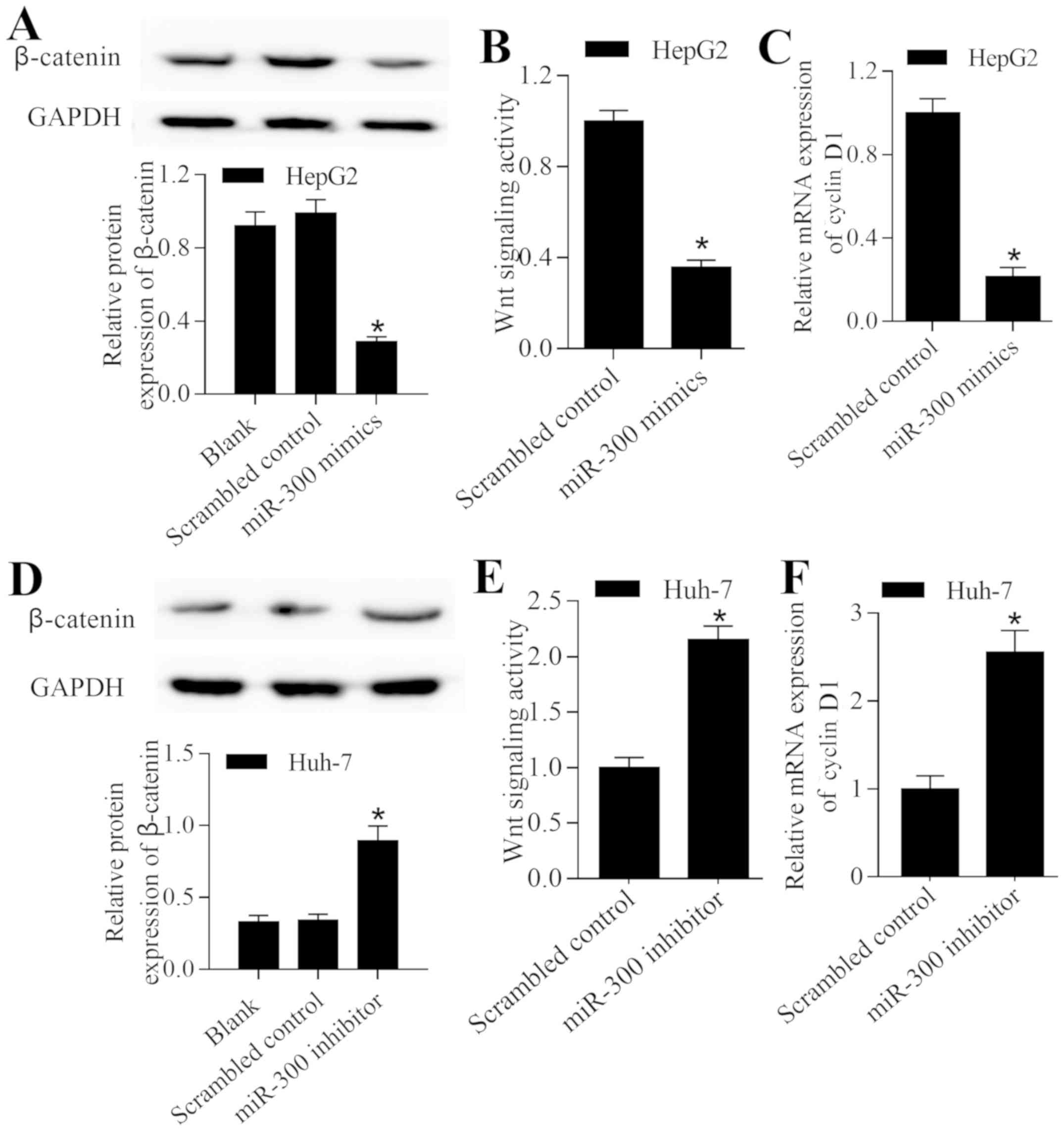
Wnt/β-catenin signaling pathway. The present study revealed that
overexpression of miR-300 significantly decreased the expression
level of β-catenin (P<0.05; Fig.
