Introduction
Liver cancer (LC) is the fifth most common type of
cancer and the third leading cause of cancer-associated mortality
worldwide (1). The clinical outcomes
of patients with liver cancer remain poor, which is largely due to
the high frequency of tumor recurrence and distant metastasis
following surgical resection. Therefore, a better understanding of
the molecular mechanisms of liver cancer is necessary to improve
the prognosis of this disease (2).
Several studies have shown that mutual interactions
between the tumor and its microenvironment contribute to tumor
progression (3); uncontrolled
inflammation also serves an important role in promoting malignant
progression and metastasis (4).
Chemokines, as an important class of non-resolving inflammatory
factors, are important in tumor progression and metastasis and
represent a potential tool for tumor detection (5,6).
Increasing experimental evidence has shown that chemokines produced
in the tumor microenvironment serve critical roles in
cancer-related inflammation, and promote invasion and metastasis in
human cancer (7–9). In particular, IL-8 is a multifunctional
CXC chemokine that affects human neutrophil functions including
chemotaxis, enzyme release and the expression of surface adhesion
molecules, and promotes tumor angiogenesis and metastasis through
binding to its receptors CXCR1 and CXCR2 (10).
CXCR1 and CXCR2, as two important members of the CXC
chemokine receptor family, were successfully cloned by Holmes et
al (11) and Murphy and Tiffany
(12), respectively. They are
located on chromosome 2q35 and have a homology of ≤77%. CXCR1 and
CXCR2 combine with their common ligand, IL-8, with a high affinity
and then induce leukocyte chemotaxis, calcium ion flow, cell
proliferation and migration, and the regulation of angiogenesis
(13). Numerous studies have
suggested that CXCR1/2 is overexpressed in a wide variety of cancer
types (14,15), and serves a critical role in the
pathogenesis, growth, invasion, metastasis, angiogenesis and drug
resistance of malignant melanoma (16), breast cancer (17), prostate cancer (18), pancreatic cancer, colon cancer
(19), gastric carcinoma (20) and epithelial ovarian cancer (21).
Previous studies have shown that IL-8 and its
receptors have an important regulatory role in the occurrence,
proliferation, invasion and metastasis of cancer, and are involved
in the regulation of tumor stem cell self-renewal. However, little
is known about the expression and functional roles of IL-8, CXCR1
and CXCR2 in the progression of liver cancer, or the downstream
signaling pathways that mediate IL-8-directed migration in liver
cancer. In order to investigate the expression and functional roles
of IL-8, CXCR1 and CXCR2 in the proliferation and invasion of liver
cancer, the present study aimed to investigate the effect of their
expression and define their roles in vitro.
Materials and methods
Human tissues
A total of 46 patients with liver cancer, who
underwent surgery conducted by the same surgical team in the
Department of the Fourth Affiliated Hospital of Anhui Medical
University (Hefei, China) between October 2014 and September 2015
and agreed to participate in the study, were recruited. There were
25 men and 20 women, with an age range of 37–71 years old. Each
subject had undergone a percutaneous liver biopsy at the Fourth
Affiliated Hospital of Anhui Medical University. The standards for
diagnosis of liver cancer have been previously described in The
Standard for Diagnosis and Treatment of Primary Hepatocellular
Carcinoma (2011 edition) (22). A
total of 30 patients with liver cirrhosis, including 17 men and 13
women, with an age range of 33–71 years old, were considered for
analysis. All cases were diagnosed using pathological, CT,
ultrasound and clinical data. Another 28 blood donors from the City
Center Blood Station with normal physical examination results, were
selected as normal controls, which comprised 16 men and 12 women,
aged between 28 and 69 years old.
Cell lines
The Huh-7 and HepG2 human liver cancer cell lines
were purchased from the Shanghai Institute of Biological Sciences,
Chinese Academy of Sciences. The HepG2 cell line used in the
present study has been authenticated by STR profiling. The human
Huh-7 and HepG2 cell lines were cultured in DMEM (Hyclone;
GEHealthcare Life Sciences) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5%
CO2. In order to observe the effect of IL-8 on the
migration and invasion of liver cancer cell lines, the Huh-7 and
HepG2 cells (1.0×106) were incubated with IL-8 at 37°C
for 24 and 48 h. In certain cases, the Huh-7 and HepG2 cells were
pretreated with 5 µM anti-CXCR1 (1:400, cat. no. ab137351; Abcam)
or anti-CXCR2(1:400; cat. no. ab14935; Abcam) for 30 min at 37°C to
inhibit the function of CXCR1 or CXCR2, and the migration and
invasion were measured in vitro via a wound healing assay
and Transwell assay.
Wound healing assay
For the wound healing migration assay,
5×105 cells/well were seeded in 6-well plates and
incubated until 90% confluence. The cell monolayer was scratched
with a fine pipette tip, and then cultured in serum-free medium
containing IL-8 for a further 24 and 48 h. Images were captured
along the scrape line using a microscope (cat. no. CKX31; Olympus
Corporation). The results are expressed as the relative scratch
width, based on the distance migrated relative to the original
scratch width. The experiment was performed in triplicate.
