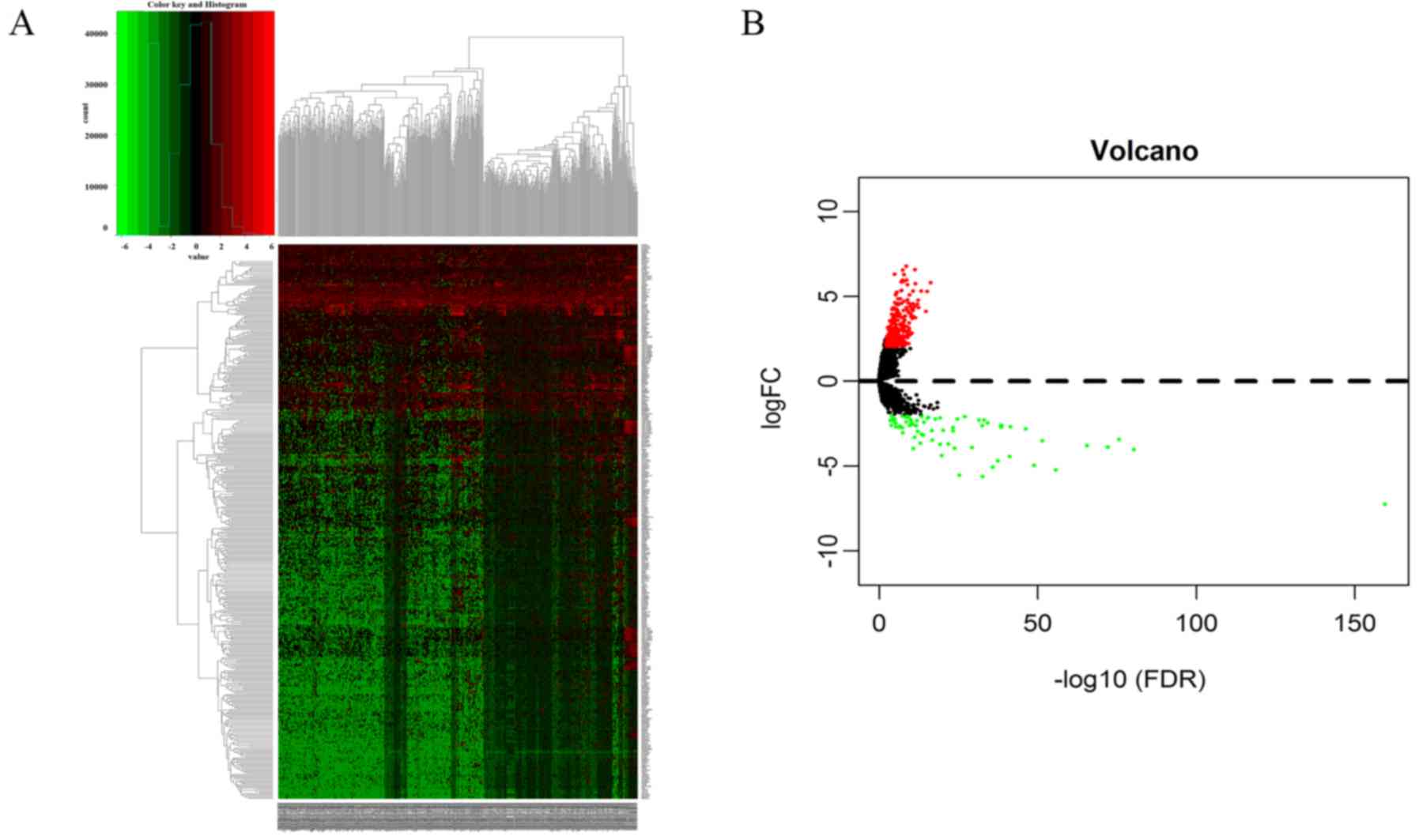
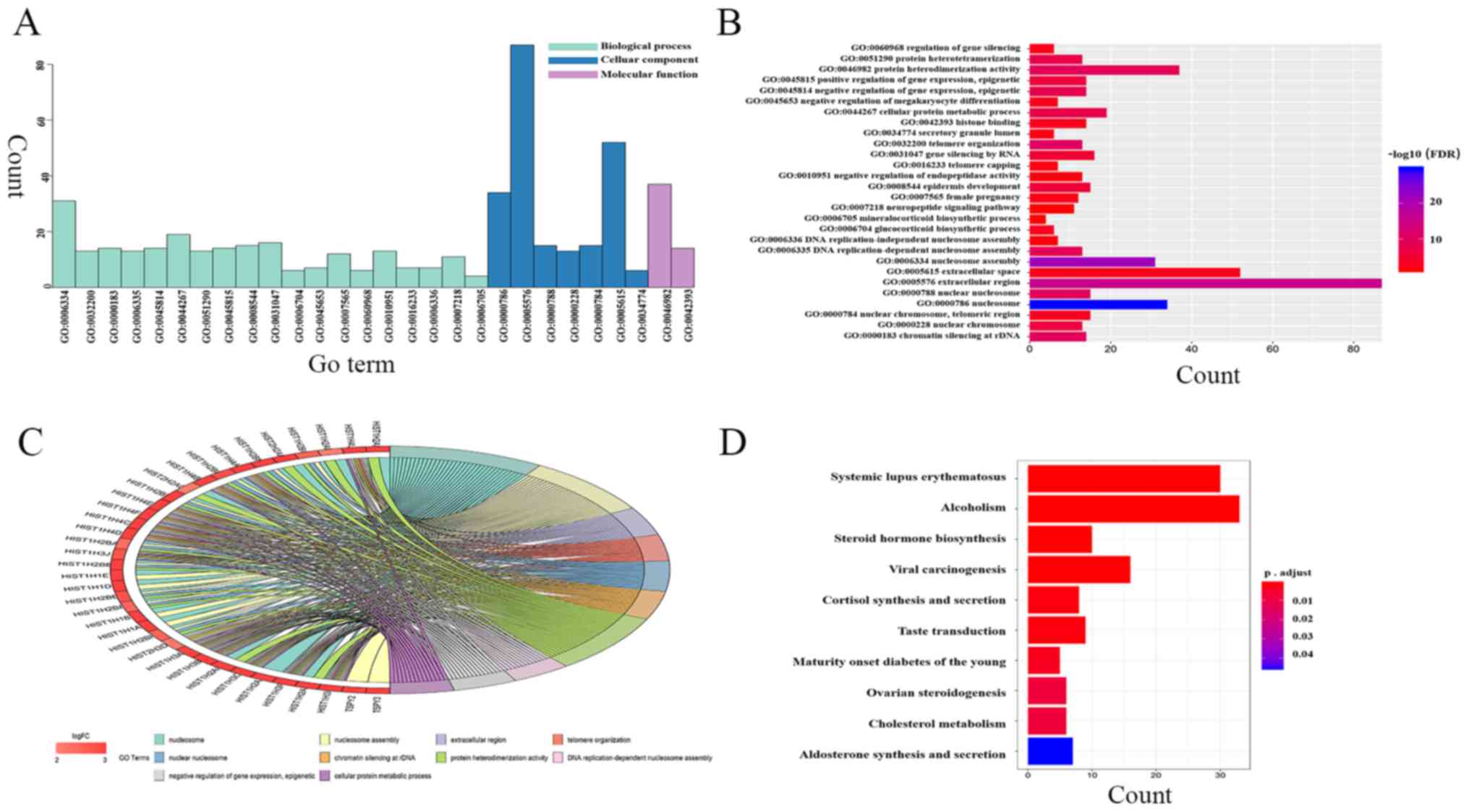
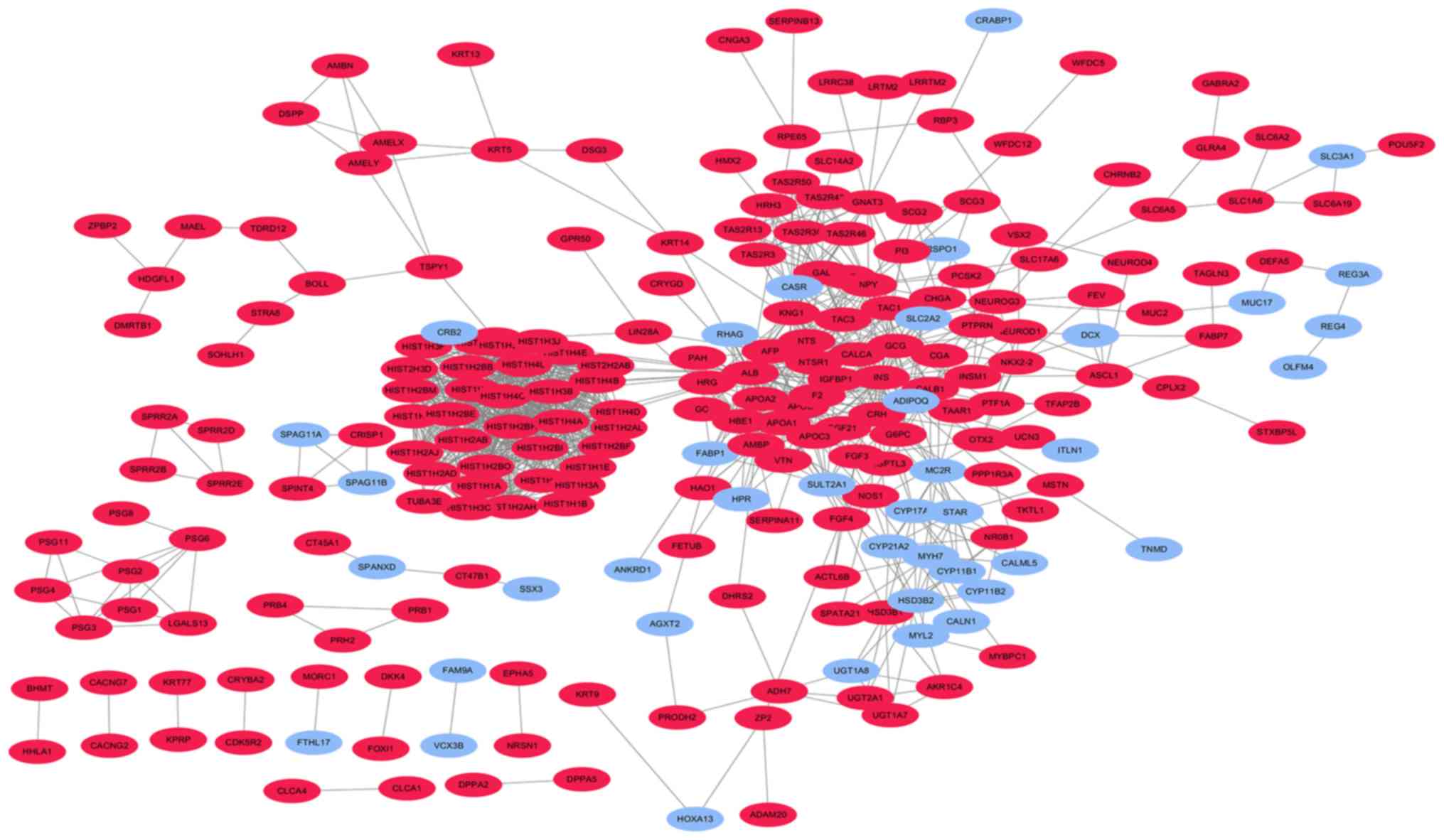
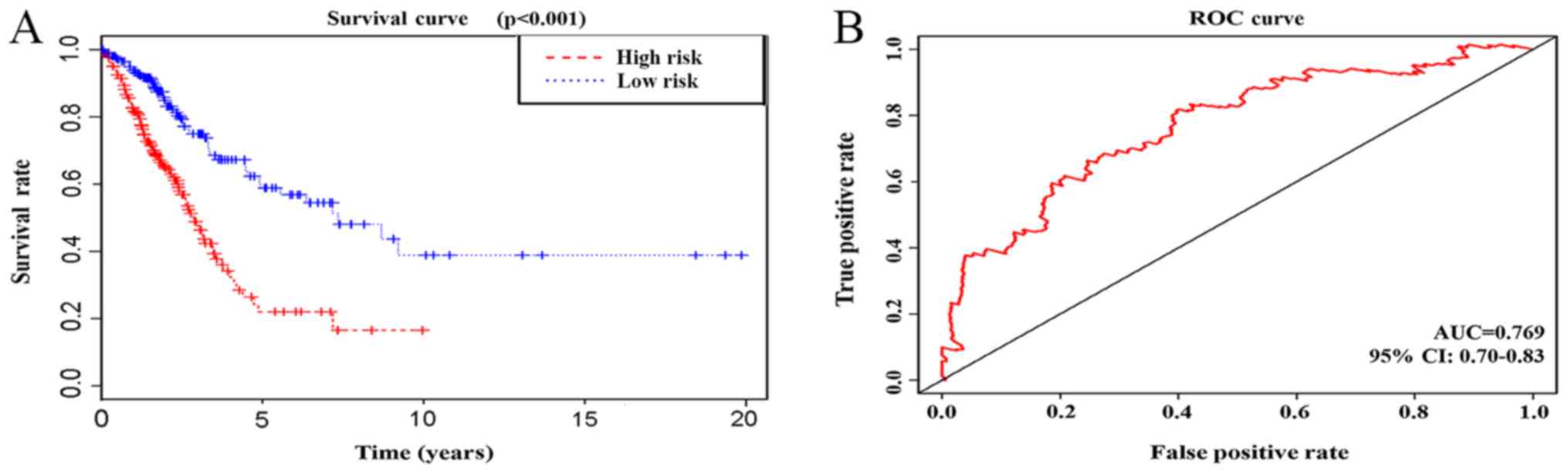
| CALB1, HIST1H4C,
HIST1H1E, HIST1H1B, POU5F2, HIST1H4B, HIST2H2AB, HIST1H4E,
HIST1H2BB, WFDC5, HIST1H4D, HIST1H1D, HIST1H2BI, PNMA5, HIST1H3B,
HIST1H2AB, WFDC12, HIST1H2AJ, TEX19, KIR2DL1, HIST1H2BL, MSTN,
HIST1H2AH, HIST1H2BE, GPR22, HIST1H3C, TAS2R30, NNAT, NTS, APOA1,
GPR52, DHRS2, HIST1H2BM, HIST2H2AC, HIST1H3F, PRH2, HIST1H4A,
HIST1H2BH, HIST1H3J, LRRC38, APOA2, AFP, HIST1H1A, HIST1H3A,
HIST1H2AL, HIST1H3I, PRB4, HIST1H2BO, HIST2H3D, NECAB2, PRB3, CHGA,