5A). Furthermore, overexpression of miR-300 significantly
inhibited Wnt/β-catenin signaling (P<0.05; Fig. 5B) and significantly decreased the
expression level of cyclin D1 (P<0.05; Fig. 5C). By contrast, inhibition of miR-300
induced the opposite effect on the Wnt/β-catenin signaling pathway
(Fig. 5D-F). These results suggest
that miR-300 exerts a regulatory effect on Wnt/β-catenin signaling
in liver cancer cells.
miR-300 inhibits the growth of liver
cancer cells and the Wnt/β-catenin signaling pathway
To investigate whether miR-300 exerts its function
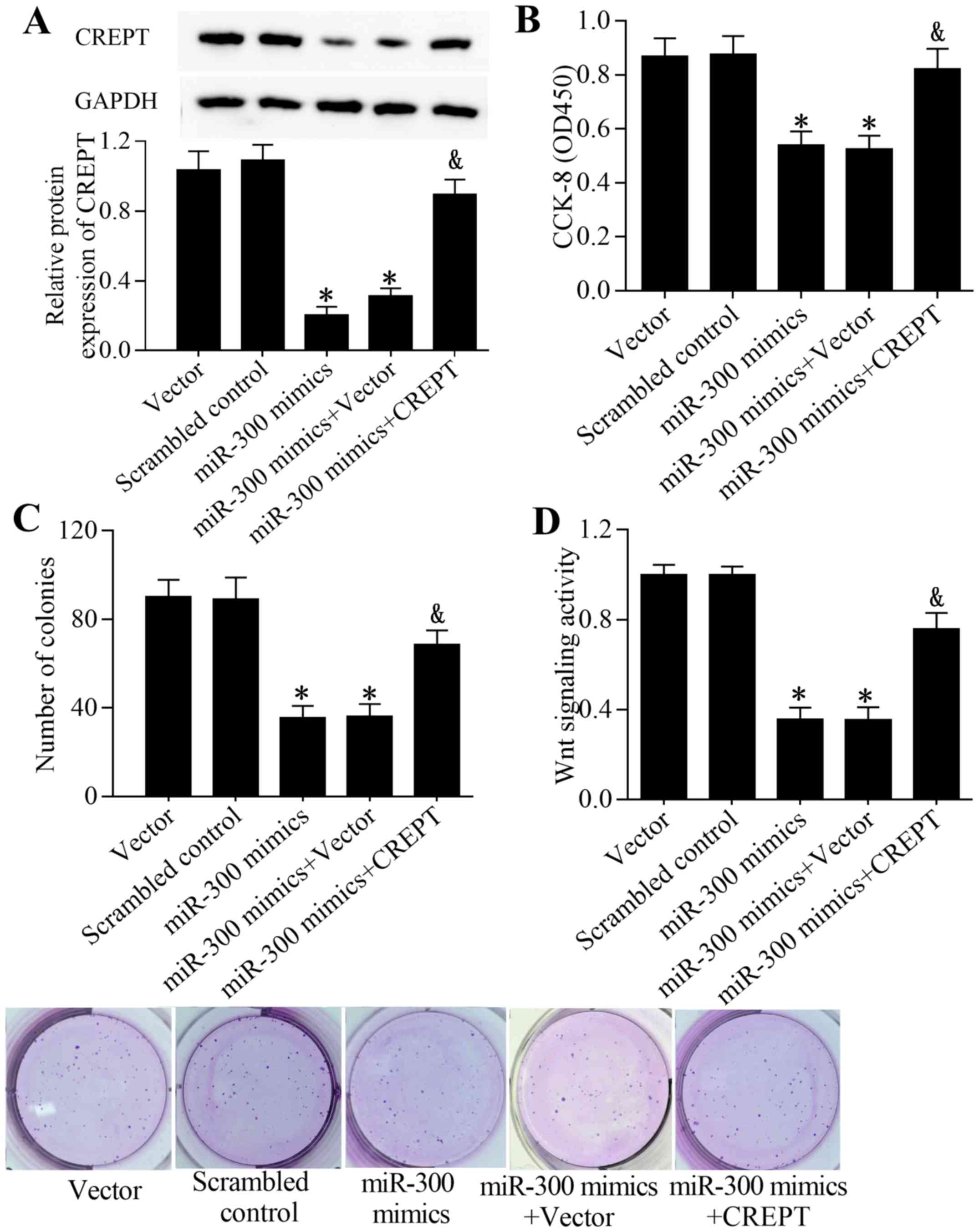
by targeting CREPT, rescue experiments were performed. Transfection
of the CREPT expression vector significantly restored the
expression level of CREPT in cells transfected with an miR-300
mimic (P<0.05; Fig. 6A).