Migration and invasion assays
The migration and invasion assays were performed
using a QCM 24-Well Cell Invasion Assay kit with 8-µm membranes
(EMD Millipore). The cells (1×105 cells/well) in 150 µl
serum-free medium were seeded into the upper chamber. The same
medium containing IL-8 was used as a chemoattractant in the lower
chamber. Following culture for 48 h at 37°C, the upper surface of
the Transwell membrane was wiped gently with a cotton swab to
remove non-migrating cells. Those cells that had invaded to the
lower surface of the membrane were fixed in methanol and stained
with 0.1% crystal violet solution, followed by image capture and
counting under an inverted microscope.
Detection of the mRNA expression
levels of CXCR1, CXCR2 and IL-8 by reverse transcription
(RT)-semi-quantitative PCR
The hepatic mRNA expression levels of CXCR1, CXCR2
and CXCL8 were assessed by RT-qPCR analysis. The details were as
follows: Total RNA of the tissue samples was extracted from cells
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. The reverse transcription reaction
was performed using the RevertAid First Strand cDNA Synthesis kit.
Equal quantities of cDNA were submitted to PCR in the presence of
SYBR Green Real-time PCR Master mix (Toboyo Life Science) and run
in a real-time PCR detection machine with ABI 7500 PRISM (Applied
Biosystems, Sunnyvale, CA, USA). The PCR conditions were as
follows: Initial denaturation at 94°C for 5 min, followed by 30
cycles of 94°C for 30 sec, annealing at a temperature in accordance
with the primer sequence for 30 sec, and then 72°C for 30 sec, with
a final extension at 72°C for 10 min. The PCR products were
analyzed using 2% agarose gel electrophoresis, stained with
ethidium bromide and visualized under UV illumination. Realtime
qPCR analysis was performed with specific primers for CXCR1, CXCR2
and CXCL8 (Table I). The
housekeeping gene GAPDH was used as an internal control.
Densitometry analysis was performed using Quantity One software
(version 4.62; Bio-Rad Laboratories, Inc.).
 | Table I.Primer sequences of CXCR1, CXCR2 and
GAPDH. |
Table I.
Primer sequences of CXCR1, CXCR2 and
GAPDH.
| Gene | Sense (5→3) | Antisense (5→3) | Length (bp) |
|---|
| CXCR1 |
CAGATCCACAGATGTGGGAT |
AGCAGCCAAGACAAACAAACTT | 468 |
| CXCR2 |
CTTTTCTACTAGATGCCGC |
AGATGCTGAGACATATGAATTT | 417 |
| IL-8 |
CTTTGTCCATTCCCACTTCTGA |
TCCCTAACGGTTGCCTTTGTAT | 306 |
| GAPDH |
ACCACAGTCCATGCCATCAC |
TCCACCACCCTGTTGCTGTA | 452 |
Western blot analysis
In brief, liver cancer cells were lysed using
radioimmunoprecipitation assay buffer (BIOSS) for 30 min on ice.
The cell lysate was centrifuged at 12,000 × g for 20 min at 4°C and
the supernatant was collected. Protein concentration was determined
by BCA protein assay. A total of 30 µg protein/lane was separated
by SDS-PAGE on a 10% gel and transferred onto nitrocellulose filter
membranes. Following blocking with 5% skim milk for 1 h at room
temperature, the membranes were incubated with primary antibodies
overnight at 4°C, and then with secondary antibodies for 2 h at
room temperature. The primary antibodies used were as follows:
Rabbit polyclonal anti-CXCR1 (1:2,000; cat. no. ab137351; Abcam)
and anti-CXCR2 (1:2,000; cat. no. ab14935; Abcam) antibodies;
monoclonal mouse anti-GAPDH (1:2,000; cat. no. TA-08; Zhongshan
Jinqiao Biotechnology) and β-actin (1:2,000, cat. no. TA-09;
Zhongshan Jinqiao Biotechnology, Beijing, China). The goat
anti-mouse (cat. no. ZB2305; 1:10,000) and anti-rabbit secondary
antibodies (cat. no. ZB2301; 1:10,000) were purchased from
Zhongshan Jinqiao Biotechnology. The protein bands were visualized
by ECL reagents (cat. no. C05-07003; BIOSS). Images were captured
and the intensity of the bands was quantitated with the Bio-Rad
Versa Doc imaging system (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data are shown as the mean ± standard deviation.
Student's t-test was used to compare two independent groups of
data. Differences among the three groups were analyzed using
one-way analysis of variance. Multiple comparisons between the
groups were performed using the Bonferroni correction method.
Correlation between continuous variables was determined using
Pearson's correlation coefficient. Chi-square tests were applied to
analyze the relationships among immunohistochemical staining of
IL-8, CXCR1 and CXCR2. All statistical analyses were performed
using SPSS software (version 16.0; SPSS, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Association between the mRNA
expression levels of CXCR1, CXCR2 and IL-8 and the
clinicopathological characteristics of liver cancer
The clinical analysis revealed that the mRNA levels
of CXCR1, CXCR2 and IL-8 had no significant correlation with age or
sex in patients with liver cancer (P>0.05), but were associated
with the depth of tumor infiltration, lymph node or distant
metastasis and TNM stage (P<0.05). The mRNA expression levels of
CXCR1, CXCR2 and IL-8 were significantly increased in patients with
liver cancer with deep invasion, lymph node or distant metastasis
and a late TNM stage (P<0.05; Table
II).
 | Table II.Association between the mRNA
expression of CXCR1, CXCR2 and IL-8 and the clinicopathological
parameters of patients with liver cancer. |
Table II.
