Introduction
Non-coding RNAs (ncRNAs) are protein-free RNAs
produced by genome transcription. There are 20,000-25,000 genes
encoded in the human genome, of which 40–90% are estimated to be
regulated by miRNA (1). A variety of
RNAs, including microRNAs (miRNAs/miRs) and long non-coding RNAs
(lncRNAs), regulate gene expression at the transcription,
post-transcription and epigenetic levels. Research has shown that
ncRNA inhibits the translation activity of mRNA, regulates the
expression of specific target genes, and ultimately regulates
biological signaling pathways and manipulates cell fates (2,3). In
recent years, substantial progress has been made in basic research
regarding the underlying disease mechanisms, using model organisms
and new experimental systems. For example, mir-21 was shown to
promote cell migration and invasion, and play a key role in cancer
cell infiltration and remote metastasis. These studies have
provided evidence that ncRNAs play important roles in the
development and progression of pancreatic cancer (1,4).
Pancreatic cancer ranks 4th in the world among all
malignant tumors, with high invasiveness, early metastasis, lack of
specific symptoms and high mortality. The median survival time is
only 3–6 months, with a 5-year survival rate of <5%. In China,
the incidence of pancreatic cancer has increased 6-fold in the past
20 years (5). In 2012, the incidence
of pancreatic cancer was 4.8/100,000, ranking 7th among all
malignant tumors. Even when early diagnosed, small pancreatic
cancer (<2 cm in diameter) can easily metastasize and lead
quickly to the death of the patient. Surgical resection remains the
only treatment for radical treatment. Nevertheless, only 15–20% of
pancreatic cancer patients are suitable for surgical treatment
(1,6,7). Even
with multidisciplinary comprehensive treatment, the median survival
time of patients with advanced pancreatic cancer is only ~6 months.
Furthermore, early postoperative recurrence and occult metastasis
also reduce the efficacy of surgical treatment. Even after systemic
therapy, liver metastasis occurs in 50% of patients. However, the
main reasons for the high mortality rates of pancreatic cancer are
the failure of an early diagnosis, before it moves to other organs,
and the resistance of the cancer to existing therapies (8).
In recent years, with the development of molecular
biology, especially gene sequencing technology, the mechanism of
action of lncRNAs in various types of cancer has been gradually
revealed, and the study of ncRNAs has been significantly augmented
for various types of cancer. In the case of pancreatic cancer,
various lncRNAs are involved in the biological behavior and signal
pathways of metastatic pancreatic cancer, including the cell cycle,
apoptosis, autophagy, epithelial-mesenchymal transition (EMT) and
cancer stem cells (CSCs) (6,9). The present review focuses on the
characteristics and biological roles of several ncRNAs, with
particular emphasis on their role in pancreatic cancer (Table I).
 | Table I.Specific types and functions of
different ncRNAs and their association with pancreatic cancer. |
Table I.
Specific types and functions of
different ncRNAs and their association with pancreatic cancer.
| First author
(year) | Type of RNAs | Length, nt | Function | (Refs.) |
|---|
| Fu (2017), Gao
(2018) Gao (2018) | Small nucleolar
RNAs | 70–200 | Involved in
maturation of other ncRNAs | (6,7,59) |
| Fu (2017), Gao
(2018) | Small nuclear
RNAs | 100–215 | Binding to proteins
to form splicers that control selective splicing | (6,59) |
| Gu (2017) | Small interfering
RNAs | 21–22 | Silencing specific
genes in a sequence-specific manner | (60) |
| Gao (2018), Gu
(2017) | Small ncRNAs | <200 | Carrying amino
acids that bind to the ribosome | (59,60) |
| Peng (2016) | lncRNAs | >200 | Component of
ribosomes | (61) |
| Peng (2016), Peng
(2016) | Ribosomal 28S and
18SRNA | 200-5,050 | Component of
ribosomes | (2,61) |
| Vorvis (2016), Wang
(2017) | Piwi-interacting
RNA | 25–34 | Controlling retro
transposition and regulating methylation | (3,71) |
| Xiong (2017), Zhang
(2017) | Promoter-associated
short RNAs | <200 | Regulating gene
expression through interaction with gene promoter sites | (11,63) |
| Zhou (2017) | Long intergenic
ncRNAs or long intronic ncRNAs | >200 | Various | (68) |
| Zhou (2017) | Antisense RNA | >200 | Binding and
blocking of mRNA target genes | (68) |
| Zhou (2017), Zhang
(2017) | Promoter-associated
short RNAs | >200 | Regulating gene
expression through interaction with gene promoter sites | (31,85) |
| Zhang (2017) | Telomere-associated
ncRNAs | 100 bp >9
kb | Negative telomere
regulators | (79) |
| Zhang (2017), Zhang
(2017) | Transcribed
ultra-conserved regions | 200–799 | Long-range
enhancer-like activity, maintenance of splicing factor expression
levels and transcription regulation | (79,85) |
miRNA
miRNA is the most characteristic ncRNA; it is a
short sequence of RNA with a length of ~22 nt. miRNA expression is
related to the type of tumor, as well as its occurrence and
development (10). Therefore, miRNA,
as a potential tumor marker, has been the focus of clinical
research. As miRNA binds mRNA through incomplete pairing, thereby
inhibiting translation, the same miRNA can simultaneously regulate
several target genes and signaling pathways (11). Furthermore, miRNAs are also
conservative, testable and can exist stably outside cells.