Furthermore, overexpression of CREPT partially reversed the
inhibitory effect of miR-300 on cell growth (Fig. 6B and C) and the Wnt/β-catenin
signaling pathway (Fig. 6D). In
conclusion, these results suggest that miR-300 inhibits the growth
of liver cancer cells and Wnt/β-catenin signaling in liver cancer
cells by targeting CREPT.
Discussion
A number of miRNAs have been reported to be
associated with tumorigenesis of HCC (13,30);
however, additional miRNAs remain to be identified and
characterized. The present study reported miR-300 as a novel miRNA
associated with HCC. miR-300 was revealed to inhibit the growth of
HCC cells, which indicates it functions as a tumor-suppressive
miRNA in HCC (31). Notably, it was
identified that the underlying mechanism is associated with a
regulatory effect of miR-300 on CREPT. The present study suggests
that miR-300 may be used as a therapeutic target for HCC.
Numerous studies have demonstrated that
dysregulation of miR-300 is involved in the development and
progression of cancer (24–26). A low expression level of miR-300 is
present in glioma tissues and overexpression of miR-300 inhibits
the proliferation and invasion of glioma cells in vitro
(32). Furthermore, miR-300 has been
reported to suppress the epithelial to mesenchymal transition and
metastasis of head, and neck squamous cell carcinoma and breast
cancer cells by targeting Twist (26). The expression level of miR-300 has
been revealed to be lower in laryngeal squamous cell carcinoma and
overexpression of miR-300 represses proliferation and metastasis by
targeting c-ros oncogene 1 receptor tyrosine kinase (33,34).
Inhibition of miR-300 contributes to cell proliferation and
metastasis of gallbladder carcinoma (35). Additionally, recent studies have
demonstrated that miR-300 inhibits the progression of pancreatic
cancer and osteosarcoma by targeting cullin 4B (24,36).
These findings suggest a tumor suppressive role of miR-300.
Comparable with the aforementioned studies, the present results
support a tumor suppressive role of miR-300 in tumor progression.
The current study demonstrated that the miR-300 expression level
was lower in liver cancer and overexpression of miR-300 could
inhibit proliferation and colony formation, and induce
G0/G1 cell cycle arrest of liver cancer
cells, which indicates an antitumor effect of miR-300 in HCC. By
contrast, certain studies have suggested an oncogenic role of
miR-300 in tumorigenesis. miR-300 has been reported to promote
tumorigenesis of colorectal cancer, osteosarcoma and glioma by
targeting p53 and bromodomain-containing protein 7 (37,38).
Zhang et al (39) reported
that miR-300 was upregulated in HCC tissues and promoted cancer
growth by targeting MDC1. Those controversial results indicated
that disregulation of miR-300 might not be the first event during
the pathology of HCC. Other master regulators within tumors or
tumor microenvironment might exist to regulate the expression
miR-300 as disease progress. Wang et al (31) reported that miR-300 was downregulated
in HCC tissues and cell lines. Those controversial results indicate
the complexity of miR-300 regulation during the pathology of HCC.
Disregulation of miR-300 might affect disease progress. Moreover,
HCC samples from the present study and Wang's study are from
patients who did not undergo chemotherapy or radiotherapy.
Chemotherapy and radiotherapy are reported to induce expression of
some miRNAs in cancers (40,41). It is very possible that the variation
of miR-300 expression might come from the treatment difference in
different patients. Of note, SMMC-7721 cells used in Wang's study
is reported to be Hela contaminated, which make the conclusion
unreliable (31). So, future
mechanism studies and HCC tissue from a larger patient population
are needed to draw a complete picture of miR-300 in HCC. Therefore,
the precise role of miR-300 in tumor progression remains to be
further investigated.
CREPT was initially identified as a potential
oncogene in colorectal cancer; it has been identified to be
overexpressed at the mRNA and protein levels in colorectal cancer
tissues and cell lines (16). High
CREPT expression is correlated with tumor differentiation,
metastasis and a short survival time for patients with colorectal
cancer (20). Functional experiments
demonstrated that CREPT can promote the proliferation and cell
cycle progression of colorectal cancer cells by regulating the
transcription of cell cycle-associated genes (16,20,42).