Association between the mRNA
expression of CXCR1, CXCR2 and IL-8 and the clinicopathological
parameters of patients with liver cancer.
| Variable | n | CXCR1 mRNA (fold
change) | CXCR2 mRNA (fold
change) | IL8 mRNA (fold
change) |
|---|
| Age (years) |
|
|
|
|
| ≤50 | 19 | 0.9874±0.2197 | 0.8842±0.1911 | 1.7862±0.2875 |
|
>50 | 27 | 0.9389±0.1815 | 0.9059±0.1463 | 1.8119±0.2683 |
| P-value |
| 0.418 | 0.665 | 0.758 |
| Gender |
|
|
|
|
| Male | 26 | 0.9781±0.2081 | 0.8900±0.1816 | 1.8043±0.2782 |
|
Female | 20 | 0.9340±0.1845 | 0.9059±0.1435 | 1.7974±0.2745 |
|
P-value |
| 0.459 | 0.749 | 0.934 |
| Depth of
infiltration |
|
|
|
|
|
T1-T2 | 19 | 0.8721±0.1480 | 0.8057±0.1165 | 1.6271±0.1573 |
|
T3-T4 | 27 | 1.0200±0.2069 | 0.9611±0.1647 | 1.9239±0.2731 |
|
P-value |
| 0.011 | 0.001 | <0.001 |
| Lymph node
metastasis |
|
|
|
|
| No | 21 | 0.8695±0.1559 | 0.8266±0.1203 | 1.6255±0.1590 |
|
Yes | 25 | 1.0340±0.1997 | 0.9560±0.1754 | 1.9490±0.2636 |
|
P-value |
| 0.004 | 0.005 | <0.001 |
| Distant
metastasis |
|
|
|
|
| No | 36 | 0.9056±0.1507 | 0.8528±0.1383 | 1.7004±0.1736 |
|
Yes | 10 | 1.1510±0.2322 | 1.0560±0.1585 | 2.1645±0.2638 |
|
P-value |
| 0.009 | <0.001 | <0.001 |
| TNM stage |
|
|
|
|
|
I+II | 16 | 0.8525±0.1335 | 0.7987±0.1119 | 1.6064±0.1580 |
|
III+IV | 30 | 1.0157±0.20394 | 0.9493±0.1655 | 1.9053±0.2665 |
|
P-value |
| 0.006 | 0.002 | <0.001 |
mRNA expression levels of CXCR1, CXCR2
and IL-8 are increased in the peripheral blood mononuclear cells
(PBMCs) of patients with liver cancer
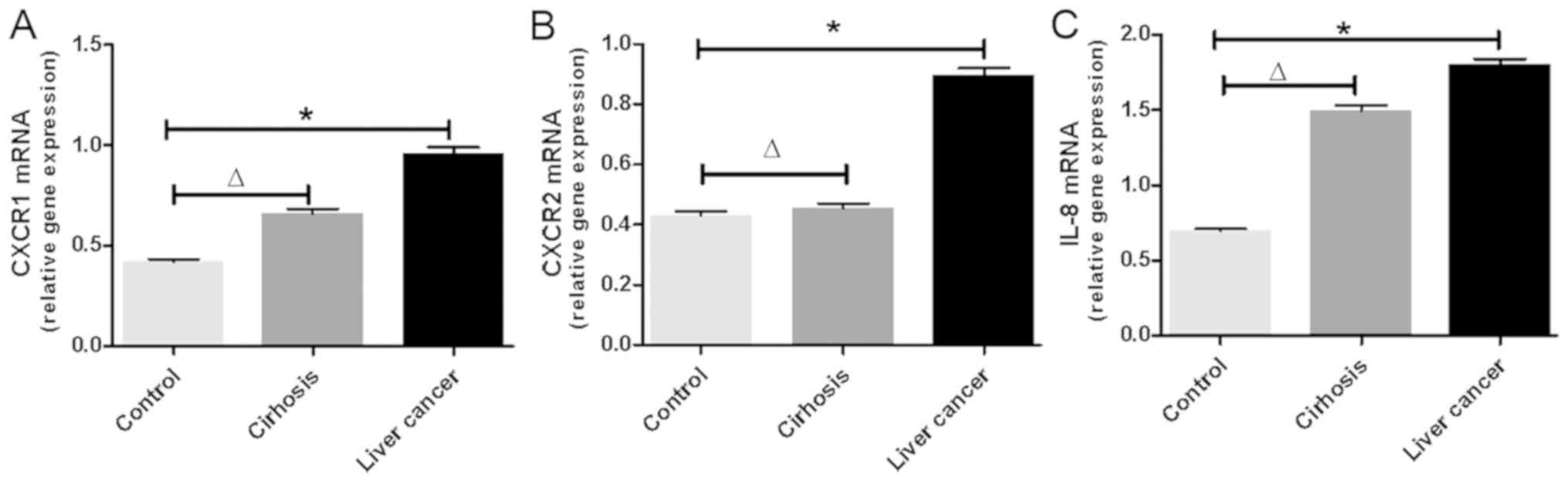
The mRNA expression levels of CXCR1, CXCR2 and IL-8
were detected in each patient with liver cancer. As shown in
Fig. 1A-C, the mRNA levels of CXCR1,
CXCR2 and IL-8 were significantly increased in the PBMCs of
patients with liver cancer compared with those of the controls
(P<0.01). Additionally, it was found that the mRNA levels of
CXCR1 and IL-8 increased with the progress of liver disease. The
mRNA levels of CXCR1 and IL-8 were significantly increased in
patients with cirrhosis compared with those in normal controls
(P<0.05). The mRNA levels of CXCR1 and IL-8 were higher in
patients with liver cancer than in those with cirrhosis. The mRNA
levels of CXCR2 did not differ significantly between the two
groups.
