Oral cancer is a common and fatal malignancy among
head and neck malignant neoplasms; the number of new cases of oral
cancer globally was 354,864 in 2018 (1). At present, principal treatments of oral
cancer include extensive exeresis of the primary carcinoma, with or
without neck dissection, and pre- or postoperative adjuvant
chemotherapy and radiotherapy (2).
However, the overall 5-year survival rate of patients with oral
cancer was 65%, and the overall 5-year survival rate of patients
with advanced oral cancer was as low as 27% between 2007 and 2013
in the USA (3). Despite the
application of reconstructive radical resection and postoperative
radiotherapy or chemotherapy, the 5-year survival rate of patients
with terminal oral cancer has not improved effectively over the
past years (4–6). Furthermore, the dysphagia,
maxillofacial malformation and dysarthria induced by the
aforementioned therapies negatively affect the quality of life and
psychology of patients (7).
Therefore, there is a requirement to identify more effective
treatment strategies to improve the survival rate and reduce
complications of patients with oral cancer.
MicroRNAs (miRNAs/miRs) have attracted increasing
attention over the past years as their roles in malignant tumors,
where they regulate target genes and downstream signaling pathways,
have been recognized (8). Non-coding
RNA, composed of 18–22 nucleotides, silences corresponding target
genes by binding to the 3′-untranslated regions (3′UTRs) of mRNA to
mediate the biological behavior of cancer cells (9). Dysregulation of miRNA in hepatoma,
lymphoma and colorectal, ovarian and pancreatic cancer has been
previously reported (10–15). In oral carcinoma, miRNAs are
associated with oral carcinomatous cell proliferation, apoptosis,
invasion, metastasis, epithelial-mesenchymal transition (EMT),
chemoresistance, radioresistance and cell cycle arrest. The
abnormal expression of miRNA detected in tumor, serum and saliva
samples obtained from patients with oral cancer has clinical
significance in prognosis prediction and the development of
effective treatments (16–19). The present review discusses recent
developments with regard to miRNAs and their potential clinical
applications in oral cancer.
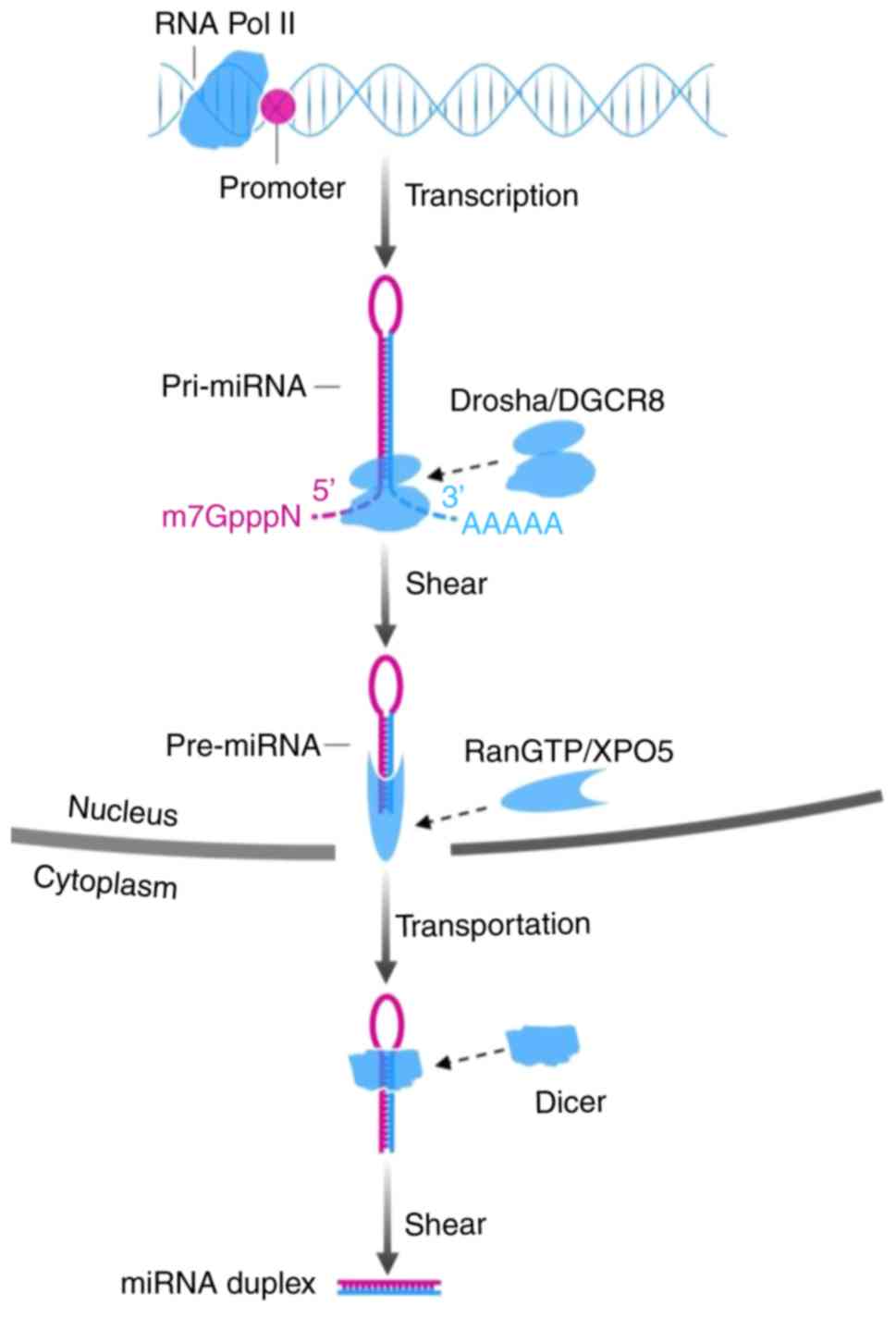
The biogenesis of miRNA involves processes in the
nucleus and cytoplasm. In the nucleus, primary miRNA (pri-miRNA), a
type of miRNA that contains 300–1,000 nucleotides, is generally
issued from the miRNA gene and is transcribed by RNA polymerase II.