CREPT overexpression has been associated with tumor stage,
histology type and depth of myometrial invasion in endometrial
cancer, and knockdown of CREPT inhibits cell proliferation and
induces G0/G1 cell cycle arrest by
downregulating cyclin D1, cell cycle dependent kinase (CDK)4 and
CDK6 in vitro (23).
Knockdown of CREPT inhibits the proliferation and migration of
non-small cell lung cancer cells, whereas overexpression of CREPT
demonstrates an oncogenic effect (21,43).
Similarly, an oncogenic function of CREPT has been observed in oral
squamous cell carcinoma and gastric cancer (22,44).
Notably, CREPT has been reported to be highly expressed in HCC
tissues and cell lines (16).
Furthermore, CREPT achieves its oncogenic effects via regulation of
HCC cell growth and cell cycle progression (16). These findings suggest that CREPT is a
novel oncogene that can be used as a target for cancer treatment.
Notably, a recent study demonstrated that CREPT expression is
regulated by miR-138, which contributes to breast cancer
progression (45). This indicates
that high expression of CREPT may be induced by dysregulated
miRNAs. However, to the best of our knowledge, the regulatory
mechanism of miRNAs against CREPT in HCC remains unknown.
The present study identified that CREPT is targeted
and regulated by miR-300 in HCC. It was revealed that miR-300 can
inhibit liver cancer cell growth by targeting CREPT, whereas
overexpression of CREPT partially reverses the antitumor effect of
miR-300. Therefore, decreased expression of miR-300 may contribute
to a high expression level of CREPT in HCC, which leads to HCC
development and progression. Therefore, the miR-300/CREPT axis may
serve an important role in the molecular pathogenesis of HCC.
CREPT has been reported to be a positive regulator
of the Wnt/β-catenin signaling pathway. Overexpression of CREPT
enhances the expression levels of β-catenin, transcription factor 4
(TCF4) and cyclin D1 in chicken fibroblast cells (28). Furthermore, CREPT has been reported
to promote Wnt/β-catenin signaling by enhancing the association of
β-catenin with TCF4 (29). Notably,
a recent study demonstrated that CREPT facilitates Wnt/β-catenin
signaling by promoting p300-mediated β-catenin acetylation and
stabilization (19). Similarly, the
present results demonstrated that inhibition of CRPET decreased the
activation of Wnt/β-catenin signaling in HCC cells. Therefore,
CREPT may serve as a novel target for inhibiting Wnt/β-catenin
signaling in tumorigenesis.
In conclusion, the current study provides promising
evidence that miR-300 acts as a tumor suppressor in HCC and
inhibits the growth of liver cancer cells by targeting and
inhibiting CREPT. The present results demonstrate that the
miR-300/CREPT axis may be involved in regulating the Wnt/β-catenin
signaling pathway, which may serve an important role in the
development and progression of HCC. As a limitation of this study,
the detail relationship between Wnt signaling and miR-300 is still
unknown. This interesting project is now ongoing in the authors'
lab. In conclusion, the current study may increase understanding of
the mechanisms involved in tumorigenesis and suggests a novel
target for HCC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JB and XZ designed the study; YG and YD conducted
the experiments; XZ contributed new reagents or analytic tools; YG
and XY analyzed the data and prepared figures; JB, XZ and YD
drafted the manuscript; all authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients. The present study was approved by the Institutional Human
Experiment and Ethics Committee of Changchun University of Chinese
Medicine, Changchun, China (approval no. CCZYFYLL2019-020).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology 127 (5
Suppl 1). S72–S78. 2004.
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu MD, Jia LH, Liu HB, Zhang KH and Guo
GH: Sorafenib in combination with transarterial chemoembolization
for hepatocellular carcinoma: A meta-analysis. Eur Rev Med
Pharmacol Sci. 20:64–74. 2016.PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei R, Huang GL, Zhang MY, Li BK, Zhang
HZ, Shi M, Chen XQ, Huang L, Zhou QM, Jia WH, et al: Clinical
significance and prognostic value of microRNA expression signatures
in hepatocellular carcinoma. Clin Cancer Res. 19:4780–4791. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, Zhang SG, Wang ZH and Liao JC:
Down-regulation of miR-342-3p in hepatocellular carcinoma tissues
and its prognostic significance. Eur Rev Med Pharmacol Sci.