Protein expression of CXCR1, CXCR2 and
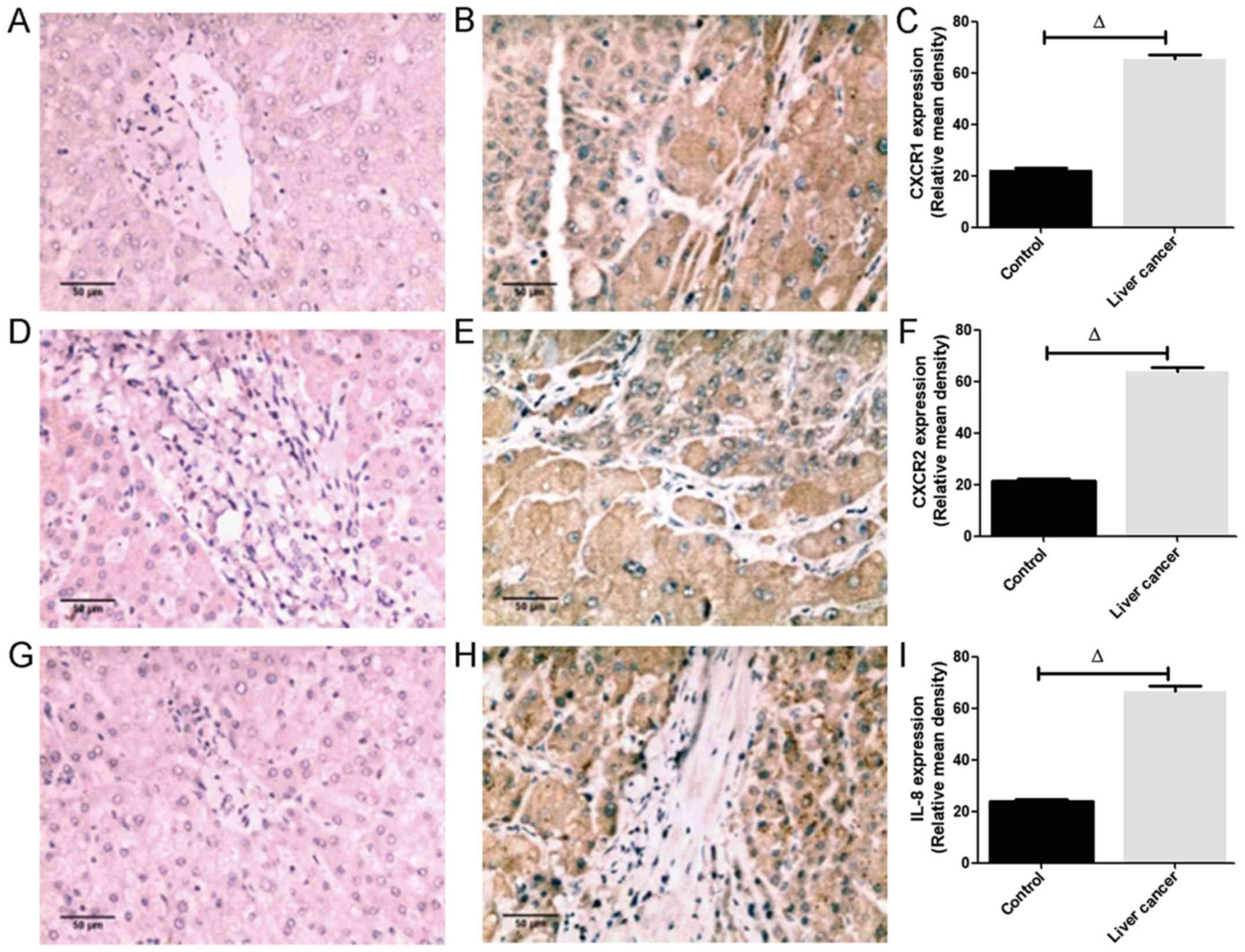
IL-8 is augmented in liver cancer
The immunohistochemical staining (Fig. 2) showed that the CXCR1 (Fig. 2B), CXCR2 (Fig. 2E) and IL-8 (Fig. 2H) proteins were apparent in almost
all hepatocytes, infiltrating inflammatory cells, vascular
endothelial cells and bile duct cells, and CXCR1, CXCR2 and IL-8
were localized in the cytoplasm. In addition, the intensity of
CXCR1, CXCR2 and IL-8 staining in liver cancer tissues was higher
than that in the controls. Image analysis of 30 liver cancer
samples revealed that the positive rate of CXCR1 expression was
86.7% (26/30), in which the proportions of weak and strong
expression accounted for 30% (9/30) and 56.7% (17/30),
respectively. The positive rate of CXCR2 expression was 76.7%
(23/30), in which the proportion of weak and strong expression
accounted for 26.7% (8/30) and 50% (15/30), respectively. The
positive rate of IL-8 expression was 93.3% (28/30), in which the
proportions of weak and strong expression accounted for 33.3%
(10/30) and 60% (18/30), respectively. The positive rates of CXCR1,
CXCR2 and IL-8 in 12 normal hepatic tissues were 33.3% (4/12), 25%
(3/12) and 41.7% (5/12), respectively. The semi-quantitative
analysis revealed that the relative mean densities of hepatic CXCR1
(Fig. 2C), CXCR2 (Fig. 2F) and IL-8 (Fig. 2I) staining in liver cancer were
significantly increased compared with those in normal liver tissues
(P<0.05; Fig. 2A-I).