Subsequently the long transcript, pri-miRNA, is cleaved by the
drosha ribonuclease III (DROSHA)/DiGeorge syndrome chromosomal
region 8 (DGCR8) complex into a ~70-nt structure termed precursor
miRNA (pre-miRNA), which has lost a 7-methyl guanine nucleoside in
the 5′-capped end and a 3′poly-(A) tail, but has a conserved
stem-loop. Subsequently, the aforementioned stem-loop is sheared by
RNA III enzyme Dicer and double-strand RNA-binding domain protein
after the GTP-binding nuclear protein Ran/exportin-5 (RanGTP/XPO5)
complex carries pre-miRNA to the cytoplasm, to form a double-strand
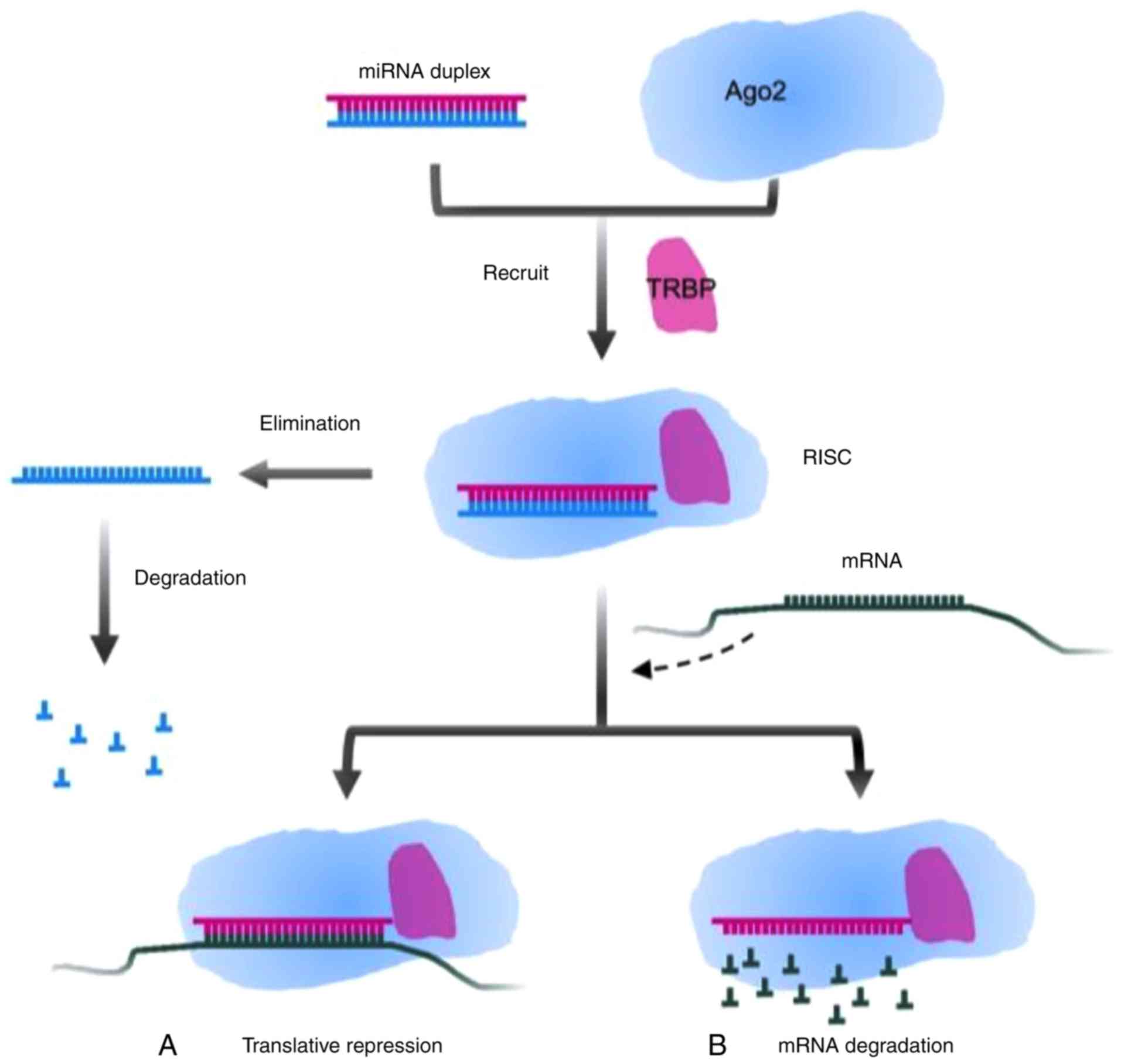
miRNA molecule consisting of 22 nucleotides. Transactivation
response element RNA binding protein (TRBP) recruits the mature
miRNA to RNA-induced silencing complex (RISC) together with
argonaute 2 (Ago2) that has endonuclease activity. A single strand
of the double-strand miRNA is conserved in the RISC, and another
one is degraded (20,21). Partial or complementary pairing
between the RISC and the 3′-UTR of target mRNA functionally plays a
role in repressing the translation of mRNA or degrading target mRNA
(22,23). The mechanisms are presented in
Figs. 1 and 2.
Previous studies suggested that common potentially
malignant disorders and precancerous conditions, including oral
leukoplakia (OL) and oral lichen planus (OLP), were correlated with
aberrant miRNA, but the mechanism of malignant transformation by
miRNA remains unclear (24,25). However, investigating unknown
neoplastic transformations for precancerous lesions or conditions
may provide novel insight into the mechanisms of tumorigenesis. In
addition, the significant differential expression of miRNA between
normal tissue, potential malignant disorders and oral cancerous
specimens indicates that miRNA may be used as an independent
prognostic marker (26,27). Ultimately, a minimally invasive or
non-invasive method, such as the detection of miRNA in saliva or
serum, may be applied in preoperative prediction and postoperative
follow-up (28–30). Indeed, a number of recent studies
demonstrated that the aberrant expression of different miRNAs, such
as miR-195-5p, miR-375, miR-143, miR-26b, miR-155-5p and
miR-483-5p, was associated with oral cancer (31–36).
Therefore, miRNAs may have prognostic and diagnostic value in oral
cancer and may serve as targets for novel therapeutic
strategies.
Previous studies identified that the miR-31
expression level in oral potential malignant disorder (OPMD) is
higher than that in normal oral mucosa, which is correlated with
higher expression of vascular endothelial growth factor and lower
expression of E-cadherin in OPMD. miR-31 expression was further
upregulated in patients with recurrent OPMD and malignant
transformation (37). Another
previous study suggested that aberrant expression of miR-200c was
associated with oral submucous fibrosis (OSF). The overexpression
of miR-200c inhibited collagen gel contraction and migration and
invasion of fibrotic buccal mucosal fibroblasts induced by
arecoline via inhibition of zinc finger E-box binding homeobox 1
(ZEB1). Additionally, reverse transcription-quantitative PCR
(RT-qPCR) analysis revealed that miR-200c was downregulated in 25
OSF samples compared with 25 normal mucosae (38). Brito et al (39) concluded that higher expression of
miR-21 in OL was associated with increased mitotic figures,
incremental nuclear/cytoplasmic ratio and hyperchromasia. Nylander
et al (40) found that miR-21
was upregulated in 30 patients diagnosed with multifocal OLP
compared with 10 healthy subjects, and in agreement, another study
demonstrated that upregulation of miR-21 served a tumor-promoting
role in oral cancer and upregulation of miR-21 was observed in 60
of 79 individuals with the disease (41). Aghbari et al (26) identified that miR-27b and miR-137
levels were downregulated in tissue and saliva samples of patients
with OLP compared with those in normal controls. Among OLP
subgroups, it was demonstrated that miR-137 exhibited the lowest
expression level in the erosive type, suggesting that it serve as a
biomarker for monitoring potential malignant transformation. The
level of miR-375 in progressive lesions was significantly
downregulated compared with that in non-progressive control
lesions, and miR-375 expression was significantly downregulated in
tissues following the transformation of premalignant lesions
(including verrucous hyperplasia and verrucopapillary
hyperkeratosis) into carcinoma, by comparison of premalignant
lesions and oral carcinoma in situ (27). While only 2–3 and 0.4–2% of patients
with OL and OLP, respectively, exhibit malignant transformation
(42,43), further investigation of the roles of
miR-21, miR-27b, miR-137, miR-200c and miR-375 may provide novel
insights into pathways involved in the development of oral
cancer.
Dysregulation of specific miRNAs has been previously
reported in oral cancer. Wang et al (31) reported that miR-195-5p was
significantly downregulated in 40 oral cancerous tissues compared
with non-tumor tissues. Another study reported a distinct
downregulation of miR-375 in 44 cancerous tissues compared with
that in normal mucosae (32).
Furthermore, a previous study revealed that miR-143 was
downregulated in cancerous tissues compared with that in
corresponding adjacent non-cancerous tissues in 81.6% (40/49) of
patients (33). miR-802 was
downregulated in 60.0% (12/20) of tongue squamous cell carcinoma
(TSCC) cases compared with that in normal tissues (44). The expression levels of miR-137
(n=25) and miR-204-5p (n=52) were downregulated in oral cancer
samples compared with those in matched normal tissues (45,46).