HRG, INSM1, TAC3, IFNK, MYT1, MAEL, SCG2, HIST1H4F, PRSS48, ACTN3,
HIST1H4L, C10orf113, NSG2, HIST1H2BF, VTN, IRX4, SPIC, LRRTM2,
TAS2R13, GAL, DPPA2, PSG11, FABP7, TKTL1, SEZ6, ZPBP2, NKX2-3,
PSG1, KCNH6, ADGRB1, GABRA2, TAS2R46, TUBA3E, ADAM20, PSG8,
STXBP5L,4-Mar, OR6T1, ANGPTL3, ZP2, PSG5, F2, TAGLN3, PSG3, HBE1,
FXYD4, SERPINB13, TDRD12, PNMA6E, SPATA21, CDK5R2, BOLL, RPE65,
SPINK4, HIST1H2AD, PTPRN, HMX2, SPRR2E, PBOV1, SLC14A2, SPRR2G,
MAB21L2, CT45A1, AKR1C4, RNF113B, BHMT, PSG2, AMBP, PRSS56, HRH3,
PI3, KRT14, TSPYL6, SLC1A6, CHRNB2, RBM46, TDRD15, MPC1L, XKR7,
ACTL6B, NOS1, CLCA4, PSG7, FGF4, LIPF, KIR3DL2, EPHA5, KRT13,
KCNJ13, C12orf40, OR4A16, FEV, GC, SBSN, DPPA5, CXorf67, LRTM2,
CGA, APOC3, TSPY2, PSG6, KNG1, NEUROD4, FRG2C, NKX2-2, TAS2R50,
CNGA3, KRT5, TAS2R3, CDH9, GCG, APOB, HHLA1, HEPACAM2, KLK13, VSX2,
KRT31, NEUROG3, NTSR1, ADH7, CA6, SLC7A14, MSMB, KRT33A, C6orf10,
FOXI1, VGLL2, SNX31, PTF1A, DKK4, LGALS14, UGT2A1, CLEC2A, TSPY3,
DEFA5, KRT83, BANF2, FETUB, PRB1, TMIGD1, LCE3D, KRT77, TEX13B,
CBLN1, OR51B5, CRISP1, SERPINA11, FAM83C, MYBPC1, NRSN1, RAX,
SPRR2A, KPRP, H3.Y, SCG3, NPY, NLRP11, PPP1R3A, CALY, PAH, FGF3,