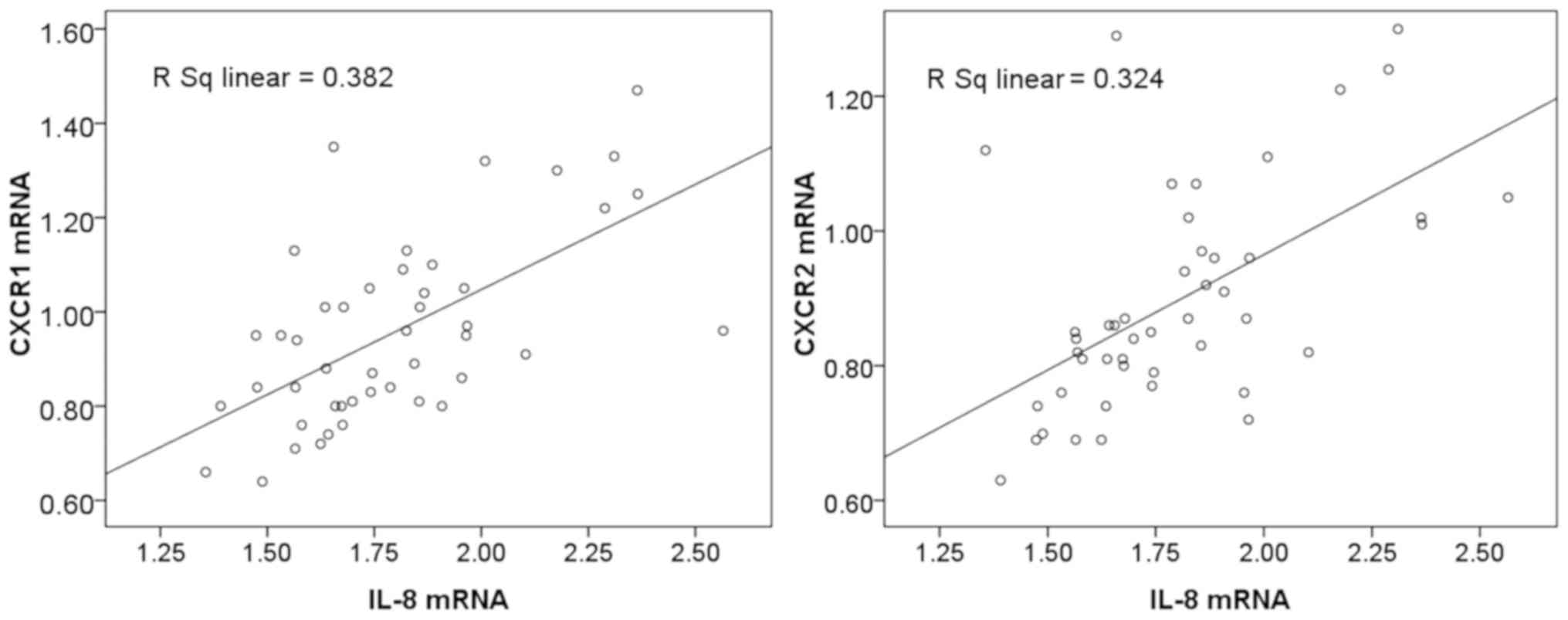
Expression of IL-8 is positively
associated with the expression of CXCR1 and CXCR2 in liver
cancer
The clinical findings in the present study suggested
that the mRNA levels of IL-8, CXCR1 and CXCR2 were increased in
patients with liver cancer, therefore the relationship between the
expression of IL-8 and the expression of CXCR1 and CXCR2 was
further examined. Statistical analysis revealed that the mRNA
expression of IL-8 was positively associated with that of CXCR1
(r=0.618; P<0.05) and CXCR2 (r=0.569; P<0.05). CXCR1 and
CXCR2 mRNA gradually increased with the increased expression of
IL-8 in liver cancer (Fig. 3).