Moreover, upregulation of specific miRNAs was observed in oral
cancer tissues. Upregulation of miR-183 was identified in 68.3%
(41/60) of TSCC tissues compared with adjacent non-cancerous
tissues (47). The miR-373-3p
expression level in oral cancerous tissues (n=63) was increased
compared with that in adjacent non-cancerous tissues (48). The miR-155 expression level was
upregulated in oral squamous cell carcinoma (OSCC) tissues (n=46)
compared with that in normal oral mucosa, and the expression level
was increased with increasing Tumor-Node-Metastasis (TNM) stage
(49). miR-31, miR-182, miR-200a and
miR-141 were significantly upregulated in cancerous tissues in 10
patients compared with adjacent non-cancerous tissues (50). The expression of miR-24 was
significantly increased in TSCC tissues of 84 patients compared
with adjacent non-cancerous tissues (51). Liu et al (52) demonstrated that 67% (10/15) of
patients with primary oral cancer had increased miR-1275 expression
in tumor tissues compared with that in adjacent tissues.
Previous studies showed that RNA in saliva is
protected from degradation by binding to macromolecules such as
apoptotic bodies and RISC, a mechanism also observed in plasma and
serum RNAs (53–55). Park et al (56) analyzed saliva by immunoblotting
analysis using an antibody against Ago2, and demonstrated that Ago2
was present in saliva, where it may confer stability to miRNAs.
Furthermore, Park et al (56)
found lower levels of miR-125a and miR-200a in the saliva of
patients with oral cancer (n=12) compared with those in healthy
controls (n=12), suggesting that the aforementioned miRNAs may
serve as stable biomarkers of the disease. Liu et al
(28) reported that the level of
miR-31 in the saliva of patients with OSCC (n=45) prior to surgery
was significantly increased compared with that in healthy subjects
(n=24). Moreover, the miR-31 level in saliva samples was higher
compared with that in plasma samples. The upregulation of miR-31
was detected with high sensitivity even in very small tumors, and
the ability to detect miR-31 levels in the saliva of patients with
small tumors was not significantly different compared with patients
with advanced tumors, suggesting that salivary miR-31 may be
utilized to detect and diagnose oral cancer lesions in high-risk
populations. Zahran et al (29) reported a significant upregulation in
salivary miR-21 and miR-184 levels in patients with oral cancer
compared with those in healthy controls. Specifically, a four-fold
increase in miR-21, with 65% specificity and 65% sensitivity, and a
three-fold increase in miR-184, with 75% specificity and 80%
sensitivity, were observed. The expression of salivary miR-145 was
significantly decreased in patients with OSCC compared with
clinically healthy controls, with 70% specificity and 60%
sensitivity. Ries et al (30)
reported that miR-3651 and miR-494 levels were upregulated, while
the miR-186 level was significantly downregulated in whole blood
samples of patients with recurrent tumors compared with
non-recurrent controls. Therefore, miRNAs may be promising
candidates for the development of diagnostic tools for oral
cancer.
Certain miRNAs are known to be downregulated in
tumor tissues compared with noncancerous tissues. Lower levels of
miR-195-5p were associated with higher pathological differentiation
grade (31). In comparison with
patients without lymph node metastases (n=26), patients with lymph
node metastases (n=18) had significantly downregulated miR-375.
Furthermore, the overall survival of patients with oral cancer with
low miR-375 expression (n=19) was lower than that of patients with
high miR-375 expression (n=25) (32). Cao et al (34) revealed that advanced clinical stage
and large tumor size of oral cancer were associated with low
miR-26b expression. The 5-year survival rates of patients with low
and high miR-26b levels were 26.7 and 53.3%, respectively.
On the other hand, certain miRNAs are upregulated in
tumor tissues compared with adjacent non-cancerous tissues.
Upregulation of miR-183 in patients with oral cancer markedly
shortened the overall survival time and increased the risk of poor
prognosis by 5.666 times. Upregulation of miR-21 resulted in a
higher risk of short survival time (47). The expression of miR-373-3p was
higher in primary tumors with metastases compared with that in
tumors with no metastases (48). The
upregulation of miR-24 was associated with advanced clinical stage
in patients with oral cancer (51).
A positive correlation between a high miR-155-5p level and cervical
lymphatic metastases was observed, and the survival analysis of
carcinomatous recurrence and metastasis identified an association
between high miR-155-5p expression and a poor survival rate (n=73);
miRNA-155-5p may be considered as a specific factor resulting in a
worse prognosis (35).
Serum miR-483-5p was higher in patients with oral
cancer (n=101) compared with healthy controls (n=103); the survival
rates of patients with high miR-483-5p serum level (n=43;
>3.23-fold higher compared with healthy controls) were decreased
compared with that of patients with lower miR-483-5p serum levels
(n=42; <3.23-fold higher compared with healthy controls), and
multivariate analyses for overall survival suggested that a high
miR-483-5p serum level was an independent prognostic indicator
(36). Furthermore, patient
follow-up revealed that patients with higher blood miR-372 levels
had more extensive primary tumors, a greater tendency of node
metastases, a more terminal stage and higher mortality rates
(57). Additionally, miR-372 was
downregulated in plasma and saliva among postoperative patients
compared with preoperative patients (57). By measuring salivary miR-31, it was
determined that 86.4% (19/22) of patients exhibited a significant
decrease in miR-31 levels following tumor resection (28). miR-372 and miR-31 may therefore serve
as biomarkers for the evaluation of surgical efficacy (28,57). Sun
et al (58) suggested that
serum miR-9 is an independent prognostic factor for oral cancer, as
downregulated miR-9 was associated with lymph node metastases,
advanced TNM stage and poor prognosis. Patients with low serum
miR-9 expression had a worse disease-free survival rate compared
with patients with high miR-9 expression (26.5 and 54.6%,
respectively). The overall survival rate of patients with low and
high miR-9 expression was 42.9 and 67.3%, respectively. By
determining levels of miRNAs in the serum or saliva of patients,
miRNAs may be used in minimally or non-invasive methods to predict
lymph node metastasis and assess the prognosis of patients with
oral cancer. Table I presents the
aberrant levels of other miRNAs in the saliva, blood, serum and
plasma (28–30,59–68).
As a salivary gland-derived malignancy,
mucoepidermoid carcinoma may occur in the oral cavity. Shin et
al (69) reported that the
overexpression of miR-127-3p led to an increase in the number of
cells in the G1 phase, indicating that miR-127-3p
resulted in G1/S cell cycle arrest in vitro.
miR-127-3p-induced cell cycle arrest in mucoepidermoid carcinoma
MC-3 cells was associated with the increase of cyclin-dependent
kinase inhibitor 1A and interferon α inducible protein 27
expression via the regulation of Sp1 transcription factor (69). Binmadi et al (70) found that miR-302a was significantly
increased in mucoepidermoid carcinoma tissues compared with normal
tissues. Furthermore, upregulated miR-302a induced invasion of
mucoepidermoid carcinoma cells in vitro.
Adenoid cystic carcinoma (ACC) generally occurs in
the minor salivary glands and has a poor long-term prognosis due to
perineural invasion and lung metastasis (71). Wang et al (72) analyzed the expression of miR-130a in
21 patients with ACC and corresponding normal salivary gland
tissues. Compared with that in normal salivary gland tissues, the
expression of miR-130a in ACC tissues increased by 1.58–29.1 times.