21:2098–2102. 2017.PubMed/NCBI
|
|
15
|
Zhao XQ, Liang B, Jiang K and Zhang HY:
Down-regulation of miR-655-3p predicts worse clinical outcome in
patients suffering from hepatocellular carcinoma. Eur Rev Med
Pharmacol Sci. 21:748–752. 2017.PubMed/NCBI
|
|
16
|
Lu D, Wu Y, Wang Y, Ren F, Wang D, Su F,
Zhang Y, Yang X, Jin G, Hao X, et al: CREPT accelerates
tumorigenesis by regulating the transcription of cell-cycle-related
genes. Cancer Cell. 21:92–104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carvalho B, Postma C, Mongera S, Hopmans
E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthäi A,
Cuesta MA, et al: Multiple putative oncogenes at the chromosome 20q
amplicon contribute to colorectal adenoma to carcinoma progression.
Gut. 58:79–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng G, Yu M, Chen LC, Moore D, Kurisu W,
Kallioniemi A, Waldman FM, Collins C and Smith HS: Amplifications
of oncogene erbB-2 and chromosome 20q in breast cancer determined
by differentially competitive polymerase chain reaction. Breast
Cancer Res Treat. 40:271–281. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Wang S, Kang W, Liu C, Dong Y,
Ren F, Wang Y, Zhang J, Wang G, To KF, et al: CREPT facilitates
colorectal cancer growth through inducing Wnt/β-catenin pathway by
enhancing p300-mediated β-catenin acetylation. Oncogene.
37:3485–3500. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng G, Li W, Zuo B, Guo Z, Xi W, Wei M,
Chen P, Wen W and Yang AG: High expression of CREPT promotes tumor
growth and is correlated with poor prognosis in colorectal cancer.
Biochem Biophys Res Commun. 480:436–442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li W, Zheng G, Xia J, Yang G, Sun J, Wang
X, Wen M, Sun Y, Zhang Z and Jin F: Cell cycle-related and
expression-elevated protein in tumor overexpression is associated
with proliferation behaviors and poor prognosis in non-small-cell
lung cancer. Cancer Sci. 109:1012–1023. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma J, Ren Y, Zhang L, Kong X, Wang T, Shi
Y and Bu R: Knocking-down of CREPT prohibits the progression of
oral squamous cell carcinoma and suppresses cyclin D1 and c-Myc
expression. PLoS One. 12:e01743092017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Qiu H, Hu W, Li S and Yu J: RPRD1B
promotes tumor growth by accelerating the cell cycle in endometrial
cancer. Oncol Rep. 31:1389–1395. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Z, Zhang W, Jiang K, Chen B, Wang K,
Lao L, Hou C, Wang F, Zhang C and Shen H: MicroRNA-300 regulates
the ubiquitination of PTEN through the CRL4BDCAF13 E3
ligase in osteosarcoma cells. Mol Ther Nucleic Acids. 10:254–268.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He J, Feng X, Hua J, Wei L, Lu Z, Wei W,
Cai H, Wang B, Shi W, Ding N, et al: miR-300 regulates cellular
radiosensitivity through targeting p53 and apaf1 in human lung
cancer cells. Cell Cycle. 16:1943–1953. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu J, Xie F, Bao X, Chen W and Xu Q:
miR-300 inhibits epithelial to mesenchymal transition and
metastasis by targeting Twist in human epithelial cancer. Mol
Cancer. 13:1212014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin K, Chen H, Zuo Q, Huang C, Zhao R, Yu
X, Wang Y, Zhang Y, Chang Z and Li B: CREPT and p15RS regulate cell
proliferation and cycling in chicken DF-1 cells through the
Wnt/β-catenin pathway. J Cell Biochem. 119:1083–1092. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Liu C, Duan X, Ren F, Li S, Jin
Z, Wang Y, Feng Y, Liu Z and Chang Z: CREPT/RPRD1B, a recently
identified novel protein highly expressed in tumors, enhances the
β-catenin. TCF4 transcriptional activity in response to Wnt
signaling. J Biol Chem. 289:22589–22599. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan XP, Wang HX, Tong DM, Li Y, Huang LH
and Wang C: miRNA-370 acts as a tumor suppressor via the
downregulation of PIM1 in hepatocellular carcinoma. Eur Rev Med
Pharmacol Sci. 21:1254–1263. 2017.PubMed/NCBI
|
|
31
|
Wang R, Yu Z, Chen F, Xu H, Shen S, Chen
W, Chen L, Su Q, Zhang L, Bi J, et al: miR-300 regulates the
epithelial-mesenchymal transition and invasion of hepatocellular
carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed
Pharmacother. 103:1632–1642. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou F, Li Y, Hao Z, Liu X, Chen L, Cao Y,
Liang Z, Yuan F, Liu J, Wang J, et al: MicroRNA-300 inhibited
glioblastoma progression through ROCK1. Oncotarget. 7:36529–36538.