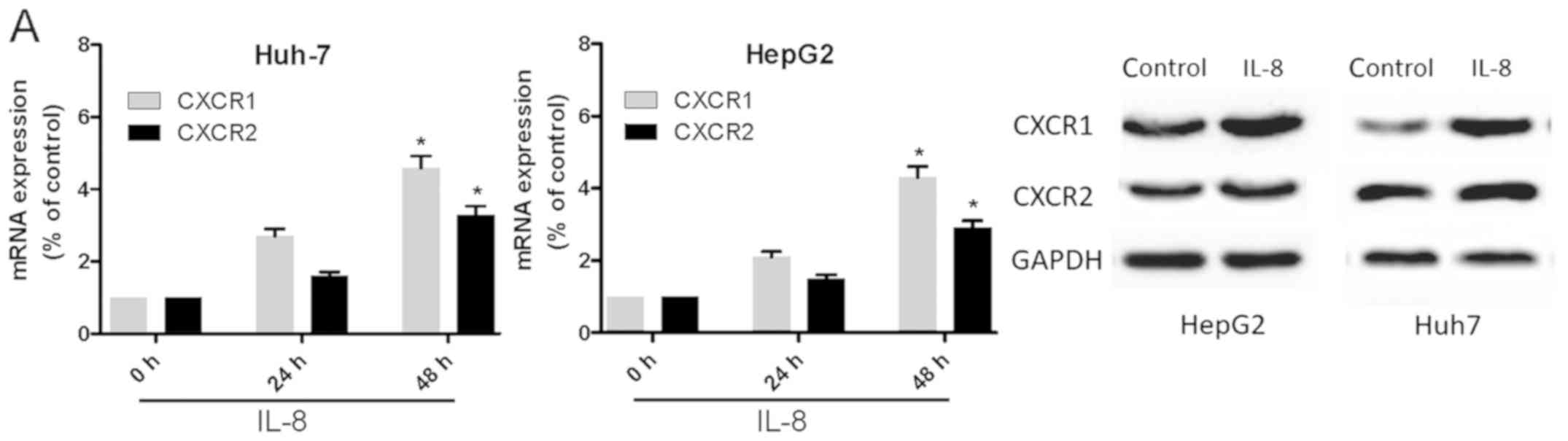
IL-8 promotes human liver cancer cell
invasion and metastasis through CXCR1/2 receptors
To determine whether IL-8 induces the expression of
CXCR1 or CXCR2 in Huh-7 and HepG2 cells, the cell lines were
treated with IL-8 for 24 h and then subjected to RT-qPCR and
western blot analyses (Fig. 4A). It
was found that IL-8 significantly increased the mRNA expression of
CXCR1 and CXCR2. It has been shown that IL-8 directs the migration
and invasion of human liver cancer cells. In the present study, the
effect of IL-8 on the motility of Huh-7 and HepG2 cells was
verified. The wound-healing activity (Fig. 4B and C) and the migration and
invasion of Huh-7 and HepG2 cells (Fig.
4D) were increased by IL-8 after 24 and 48 h. Pretreatment of
the cells with anti-CXCR1 or anti-CXCR2 (5 µM) for 30 min markedly
inhibited IL-8-directed cell migration and invasion (Fig. 4E). This suggested that IL-8 promoted
liver cancer cell migration via the CXCR1 and CXCR2 receptors.
Discussion
Treatment for many patients with early-stage liver
cancer is successful, however, patients with distant metastasis
have a poor prognosis with conventional therapy. When the disease
is recurrent or metastatic, conventional chemotherapy or
radiotherapy has limited benefit. Increasing epidemiologic and
experimental evidence has shown that inflammation serves an
important role in promoting the malignant progression and
metastasis of liver cancer. Chemokines and their receptors produced
in the tumor microenvironment serve critical roles in
cancer-related inflammation and promote invasion and metastasis in
human cancer.
CXCR1 and CXCR2, as two important members of the CXC
chemokine receptor family, have been shown to contribute to human
tumor growth invasion and metastasis through binding their common
ligand IL-8. Considerable data demonstrate that tumor cells express
widely functional chemokine receptors to facilitate tumor
progression. However, there is no direct in vivo evidence
that CXCR1 and CXCR2 are expressed in liver cancer.
In the present study, the mRNA levels of CXCR1,
CXCR2 and IL-8 in PBMCs from 46 patients with liver cancer were
detected by RT-qPCR. It was found that the mRNA levels of CXCR1,
CXCR2 and IL-8 were highest in patients with liver cancer and
lowest in the normal control group (P<0.05). With the
progression of liver cancer, the mRNA levels of CXCR1, CXCR2 and
IL-8 increased gradually, suggesting that these expression changes
were significantly associated with advanced liver cancer
progression. Correlation analysis was performed between IL-8 and
CXCR1/2 in liver cancer. The results revealed that the mRNA
expression of IL-8 was positively associated with that of CXCR1
(r=0.618; P<0.05) and CXCR2 (r=0.569; P<0.05). The mRNA
expression levels of CXCR1 and CXCR2 gradually increased as the
expression of IL-8 was elevated in liver cancer. In addition,
clinical analysis revealed that the mRNA expression levels of
CXCR1, CXCR2 and IL-8 did not significantly correlate with the age
or sex of patients with liver cancer (P>0.05), but were
associated with the depth of tumor infiltration, lymph node or
distant metastasis and TNM stage (P<0.05). The mRNA expression
levels of CXCR1, CXCR2 and IL-8 were significantly increased in
patients with liver cancer with T3-T4, lymph node metastasis,
distant metastasis and advanced tumor stage (pTNM stages III–IV),
suggesting that CXCR1, CXCR2 and IL-8 were associated with the
clinical stage, lymph node metastasis and distant metastasis of
liver cancer. These findings were concordant with those of Ren
et al (23), who detected the
serum levels of IL-8 in 59 patients with liver cancer and 15
healthy subjects using ELISAs. The results showed that the serum
level of IL-8 was significantly elevated in patients with liver
cancer and associated with larger tumor size (>5 cm), absence of
a tumor capsule, presence of venous invasion and advanced
pathological tumor-node-metastasis stage. Akiba et al
(24) provided evidence that HCC
cells are a major producer of IL-8 in tissues, and cases with a
high level of IL-8 in cancerous tissues had a significantly higher
frequency of portal vein invasion, venous invasion and bile duct
invasion. This suggests that IL-8 may serve an important role in
the invasion and metastasis of liver cancer.