In addition, the level of miR-130a was negatively correlated with
N-myc downstream-regulated gene 2 in ACC tissues. Chen et al
(73) analyzed miRNAs during the
metastasis of ACC cells and found that the expression levels of
miR-4487, miR-4430 and miR-486-3p were upregulated, and the
expression levels of miR-5191, miR-3131 and miR-211-3p were
downregulated. Andreasen et al (74) found that high expression levels of
miR-21, miR-181a-2 and miR-152 in patients with ACC was associated
with a decreased overall survival rate, and high expression of
miR-374c was associated with an improved relapse-free survival
rate. Wang et al (75) found
that an miR-21 inhibitor significantly reduced the resistance of
lung metastatic salivary adenoid cystic carcinoma cells (SACC-LM)
to simvastatin. Furthermore, the combination of simvastatin and
miR-21 inhibitor decreased the proliferation of SACC-LM cells
(75).
Bioinformatics analysis may be used to predict the
pairing sequences of miRNAs and target genes, which may be verified
by luciferase reporter assays. Western blotting and RT-qPCR may
subsequently be used to detect protein and miRNA expression,
respectively. Frequently used oral cancer cell lines include SCC-9,
SCC-4, Tca-8113 and Cal27 (45,48,49,51).
Cell experiments demonstrated that specific miRNAs served an
anticancer role such as miR-138, miR-200c, miR-15b, miR-485-5p and
miR-340 (83,87,89,91,109);
whereas other miRNAs stimulated oral cancer by regulating cellular
proliferation, apoptosis, invasion, EMT, metastasis,
radiosensitivity, chemosensitivity and glucose metabolism, such as
miR-221, miR-455-5p, miR-27a-3p, miR-21 and miR-10a (114,117,126,128,130).
The results obtained from in vitro cell experiments may
provide novel therapeutic targets for potential clinical
application.
miRNAs are promising therapeutic targets, as they
serve important roles in cancer. Theoretically, by silencing
tumor-promoting miRNAs and inducing the expression of
tumor-suppressing miRNAs synchronously, it is possible to treat
oral carcinoma. Specific miRNAs have multiple target genes, for
example, miR-375 targets IGF1R (32), platelet-derived growth factor subunit
A (131), Kruppel-like factor 5
(80) and solute carrier family 7
member 11 (81); miR-138 targets AKT
serine/threonine kinase 1 (82) and
yes-associated protein 1 (83);
miR-203 targets B lymphoma Mo-MLV insertion region 1 homolog
(84), semaphorin 6A (85) and
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(86); and miR-221 targets tissue
inhibitor of metalloproteinase 3 (114) and phosphatase and tensin homolog
(PTEN) (115). A possible treatment
strategy may involve anticancer drugs that selectively regulate the
expression of such miRNAs. Certain genes or signaling pathways are
regulated by two or more miRNAs, for instance, tripartite motif
containing 14 is targeted by miR-195-5p (31) and miR-15b (89); IGF1R is targeted by miR-98 (132) and miR-375 (32); ZEB1 is targeted by miR-205 (79) and miR-200c (87); and PTEN is targeted by miR-221/222
(115) and miR-24 (116). Therefore, the combination of more
specific inhibitors or activators of cancer-associated genes or
signaling pathways may be a suitable therapeutic strategy. With
regard to controversial miRNAs, including miR-222, which served an
ambivalent role in disparate experiments, more research is required
to verify their roles in oral cancer (88,113,115).
On the basis of current research, the aberrant
expression of miRNAs has been demonstrated to be significantly
associated with oral cancer. As either a tumor marker or a
therapeutic target, miRNA has potential to diagnose or treat oral
cancer and improve survival. miRNA serves important roles in the
occurrence, development, therapy and prognosis of oral cancer, and
is a promising target for clinical application. In terms of the
mechanism of malignant transformation or oncogenicity, prognostic
and diagnostic value, and potential as a therapeutic target, miR-31
seems to be a promising candidate for clinical application. miR-31
is differentially expressed in normal mucosa, OPMD and oral cancer,
and may be detected with high sensitivity in tissue, saliva and
plasma. Furthermore, miR-31 may be used to evaluate surgical
efficacy. However, miR-375 and miR-203 may be superior therapeutic
targets, as they target multiple genes that regulate additional
factors and malignant biological properties in oral cancer.
There are a number of challenges in the experimental
research and clinical application of miRNA. The transcriptional
activation of miRNA and the regulation of indispensable components
(containing Drosha/DGCR8, Dicer, XPO5 and TRBP) in the maturation
process of oncogenic or antineoplastic miRNA through signaling
pathways, and the interference of signaling pathways by mature
miRNA, form a series of feedback loops, which may either contribute
to tumorigenesis or be used for effective treatments. Further
clinical trials that explore specific or highly sensitive miRNA
closely associated with oral cancer are required to identify
biomarkers with prognostic value. Combining multiple miRNAs for
diagnosis and therapy is also a promising strategy that requires
further examination, and investigating the association between oral
cancer subtypes and miRNA may facilitate the development of
targeted medicine in oral cancer. Identifying the detection
threshold of different miRNAs in serum, specimen and saliva may aid
in predicting the risk of malignant transformations and in
evaluating the risk of tumor metastasis or relapse. In addition to
further elucidating the mechanisms and anticancer strategies
targeted at miRNA, the potential resistance and complications of
new antitumor drugs are novel challenges to overcome in order to
identify more effective treatments for oral cancer.
Not applicable.
The present study was supported by the Chongqing
Municipal Health Bureau (grant no. 2017HBRC004) and the Natural
Science Foundation Project of CQ CSTC (grant no.
cstc2018jcyjAX0763).
Not applicable.
YL designed the review and revised the manuscript.
CF wrote the manuscript. Both authors reviewed the final version
and approved it for publication.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Montero PH and Patel SG: Cancer of the
oral cavity. Surg Oncol Clin N Am. 24:491–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhong LP, Zhang CP, Ren GX, Guo W, William
WN Jr, Sun J, Zhu HG, Tu WY, Li J, Cai YL, et al: Randomized phase
III trial of induction chemotherapy with docetaxel, cisplatin, and
fluorouracil followed by surgery versus up-front surgery in locally
advanced resectable oral squamous cell carcinoma. J Clin Oncol.
31:744–751. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sadighi S, Keyhani A, Harirchi I, Garajei
A, Aghili M, Kazemian A, Motiee Langroudi M, Zendehdel K and
Nikparto N: Neoadjuvant chemotherapy for locally advanced squamous
carcinoma of oral cavity: A pilot study. Acta Med Iran. 53:380–386.
2015.PubMed/NCBI
|
|
6
|
Bossi P, Lo Vullo S, Guzzo M, Mariani L,
Granata R, Orlandi E, Locati L, Scaramellini G, Fallai C and
Licitra L: Preoperative chemotherapy in advanced resectable OCSCC:
Long-term results of a randomized phase III trial. Ann Oncol.
25:462–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Valdez JA and Brennan MT: Impact of oral
cancer on quality of life. Dent Clin North Am. 62:143–154. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Y, Cao XY, Li YN, Qiu YY, Li YN, Li W
and Wang H: Reversal of cisplatin resistance by
microRNA-139-5p-independent RNF2 downregulation and MAPK inhibition
in ovarian cancer. Am J Physiol Cell Physiol. 315:C225–C235. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gong R, Lv X and Liu F: MiRNA-17 encoded
by the miR-17-92 cluster increases the potential for steatosis in
hepatoma cells by targeting CYP7A1. Cell Mol Biol Lett. 23:162018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ruhl R, Rana S, Kelley K, Espinosa-Diez C,
Hudson C, Lanciault C, Thomas CR Jr, Liana Tsikitis V and Anand S:
MicroRNA-451a regulates colorectal cancer proliferation in response
to radiation. BMC Cancer. 18:5172018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Y, Sun Y, Wang H, Li H, Zhang M, Zhou
L, Meng X, Wu Y, Liu P, Liu X, et al: MicroRNA-221 induces
autophagy through suppressing HDAC6 expression and promoting
apoptosis in pancreatic cancer. Oncol Lett. 16:7295–7301.