DSPP, PSG4, MUC2, CACNG7, AMBN, SOHLH1, INS, SLC6A2, TUNAR,
FAM205C, GPR50, BPIFB4, IGFBP1, G6PC, SPINT4, TAS2R43, KRT9,
TMPRSS11A, ALB, CRYBA2, GMNC, HSD3B1, SLC6A19, ADAMTS19, MORC1,
SLC6A5, RBP3, ADGRG7, SULT1C3, PNMA6F, PAQR9, PRLHR, UCN3, NEUROD1,
HDGFL1, SPRR2D, SRARP, TLE7, FGF21, CERS3, CT45A10, LUZP4, CLCA1,
TAC1, FRG2, S100A7, ZNF560, ZMAT4, SAGE1, SLC17A6, HIST1H2BA,
CACNG2, UGT3A1, AMELY, NTSR2, LCN9, LIN28A, C10orf99, TFAP2B,
OR13H1, GNAT3, UGT1A7, HAO1, TAAR1, LGALS13, DSG3, MAGEA11, CPLX2,
OTX2, RBFOX1, CRH, STRA8, TSPY1, GLRA4, NR0B1, PCSK2, ST8SIA3,
ASCL1, NLRP13, BLID, KRT76, CRYGD, AMELX, PRODH2, DMRTB1, CT47B1,
SPRR2B, CALCA, AC187653.1, OR56A3 |
| ITLN1, PRG4, MYRFL,
CYP17A1, STAR, HSD3B2, MYL2, TNMD, PKHD1L1, ASIC2, FAM9C, BMX,
C21orf62, EBF3, GPR26, FAM9A, PDZRN4, RSPO1, CYP11B1, SLC3A1, CRB2,
CYP4F8, AXDND1, SPAG11B, CYP21A2, CYP11B2, SERTM1, MYH7, RHAG,
MC2R, SSX3, ANKRD1, FABP1, FBN2, EMX2, CALN1, HPR, STAC2, SORCS3,
PCDH8, TUSC5, BARHL2, PRSS38, CEACAM18, OLFM4, DCX, SULT2A1,
SCGB2A2, SPAG11A, AGXT2, CASR, C1orf94, BTNL3, HOXA13, VCX3B, BNC1,
CRABP1, SNTG1, REG3A, DPCR1, REG3G, REG4, SPANXD, SPANXC, MUC17,
ADIPOQ, UGT1A8, SLC2A2, CALML5, TRIM48, FTHL17 |