The present study also detected the protein
expression of CXCR1, CXCR2 and IL-8 using immunohistochemical
methods. The results showed that the CXCR1, CXCR2 and IL-8 proteins
were mainly expressed in the cytoplasm of hepatoma cells, with
higher intensities than in the normal controls (P<0.01); this
was in accordance with the data presented by Wang et al
(25). The present study found that
the intensity of CXCR1, CXCR2 and IL-8 expressed was statistically
significant between liver cancer and normal controls. These results
further confirmed that the high level of IL-8 secreted by hepatoma
cells induced high expression of CXCR1 and CXCR2 on hepatoma and
inflammatory cells, which, in turn, is involved in the development,
invasion and metastasis of liver cancer.
In order to further verify the hypothesis,
experiments were performed using Huh-7 and HepG2 cell lines. The
cell lines were incubated with IL-8 for 24 h, and the expression of
CXCR1 and CXCR2 was detected by RT-qPCR and western blot analyses.
It was found that IL-8 significantly increased the expression of
CXCR1 and CXCR2 in Huh-7 and HepG2 cell lines. To determine whether
IL-8 promotes liver cancer migration and invasion via CXCR1 and
CXCR2, the Huh-7 and HepG2 cells were incubated with IL-8 for 24
and 48 h, following which the wound-healing activity and the
migration and invasion of Huh-7 and HepG2 cells were increased.
Pretreatment of cells with anti-CXCR1 or anti-CXCR2 (5 µM) for 30
min markedly inhibited the IL-8-directed migration of Huh-7 and
HepG2 cells. Xue et al (26)
analyzed the expression profiles of 18 chemokine receptors on four
HCC cell lines of lower to higher metastatic potentials of
(SMMC-7721, MHCC97-L, MHCC97-H and HCCLM6) using RT-PCR analysis
and found that chemokine receptors are closely associated with the
metastatic potential of HCC. Huang et al (27) demonstrated that the incubation of HCC
cells with IL-8 led to the increased expression of FOXC1 via
activation of phosphoinositide 3-kinase (PI3K) signaling to AKT and
hypoxia-inducible factor 1α. Increased expression of FOXC1 can lead
to the transactivation of CXCR1 and CCL2, promoting inflammation
and the invasive and metastatic abilities of HCC cells. Therefore,
a high level of IL-8 (CXCL8) may regulate local and systemic
inflammatory responses in patients with liver cancer through the
upregulation of CXCR1/2, and be involved in the development of
liver cancer through activation of the PI3K/Akt/HIF-1α signaling
pathway (16). However, the exact
mechanism underlying the involvement of CXCR1, CXCR2 and IL-8 in
liver cancer remains to be elucidated and requires further
investigation.
In conclusion, the results of the present study
demonstrate that the mRNA and protein levels of CXCR1, CXCR2 and
IL-8 in patients with liver cancer were increased, and were
significantly associated with the clinical stage, lymph node
metastasis and distant metastasis. Correlation analysis indicated
that the expression of IL-8was positively associated with the
expression of CXCR1 and CXCR2 in liver cancer. In vitro,
IL-8 was shown to induce Huh-7 and HepG2 cell migration and
invasion via the increased expression of CXCR1 and CXCR2. Based on
these findings, and other relevant reports, IL-8, CXCR1 and CXCR2
may be involved in the invasion and metastasis of liver cancer and
may be useful in identifying patients with more aggressive tumors
for neoadjuvant or adjuvant therapy.
Acknowledgements
Not applicable.
Funding
The current study was supported by Anhui Medical
University (grant no. 2017zhyx14) and the Fund of the Fourth
Affiliated Hospital of Anhui Medical University (grant no.
2015×kj049).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
JS designed the study. HB and YZ performed the
experiments, SW, WF, WH, LY, YX, ZC and MY analyzed the data. HB,
YZ and JS wrote the manuscript. All authors reviewed the
manuscript.
Ethics approval and consent to
participate
The present study was performed in strict accordance
with the Ethics Committee of The Fourth Affiliated Hospital of
Anhui Medical University of Anhui province and written informed
consent was provided by each patient. All methods were performed in
accordance with relevant guidelines and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu SJ: A concise review of updated
guidelines regarding the management of hepatocellular carcinoma
around the world: 2010–2016. Clin Mol Hepatol. 22:7–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Omata M, Cheng AL, Kokudo N, Kudo M, Lee
JM, Jia J, Tateishi R, Han KH, Chawla YK, Shiina S, et al:
Asia-Pacific clinical practice guidelines on the management of
hepatocellular carcinoma: A 2017 update. Hepatol Int. 11:317–370.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McAllister SS and Weinberg RA: The
tumour-induced systemic environment as a critical regulator of
cancer progression and metastasis. Nat Cell Biol. 16:717–727. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang H and Xu X: Mutation-promoting
molecular networks of uncontrolled inflammation. Tumour Biol.
39:10104283177013102017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ruffini PA, Morandi P, Cabioglu N,
Altundag K and Cristofanilli M: Manipulating the
chemokine-chemokine receptor network to treat cancer. Cancer.
109:2392–2404. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karin N: Chemokines and cancer: New immune
checkpoints for cancer therapy. Curr Opin Immunol. 51:140–145.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dayer R, Babashah S, Jamshidi S and
Sadeghizadeh M: Upregulation of CXC chemokine receptor 4-CXC
chemokine ligand 12 axis ininvasive breast carcinoma: A potent
biomarker predicting lymph node metastasis. J Cancer Res Ther.