2018.PubMed/NCBI
|
|
14
|
Anastasiadou E, Stroopinsky D, Alimperti
S, Jiao AL, Pyzer AR, Cippitelli C, Pepe G, Severa M, Rosenblatt J,
Etna MP, et al: Epstein-Barr virus-encoded EBNA2 alters immune
checkpoint PD-L1 expression by downregulating miR-34a in B-cell
lymphomas. Leukemia. 33:132–147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anastasiadou E, Faggioni A, Trivedi P and
Slack FJ: The nefarious nexus of noncoding RNAs in cancer. Int J
Mol Sci. 19:20722018. View Article : Google Scholar
|
|
16
|
Lu L, Xue X, Lan J, Gao Y, Xiong Z, Zhang
H, Jiang W, Song W and Zhi Q: MicroRNA-29a upregulates MMP2 in oral
squamous cell carcinoma to promote cancer invasion and
anti-apoptosis. Biomed Pharmacother. 68:13–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu Q, Sun Q, Zhang J, Yu J, Chen W and
Zhang Z: Downregulation of miR-153 contributes to
epithelial-mesenchymal transition and tumor metastasis in human
epithelial cancer. Carcinogenesis. 34:539–549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arantes LMRB, De Carvalho AC, Melendez ME
and Lopes Carvalho A: Serum, plasma and saliva biomarkers for head
and neck cancer. Expert Rev Mol Diagn. 18:85–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chai L, Yuan Y, Chen C, Zhou J and Wu Y:
The role of long non-coding RNA ANRIL in the carcinogenesis of oral
cancer by targeting miR-125a. Biomed Pharmacother. 103:38–45. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chendrimada TP, Gregory RI, Kumaraswamy E,
Norman J, Cooch N, Nishikura K and Shiekhattar R: TRBP recruits the
Dicer complex to Ago2 for microRNA processing and gene silencing.
Nature. 436:740–744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yeom KH, Lee Y, Han J, Suh MR and Kim VN:
Characterization of DGCR8/Pasha, the essential cofactor for Drosha
in primary miRNA processing. Nucleic Acids Res. 34:4622–4629. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen HC, Tseng YK, Chi CC, Chen YH, Yang
CM, Huang SJ, Lee YC, Liou HH, Tsai KW and Ger LP: Genetic variants
in microRNA-146a (C>G) and microRNA-1269b (G>C) are
associated with the decreased risk of oral premalignant lesions,
oral cancer, and pharyngeal cancer. Arch Oral Biol. 72:21–32. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Philipone E, Yoon AJ, Wang S, Shen J, Ko
YC, Sink JM, Rockafellow A, Shammay NA and Santella RM:
MicroRNAs-208b-3p, 204–5p, 129-2-3p and 3065-5p as predictive
markers of oral leukoplakia that progress to cancer. Am J Cancer
Res. 6:1537–1546. 2016.PubMed/NCBI
|
|
26
|
Aghbari SMH, Gaafar SM, Shaker OG, Ashiry
SE and Zayed SO: Evaluating the accuracy of microRNA-27b and
microRNA-137 as biomarkers of activity and potential malignant
transformation in oral lichen planus patients. Arch Dermatol Res.
310:209–220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harrandah AM, Fitzpatrick SG, Smith MH,
Wang D, Cohen DM and Chan EK: MicroRNA-375 as a biomarker for
malignant transformation in oral lesions. Oral Surg Oral Med Oral
Pathol Oral Radiol. 122:743–752.e1. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu CJ, Lin SC, Yang CC, Cheng HW and
Chang KW: Exploiting salivary miR-31 as a clinical biomarker of
oral squamous cell carcinoma. Head Neck. 34:219–214. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zahran F, Ghalwash D, Shaker O, Al-Johani
K and Scully C: Salivary microRNAs in oral cancer. Oral Dis.
21:739–747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ries J, Baran C, Wehrhan F, Weber M,
Neukam FW, Krautheim-Zenk A and Nkenke E: Prognostic significance
of altered miRNA expression in whole blood of OSCC patients. Oncol
Rep. 37:3467–3474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang T, Ren Y, Liu R, Ma J, Shi Y, Zhang L
and Bu R: MiR-195-5p suppresses the proliferation, migration, and
invasion of oral squamous cell carcinoma by targeting TRIM14.
Biomed Res Int. 2017:73781482017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang B, Li Y, Hou D, Shi Q, Yang S and Li
Q: MicroRNA-375 inhibits growth and enhances radiosensitivity in
oral squamous cell carcinoma by targeting insulin like growth
factor 1 receptor. Cell Physiol Biochem. 42:2105–2117. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu P, Li Y, Yang S, Yang H, Tang J and Li
M: Micro-ribonucleic acid 143 (MiR-143) inhibits oral squamous cell
carcinoma (OSCC) cell migration and invasion by downregulation of
phospho-c-Met through targeting CD44 v3. Oral Surg Oral Med Oral
Pathol Oral Radiol. 120:43–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao J, Guo T, Dong Q, Zhang J and Li Y:
MiR-26b is downregulated in human tongue squamous cell carcinoma
and regulates cell proliferation and metastasis through a
COX-2-dependent mechanism. Oncol Rep. 33:974–980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Baba O, Hasegawa S, Nagai H, Uchida F,
Yamatoji M, Kanno NI, Yamagata K, Sakai S, Yanagawa T and Bukawa H:
MicroRNA-155-5p is associated with oral squamous cell carcinoma
metastasis and poor prognosis. J Oral Pathol Med. 45:248–255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu H, Yang Y, Zhao H, Yang X, Luo Y, Ren
Y, Liu W and Li N: Serum miR-483-5p: A novel diagnostic and
prognostic biomarker for patients with oral squamous cell
carcinoma. Tumour Biol. 37:447–453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hung KF, Liu CJ, Chiu PC, Lin JS, Chang
KW, Shih WY, Kao SY and Tu HF: MicroRNA-31 upregulation predicts
increased risk of progression of oral potentially malignant
disorder. Oral Oncol. 53:42–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu MY, Yu CC, Chen PY, Hsieh PL, Peng CY,
Liao YW, Yu CH and Lin KH: MiR-200c inhibits the
arecoline-associated myofibroblastic transdifferentiation in buccal
mucosal fibroblasts. J Formos Med Assoc. 117:791–797. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Brito JA, Gomes CC, Guimarães AL, Campos K
and Gomez RS: Relationship between microRNA expression levels and
histopathological features of dysplasia in oral leukoplakia. J Oral
Pathol Med. 43:211–216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nylander E, Ebrahimi M, Wahlin YB, Boldrup
L and Nylander K: Changes in miRNA expression in sera and
correlation to duration of disease in patients with multifocal
mucosal lichen planus. J Oral Pathol Med. 41:86–89. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ren W, Wang X, Gao L, Li S, Yan X, Zhang
J, Huang C, Zhang Y and Zhi K: MiR-21 modulates chemosensitivity of
tongue squamous cell carcinoma cells to cisplatin by targeting
PDCD4. Mol Cell Biochem. 390:253–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arnaoutakis D, Bishop J, Westra W and
Califano JA: Recurrence patterns and management of oral cavity
premalignant lesions. Oral Oncol. 49:814–817. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mortazavi H, Baharvand M and Mehdipour M:
Oral potentially malignant disorders: An overview of more than 20
entities. J Dent Res Dent Clin Dent Prospects. 8:6–14.