2016.PubMed/NCBI
|
|
33
|
He FY, Liu HJ, Guo Q and Sheng JL: Reduced
miR-300 expression predicts poor prognosis in patients with
laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci.
21:760–764. 2017.PubMed/NCBI
|
|
34
|
Ge W, Han C, Wang J and Zhang Y: MiR-300
suppresses laryngeal squamous cell carcinoma proliferation and
metastasis by targeting ROS1. Am J Transl Res. 8:3903–3911.
2016.PubMed/NCBI
|
|
35
|
Ma F, Wang SH, Cai Q, Jin LY, Zhou D, Ding
J and Quan ZW: Long non-coding RNA TUG1 promotes cell proliferation
and metastasis by negatively regulating miR-300 in gallbladder
carcinoma. Biomed Pharmacother. 88:863–869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang JQ, Chen S, Gu JN, Zhu Y, Zhan Q,
Cheng DF, Chen H, Deng XX, Shen BY and Peng CH: MicroRNA-300
promotes apoptosis and inhibits proliferation, migration, invasion
and epithelial-mesenchymal transition via the Wnt/β-catenin
signaling pathway by targeting CUL4B in pancreatic cancer cells. J
Cell Biochem. 119:1027–1040. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang L and Yu P: miR-300 promotes
proliferation and EMT-mediated colorectal cancer migration and
invasion by targeting p53. Oncol Rep. 36:3225–3232. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xue Z, Zhao J, Niu L, An G, Guo Y and Ni
L: Up-regulation of miR-300 promotes proliferation and invasion of
osteosarcoma by targeting BRD7. PLoS One. 10:e01276822015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Luo H, Du J and Liu Y:
MicroRNA-300 plays as oncogene by promoting proliferation and
reducing apoptosis of liver cancer cells by targeting MDC1. Int J
Clin Exp Pathol. 9:1231–1239. 2016.
|
|
40
|
Olatunji I: Potential application of tumor
suppressor microRNAs for targeted therapy in head and neck cancer:
A mini-review. Oral Oncol. 87:165–169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lo Russo G, Tessari A, Capece M, Galli G,
de Braud F, Garassino MC and Palmieri D: MicroRNAs for the
diagnosis and management of malignant pleural mesothelioma: A
literature review. Front Oncol. 8:6502018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kuang YS, Wang Y, Ding LD, Yang L, Wang Y,
Liu SH, Zhu BT, Wang XN, Liu HY, Li J, et al: Overexpression of
CREPT confers colorectal cancer sensitivity to fluorouracil. World
J Gastroenterol. 24:475–483. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu T, Li WM, Wang WP, Sun Y, Ni YF, Xing
H, Xia JH, Wang XJ, Zhang ZP and Li XF: Inhibiting CREPT reduces
the proliferation and migration of non-small cell lung cancer cells
by down-regulating cell cycle related protein. Am J Transl Res.
8:2097–2113. 2016.PubMed/NCBI
|
|
44
|
Sun M, Si G, Sun HS and Si FC: Inhibition
of CREPT restrains gastric cancer growth by regulation of cycle
arrest, migration and apoptosis via ROS-regulated p53 pathway.
Biochem Biophys Res Commun. 496:1183–1190. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liang Z, Feng Q, Xu L, Li S and Zhou L:
CREPT regulated by miR-138 promotes breast cancer progression.
Biochem Biophys Res Commun. 493:263–269. 2017. View Article : Google Scholar : PubMed/NCBI
|