14:345–350. 2018.PubMed/NCBI
|
|
8
|
Mao TL, Fan KF and Liu CL: Targeting the
CXCR4/CXCL12 axis in treating epithelial ovarian cancer. Gene Ther.
24:621–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bronger H, Karge A, Dreyer T, Zech D,
Kraeft S, Avril S, Kiechle M and Schmitt M: Induction of cathepsin
B by the CXCR3 chemokines CXCL9 and CXCL10 in human breast cancer
cells. Oncol Lett. 13:4224–4230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hosono M, Koma YI, Takase N, Urakawa N,
Higashino N, Suemune K, Kodaira H, Nishio M, Shigeoka M, Kakeji Y
and Yokozaki H: CXCL8 derived from tumor-associated macrophages and
esophageal squamous cell carcinomas contributes to tumor
progression by promoting migration and invasion of cancer cells.
Oncotarget. 8:106071–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Holmes WE, Lee J, Kuang WJ, Rice GC and
Wood WI: Structure and functional expression of a human
interleukin-8 receptor. Science. 253:1278–1280. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murphy PM and Tiffany HL: Cloning of
complementary DNA encoding a functional human interleukin-8
receptor. Science. 253:1280–1283. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bi HJ, Wang J, Huang HS and Liu JH:
Influence of IFN on expression of chemokine receptor CXCR1, CXCR2
and their ligand IL-8 in the patients with chronic hepatitis B. Xi
Bao Yu Fen Zi Mian Yi Xue Za Zhi. 28:422–425. 2012.(In Chinese).
PubMed/NCBI
|
|
14
|
Ha H, Debnath B and Neamati N: Role of the
CXCL8-CXCR1/2 axis in cancer and inflammatory diseases.
Theranostics. 7:1543–1588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singh S, Sadanandam A, Varney ML, Nannuru
KC and Singh RK: Small interfering RNA-mediated CXCR1 or CXCR2
knock-down inhibits melanoma tumor growth and invasion. Int J
Cancer. 126:328–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu H, Gu Y, Xue Y, Yuan M, Cao X and Liu
Q: CXCR2+ MDSCs promote breast cancer progression by
inducing EMT and activated T cell exhaustion. Oncotarget.
8:114554–114567. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Franz JM, Portela P, Salim PH, Berger M,
Fernando Jobim L, Roesler R, Jobim M and Schwartsmann G: CXCR2
+1208 CT genotype may predict earlier clinical stage at diagnosis
in patients with prostate cancer. Cytokine. 97:193–200. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li A, Varney ML and Singh RK: Expression
of interleukin 8 and its receptors in human colon carcinoma cells
with different metastatic potentials. Clin Cancer Res. 7:3298–3304.
2001.PubMed/NCBI
|
|
20
|
Li Z, Wang Y, Dong S, Ge C, Xiao Y, Li R,
Ma X, Xue Y, Zhang Q, Lv J, et al: Association of CXCR1 and 2
expressions with gastric cancer metastasis in ex vivo and tumor
cell invasion in vitro. Cytokine. 69:6–13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yung MM, Tang HW, Cai PC, Leung TH, Ngu
SF, Chan KK, Xu D, Yang H, Ngan HY and Chan DW: GRO-α and IL-8
enhance ovarian cancer metastatic potential via the CXCR2-mediated
TAK1/NFκB signaling cascade. Theranostics. 8:1270–1285. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ministry of Health of the People's
Republic of China, . Standard for diagnosis and treatment of
primary hepatocellular carcinoma (2011 edition). Chin J Hepatol.
20:419–426. 2012.(In Chinese).
|
|
23
|
Ren Y, Poon RT, Tsui HT, Chen WH, Li Z,
Lau C, Yu WC and Fan ST: Interleukin-8 serum levels in patients
with hepatocellular carcinoma: Correlations with
clinicopathological features and prognosis. Clin Cancer Res.
9:5996–6001. 2003.PubMed/NCBI
|
|
24
|
Akiba J, Yano H, Ogasawara S, Higaki K and
Kojiro M: Expression and function of interleukin-8 in human
hepatocellular carcinoma. Int J Oncol. 18:257–264. 2001.PubMed/NCBI
|
|
25
|
Wang J, Han Z and Zhou N: Expression of
CXCL8 and its receptors (CXCR1 and CXCR2) in peripheral blood
neutrophils of chronic hepatitis B. Chin J Immunol. 375–379.
383:2015.
|
|
26
|
Xue TC, Chen RX, Ye SL, Sun RX, Chen J and
Tang ZY: Different expressions of chemokine receptors in human
hepatocellular carcinoma cell lines with different metastatic
potentials. Zhonghua Gan Zang Bing Za Zhi. 15:261–265. 2007.(In
Chinese). PubMed/NCBI
|
|
27
|
Huang W, Chen Z, Zhang L, Tian D, Wang D,
Fan D, Wu K and Xia L: Interleukin-8 induces expression of FOXC1 to
promote transactivation of CXCR1 and CCL2 in hepatocellular
carcinoma cell lines and formation of metastases in mice.
Gastroenterology. 149:1053–1067.e14. 2015. View Article : Google Scholar : PubMed/NCBI
|