2014.PubMed/NCBI
|
|
44
|
Wu X, Gong Z, Sun L, Ma L and Wang Q:
MicroRNA-802 plays a tumour suppressive role in tongue squamous
cell carcinoma through directly targeting MAP2K4. Cell Prolif. Mar
20–2017.(Epub ahead of print). doi: 10.1111/cpr.12336. View Article : Google Scholar
|
|
45
|
Sun L, Liang J, Wang Q, Li Z, Du Y and Xu
X: MicroRNA-137 suppresses tongue squamous carcinoma cell
proliferation, migration and invasion. Cell Prolif. 49:628–635.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang X, Li F and Zhou X: MiR-204-5p
regulates cell proliferation and metastasis through inhibiting
CXCR4 expression in OSCC. Biomed Pharmacother. 82:202–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Supic G, Zeljic K, Rankov AD, Kozomara R,
Nikolic A, Radojkovic D and Magic Z: MiR-183 and miR-21 expression
as biomarkers of progression and survival in tongue carcinoma
patients. Clin Oral Investig. 22:401–409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Weng J, Zhang H, Wang C, Liang J, Chen G,
Li W, Tang H and Hou J: MiR-373-3p targets DKK1 to promote
EMT-induced metastasis via the Wnt/β-catenin pathway in tongue
squamous cell carcinoma. Biomed Res Int. 2017:60109262017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fu S, Chen HH, Cheng P, Zhang CB and Wu Y:
MiR-155 regulates oral squamous cell carcinoma Tca8113 cell
proliferation, cycle, and apoptosis via regulating p27Kip1. Eur Rev
Med Pharmacol Sci. 21:937–944. 2017.PubMed/NCBI
|
|
50
|
Wang J, Wang W, Li J, Wu L, Song M and
Meng Q: MiR-182 activates the Ras-MEK-ERK pathway in human oral
cavity squamous cell carcinoma by suppressing RASA1 and SPRED1.
Onco Targets Ther. 10:667–679. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao J, Hu C, Chi J, Li J, Peng C, Yun X,
Li D, Yu Y, Li Y, Gao M and Zheng X: MiR-24 promotes the
proliferation, migration and invasion in human tongue squamous cell
carcinoma by targeting FBXW7. Oncol Rep. 36:1143–1149. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu MD, Wu H, Wang S, Pang P, Jin S, Sun
CF and Liu FY: MiR-1275 promotes cell migration, invasion and
proliferation in squamous cell carcinoma of head and neck via
up-regulating IGF-1R and CCR7. Gene. 646:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
El-Hefnawy T, Raja S, Kelly L, Bigbee WL,
Kirkwood JM, Luketich JD and Godfrey TE: Characterization of
amplifiable, circulating RNA in plasma and its potential as a tool
for cancer diagnostics. Clin Chem. 50:564–573. 2014. View Article : Google Scholar
|
|
54
|
Park NJ, Li Y, Yu T, Brinkman BM and Wong
DT: Characterization of RNA in saliva. Clin Chem. 52:988–994. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tsui NB, Ng EK and Lo YM: Stability of
endogenous and added RNA in blood specimens, serum, and plasma.
Clin Chem. 48:1647–1653. 2002.PubMed/NCBI
|
|
56
|
Park NJ, Zhou H, Elashoff D, Henson BS,
Kastratovic DA, Abemayor E and Wong DT: Salivary microRNA:
Discovery, characterization, and clinical utility for oral cancer
detection. Clin Cancer Res. 15:5473–5477. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yeh LY, Liu CJ, Wong YK, Chang C, Lin SC
and Chang KW: MiR-372 inhibits p62 in head and neck squamous cell
carcinoma in vitro and in vivo. Oncotarget. 6:6062–6075. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun L, Liu L, Fu H, Wang Q and Shi Y:
Association of decreased expression of serum miR-9 with poor
prognosis of oral squamous cell carcinoma patients. Med Sci Monit.
22:289–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH,
Cheng HW and Lin SC: miR-181 as a putative biomarker for lymph-node
metastasis of oral squamous cell carcinoma. J Oral Pathol Med.
40:397–404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2018. View Article : Google Scholar
|
|
61
|
Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH,
Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, et al: Oncogenic
function and early detection potential of miRNA-10b in oral cancer
as identified by microRNA profiling. Cancer Prev Res (Phila).
5:665–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kao YY, Tu HF, Kao SY, Chang KW and Lin
SC: The increase of oncogenic miRNA expression in tongue
carcinogenesis of a mouse model. Oral Oncol. 51:1103–1112. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW
and Lin SC: MiR-196a overexpression and miR-196a2 gene polymorphism
are prognostic predictors of oral carcinomas. Ann Surg Oncol. 20
(Suppl 3):S406–S414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS
and Chang KW: MiR-24 up-regulation in oral carcinoma: Positive
association from clinical and in vitro analysis. Oral Oncol.
46:204–208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lu YC, Chang JT, Huang YC, Huang CC, Chen
WH, Lee LY, Huang BS, Chen YJ, Li HF and Cheng AJ: Combined
determination of circulating miR-196a and miR-196b levels produces
high sensitivity and specificity for early detection of oral
cancer. Clin Biochem. 48:115–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu CJ, Lin JS, Cheng HW, Hsu YH, Cheng CY
and Lin SC: Plasma miR-187* is a potential biomarker for oral
carcinoma. Clin Oral Investig. 21:1131–1138. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lo WY, Wang HJ, Chiu CW and Chen SF:
MiR-27b-regulated TCTP as a novel plasma biomarker for oral cancer:
From quantitative proteomics to post-transcriptional study. J
Proteomics. 77:154–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ries J, Vairaktaris E, Kintopp R, Baran C,
Neukam FW and Nkenke E: Alterations in miRNA expression patterns in
whole blood of OSCC patients. In Vivo. 28:851–861. 2014.PubMed/NCBI
|
|
69
|
Shin JA, Li C, Choi ES, Cho SD and Cho NP:
High expression of microRNA-127 is involved in cell cycle arrest in
MC-3 mucoepidermoid carcinoma cells. Mol Med Rep. 7:708–712. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Binmadi NO, Basile JR, Perez P, Gallo A,
Tandon M, Elias W, Jang SI and Alevizos I: miRNA expression profile
of mucoepidermoid carcinoma. Oral Dis. 24:537–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ramer N, Wu H, Sabo E, Ramer Y, Emanuel P,
Orta L and Burstein DE: Prognostic value of quantitative p63
immunostaining in adenoid cystic carcinoma of salivary gland
assessed by computerized image analysis. Cancer. 116:77–83.
2010.PubMed/NCBI
|
|
72
|
Wang Y, Zhang CY, Xia RH, Han J, Sun B,
Sun SY and Li J: The MYB/miR-130a/NDRG2 axis modulates tumor
proliferation and metastatic potential in salivary adenoid cystic
carcinoma. Cell Death Dis. 9:9172018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen W, Zhao X, Dong Z, Cao G and Zhang S:
Identification of microRNA profiles in salivary adenoid cystic
carcinoma cells during metastatic progression. Oncol Lett.
7:2029–2034. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Andreasen S, Tan Q, Agander TK, Hansen
TVO, Steiner P, Bjørndal K, Høgdall E, Larsen SR, Erentaite D,
Olsen CH, et al: MicroRNA dysregulation in adenoid cystic carcinoma
of the salivary gland in relation to prognosis and gene fusion
status: A cohort study. Virchows Arch. 473:329–340. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang C, Li T, Yan F, Cai W, Zheng J, Jiang
X and Sun J: Effect of simvastatin and microRNA-21 inhibitor on
metastasis and progression of human salivary adenoid cystic
carcinoma. Biomed Pharmacother. 105:1054–1061. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yang X, Ruan H, Hu X, Cao A and Song L:
miR-381-3p suppresses the proliferation of oral squamous cell
carcinoma cells by directly targeting FGFR2. Am J Cancer Res.
7:913–922. 2017.PubMed/NCBI
|
|
77
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wong TS, Gao W and Chan JY: Transcription
regulation of E-cadherin by zinc finger E-box binding homeobox
proteins in solid tumors. Biomed Res Int. 2014:9215642014.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hashiguchi Y, Kawano S, Goto Y, Yasuda K,
Kaneko N, Sakamoto T, Matsubara R, Jinno T, Maruse Y, Tanaka H, et
al: Tumor-suppressive roles of ΔNp63β-miR-205 axis in
epithelial-mesenchymal transition of oral squamous cell carcinoma
via targeting ZEB1 and ZEB2. J Cell Physiol. 233:6565–6577. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shi W, Yang J, Li S, Shan X, Liu X, Hua H,
Zhao C, Feng Z, Cai Z, Zhang L, et al: Potential involvement of
miR-375 in the premalignant progression of oral squamous cell
carcinoma mediated via transcription factor KLF5. Oncotarget.
6:40172–40185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wu Y, Sun X, Song B, Qiu X and Zhao J:
MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma
proliferation and invasion. Cancer Med. 6:1686–1697. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ji M, Wang W, Yan W, Chen D, Ding X and
Wang A: Dysregulation of AKT1, a miR-138 target gene, is involved
in the migration and invasion of tongue squamous cell carcinoma. J
Oral Pathol Med. 46:731–737. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xu R, Zeng G, Gao J, Ren Y, Zhao Z, Zhang
J, Tao H and Li D: miR-138 suppresses the proliferation of oral
squamous cell carcinoma cells by targeting Yes-associated protein
1. Oncol Rep. 34:2171–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Kim JS, Choi DW, Kim CS, Yu SK, Kim HJ, Go
DS, Lee SA, Moon SM, Kim SG, Chun HS, et al: MicroRNA-203 induces
apoptosis by targeting Bmi-1 in YD-38 oral cancer cells. Anticancer
Res. 38:3477–3485. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lim HS, Kim CS, Kim JS, Yu SK, Go DS, Lee
SA, Moon SM, Chun HS, Kim S and Kim DK: Suppression of oral
carcinoma oncogenic activity by microRNA-203 via down-regulation of
SEMA6A. Anticancer Res. 37:5425–5433. 2017.PubMed/NCBI
|
|
86
|
Lin J, Lin Y, Fan L, Kuang W, Zheng L, Wu
J, Shang P, Wang Q and Tan J: miR-203 inhibits cell proliferation
and promotes cisplatin induced cell death in tongue squamous
cancer. Biochem Biophys Res Commun. 473:382–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xie NN, Liu ZX, Wu C, Wang PL, Song GT and
Chen Z: MicroRNA-200c suppresses tumor metastasis in oral squamous
carcinoma by inhibiting epithelial-mesenchymal transition. Eur Rev
Med Pharmacol Sci. 22:3415–3422. 2018.PubMed/NCBI
|
|
88
|
Zhao L, Ren Y, Tang H, Wang W, He Q, Sun
J, Zhou X and Wang A: Deregulation of the miR-222-ABCG2 regulatory
module in tongue squamous cell carcinoma contributes to
chemoresistance and enhanced migratory/invasive potential.
Oncotarget. 6:44538–44550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wang X, Guo H, Yao B and Helms J: miR-15b
inhibits cancer-initiating cell phenotypes and chemoresistance of
cisplatin by targeting TRIM14 in oral tongue squamous cell cancer.
Oncol Rep. 37:2720–2726. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li X, Fan Q, Li J, Song J and Gu Y:
MiR-124 down-regulation is critical for cancer associated
fibroblasts-enhanced tumor growth of oral carcinoma. Exp Cell Res.
351:100–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lin XJ, He CL, Sun T, Duan XJ, Sun Y and
Xiong SJ: Hsa-miR-485-5p reverses epithelial to mesenchymal
transition and promotes cisplatin-induced cell death by targeting
PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med.
40:83–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lin Z, Sun L, Chen W, Liu B, Wang Y, Fan
S, Li Y and Li J: miR-639 regulates transforming growth factor
beta-induced epithelial-mesenchymal transition in human tongue
cancer cells by targeting FOXC1. Cancer Sci. 105:1288–1298. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liu B, Chen W, Cao G, Dong Z, Xu J, Luo T
and Zhang S: MicroRNA-27b inhibits cell proliferation in oral
squamous cell carcinoma by targeting FZD7 and Wnt signaling
pathway. Arch Oral Biol. 83:92–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Min A, Zhu C, Peng S, Shuai C, Sun L, Han
Y, Qian Y, Gao S and Su T: Downregulation of MicroRNA-148a in
cancer-associated fibroblasts from oral cancer promotes cancer cell
migration and invasion by targeting Wnt10b. J Biochem Mol Toxicol.
30:186–191. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Nagai H, Hasegawa S, Uchida F, Terabe T,
Ishibashi Kanno N, Kato K, Yamagata K, Sakai S, Kawashiri S, Sato
H, et al: MicroRNA-205-5p suppresses the invasiveness of oral
squamous cell carcinoma by inhibiting TIMP2 expression. Int J
Oncol. 52:841–850. 2018.PubMed/NCBI
|
|
96
|
Qiao B, Cai JH, King-Yin Lam A and He BX:
MicroRNA-542-3p inhibits oral squamous cell carcinoma progression
by inhibiting ILK/TGF-β1/Smad2/3 signaling. Oncotarget.
8:70761–70776. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Qiu K, Huang Z, Huang Z, He Z and You S:
miR-22 regulates cell invasion, migration and proliferation in
vitro through inhibiting CD147 expression in tongue squamous cell
carcinoma. Arch Oral Biol. 66:92–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Rastogi B, Kumar A, Raut SK, Panda NK,
Rattan V, Joshi N and Khullar M: Downregulation of miR-377 promotes
oral squamous cell carcinoma growth and migration by targeting
HDAC9. Cancer Invest. 35:152–162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ruan P, Tao Z and Tan A: Low expression of
miR-30a-5p induced the proliferation and invasion of oral cancer
via promoting the expression of FAP. Biosci Rep.
38:BSR201710272018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Sakha S, Muramatsu T, Ueda K and Inazawa
J: Exosomal microRNA miR-1246 induces cell motility and invasion
through the regulation of DENND2D in oral squamous cell carcinoma.
Sci Rep. 6:387502014. View Article : Google Scholar
|
|
101
|
Shang A, Lu WY, Yang M, Zhou C, Zhang H,
Cai ZX, Wang WW, Wang WX and Wu GQ: miR-9 induces cell arrest and
apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway.
Artif Cells Nanomed Biotechnol. 46:1754–1762. 2018.PubMed/NCBI
|
|
102
|
Shi Z, Johnson JJ, Jiang R, Liu Y and
Stack MS: Decrease of miR-146a is associated with the
aggressiveness of human oral squamous cell carcinoma. Arch Oral
Biol. 60:1416–1427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wang L and Liu H: microRNA-188 is
downregulated in oral squamous cell carcinoma and inhibits
proliferation and invasion by targeting SIX1. Tumour Biol.
37:4105–4113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang K, Jin J, Ma T and Zhai H: MiR-139-5p
inhibits the tumorigenesis and progression of oral squamous
carcinoma cells by targeting HOXA9. J Cell Mol Med. 21:3730–3740.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang K, Jin J, Ma T and Zhai H:
MiR-376c-3p regulates the proliferation, invasion, migration, cell
cycle and apoptosis of human oral squamous cancer cells by
suppressing HOXB7. Biomed Pharmacother. 91:517–525. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang Q, Lv L, Li Y and Ji H: MicroRNA-655
suppresses cell proliferation and invasion in oral squamous cell
carcinoma by directly targeting metadherin and regulating the
PTEN/AKT pathway. Mol Med Rep. 18:3106–3114. 2018.PubMed/NCBI
|
|
107
|
Wang Z, Yan J, Zou T and Gao H:
MicroRNA-1294 inhibited oral squamous cell carcinoma growth by
targeting c-Myc. Oncol Lett. 16:2243–2250. 2018.PubMed/NCBI
|
|
108
|
Weng JH, Yu CC, Lee YC, Lin CW, Chang WW
and Kuo YL: miR-494-3p induces cellular senescence and enhances
radiosensitivity in human oral squamous carcinoma cells. Int J Mol
Sci. 17(pii): E10922016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Xu P, Li Y, Zhang H, Li M and Zhu H:
MicroRNA-340 mediates metabolic shift in oral squamous cell
carcinoma by targeting glucose transporter-1. J Oral Maxillofac
Surg. 74:844–850. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zeng G, Xun W, Wei K, Yang Y and Shen H:
MicroRNA-27a-3p regulates epithelial to mesenchymal transition via
targeting YAP1 in oral squamous cell carcinoma cells. Oncol Rep.
36:1475–1482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li X, He J, Shao M, Cui B, Peng F, Li J,
Ran Y, Jin D, Kong J, Chang J, et al: Downregulation of miR-218-5p
promotes invasion of oral squamous cell carcinoma cells via
activation of CD44-ROCK signaling. Biomed Pharmacother.
106:646–654. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhuang Z, Hu F, Hu J, Wang C, Hou J, Yu Z,
Wang TT, Liu X and Huang H: MicroRNA-218 promotes cisplatin
resistance in oral cancer via the PPP2R5A/Wnt signaling pathway.
Oncol Rep. 38:2051–2061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Jiang F, Zhao W, Zhou L, Liu Z, Li W and
Yu D: MiR-222 targeted PUMA to improve sensitization of UM1 cells
to cisplatin. Int J Mol Sci. 15:22128–22141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Du L, Ma S, Wen X, Chai J and Zhou D: Oral
squamous cell carcinoma cells are resistant to doxorubicin through
upregulation of miR-221. Mol Med Rep. 16:2659–2667. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhou L, Jiang F, Chen X, Liu Z, Ouyang Y,
Zhao W and Yu D: Downregulation of miR-221/222 by a microRNA sponge
promotes apoptosis in oral squamous cell carcinoma cells through
upregulation of PTEN. Oncol Lett. 12:4419–4426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zheng X, Li J, Peng C, Zhao J, Chi J, Meng
X, Yun X, Li D, Yun Y, Gao M and Li Y: MicroRNA-24 induces
cisplatin resistance by targeting PTEN in human tongue squamous
cell carcinoma. Oral Oncol. 51:998–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Cheng CM, Shiah SG, Huang CC, Hsiao JR and
Chang JY: Up-regulation of miR-455-5p by the TGF-β-SMAD signalling
axis promotes the proliferation of oral squamous cancer cells by
targeting UBE2B. J Pathol. 240:38–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Guo Y, Ren MS, Shang C, Zhu L and Zhong M:
MTSS1 gene regulated by miR-96 inhibits cell proliferation and
metastasis in tongue squamous cellular carcinoma Tca8113 cell line.
Int J Clin Exp Med. 8:15441–15449. 2015.PubMed/NCBI
|
|
119
|
Hu J, Xu JF and Ge WL: MiR-497 enhances
metastasis of oral squamous cell carcinoma through SMAD7
suppression. Am J Transl Res. 8:3023–3031. 2016.PubMed/NCBI
|
|
120
|
Kawakubo-Yasukochi T, Morioka M, Hazekawa
M, Yasukochi A, Nishinakagawa T, Ono K, Kawano S, Nakamura S and
Nakashima M: MiR-200c-3p spreads invasive capacity in human oral
squamous cell carcinoma microenvironment. Mol Carcinog. 57:295–302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li N, Nan CC, Zhong XY, Weng JQ, Fan HD,
Sun HP, Tang S, Shi L and Huang SX: miR-182-5p promotes growth in
oral squamous cell carcinoma by inhibiting CAMK2N1. Cell Physiol
Biochem. 49:1329–1341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lin SC, Kao SY, Chang JC, Liu YC, Yu EH,
Tseng SH, Liu CJ and Chang KW: Up-regulation of miR-187 modulates
the advances of oral carcinoma by targeting BARX2 tumor suppressor.
Oncotarget. 7:61355–61365. 2016.PubMed/NCBI
|
|
123
|
Liu Z, Diep C, Mao T, Huang L, Merrill R,
Zhang Z and Peng Y: MicroRNA-92b promotes tumor growth and
activation of NF-κB signaling via regulation of NLK in oral
squamous cell carcinoma. Oncol Rep. 34:2961–2968. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Lu M, Wang C, Chen W, Mao C and Wang J:
miR-654-5p targets GRAP to promote proliferation, metastasis, and
chemoresistance of oral squamous cell carcinoma through Ras/MAPK
signaling. DNA Cell Biol. 37:381–388. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Peng SY, Tu HF, Yang CC, Wu CH, Liu CJ,
Chang KW and Lin SC: MiR-134 targets PDCD7 to reduce E-cadherin
expression and enhance oral cancer progression. Int J Cancer.
143:2892–2904. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Qiao B, He BX, Cai JH, Tao Q and King-Yin
Lam A: microRNA-27a-3p modulates the Wnt/β-Catenin signaling
pathway to promote epithelial-mesenchymal transition in oral
squamous carcinoma stem cells by targeting SFRP1. Sci Rep.
7:446882017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhao J, Chi J, Gao M, Zhi J, Li Y and
Zheng X: Loss of PTEN expression is associated with high
microRNA-24 level and poor prognosis in patients with tongue
squamous cell carcinoma. J Oral Maxillofac Surg.
75:1449.e1–1449.e8. 2017. View Article : Google Scholar
|
|
128
|
Zheng G, Li N, Jia X, Peng C, Luo L, Deng
Y, Yin J, Song Y, Liu H, Lu M, et al: MYCN-mediated miR-21
overexpression enhances chemo-resistance via targeting CADM1 in
tongue cancer. J Mol Med (Berl). 94:1129–1141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Chen YF, Yang CC, Kao SY, Liu CJ, Lin SC
and Chang KW: MicroRNA-211 enhances the oncogenicity of
carcinogen-induced oral carcinoma by repressing TCF12 and
increasing antioxidant activity. Cancer Res. 76:4872–4886. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Chen YH, Song Y, Yu YL, Cheng W and Tong
X: MiRNA-10a promotes cancer cell proliferation in oral squamous
cell carcinoma by upregulating GLUT1 and promoting glucose
metabolism. Oncol Lett. 17:5441–5446. 2019.PubMed/NCBI
|
|
131
|
Cao ZH, Cheng JL, Zhang Y, Bo CX and Li
YL: MicroRNA-375 inhibits oral squamous cell carcinoma cell
migration and invasion by targeting platelet derived growth factor
A. Mol Med Rep. 15:922–928. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Du Y, Li Y, Lv H, Zhou S, Sun Z and Wang
M: miR-98 suppresses tumor cell growth and metastasis by targeting
IGF1R in oral squamous cell carcinoma. Int J Clin Exp Pathol.
8:12252–12259. 2015.PubMed/NCBI
|