Therefore, they have become diagnostic markers and therapeutic
targets of tumors (8).
miRNAs are usually produced as primary transcripts
for RNA polymerase 2 transcription, known as pri-miRNAs. Recent
studies have shown that miRNAs can act as intermediates, regulating
a large part of human gene transcription (12). At present, feedback loops are widely
known to exist between miRNAs and basic transcription factors
(TFs). Although there are numerous differences between them, TFs
positively or negatively regulate the expression of miRNA, while
miRNAs inhibit the translation of TF-miRNA (13). Studies have shown that, after
transfection of let-7 into pancreatic cancer cells, the
proliferation capacity of the transfected cells is significantly
inhibited, presumably related to the effect of miRNA on the
expression of the k-ras gene and the activation of
mitogen-activated protein kinase (8,14).
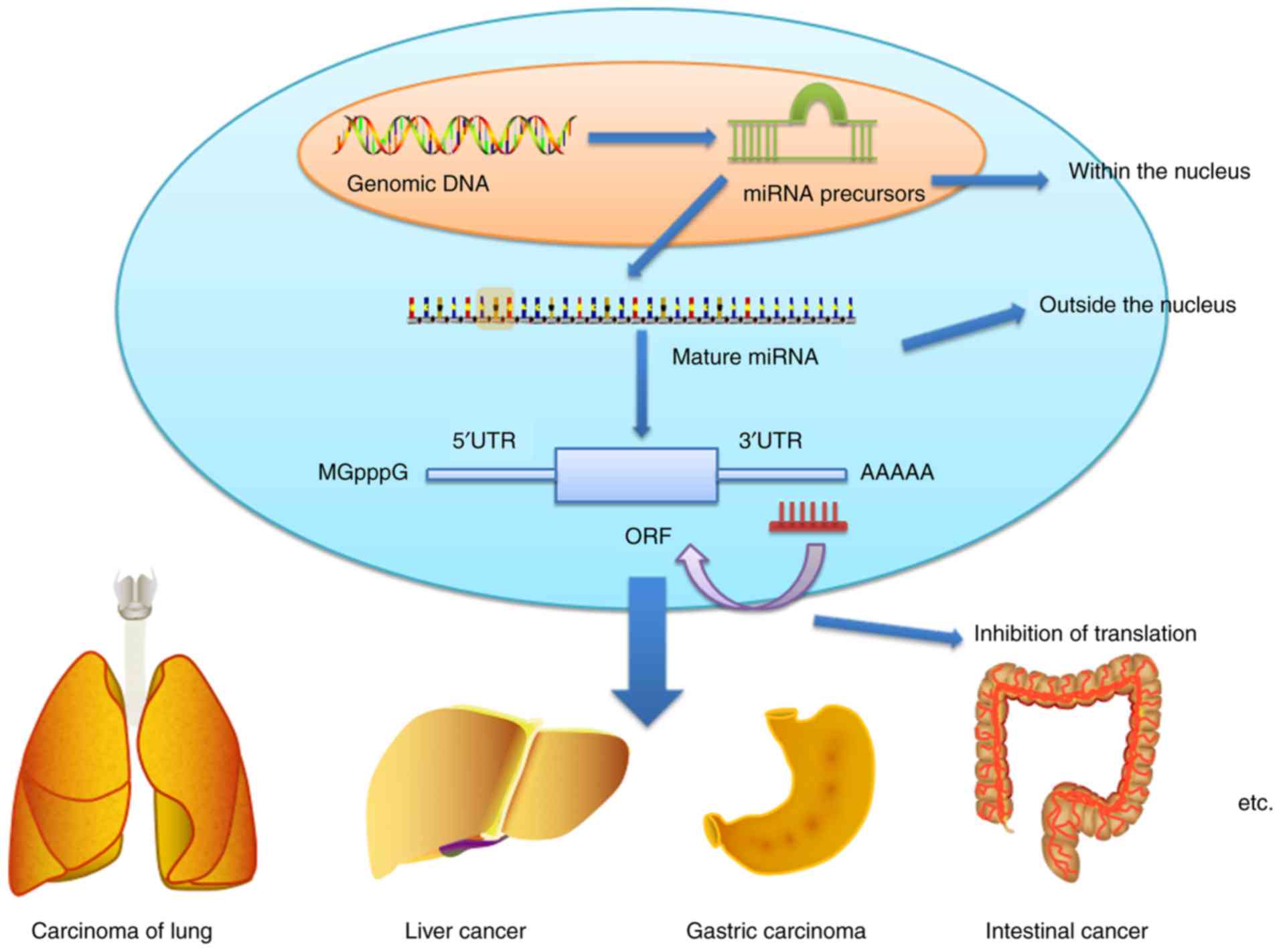
There are various ways in which miRNAs alter the
levels of gene expression. In particular, they typically bind to
the mRNA 3′UTR. Genomic DNA is transcribed into mRNA, and mRNA has
5′UTR and 3′UTR structures. miRNA binds to the mRNA 3′UTR, and then
alters the translation process after mRNA, thereby affecting the
expression results of the particular gene (Fig. 1) (15–18).
Generation of model organisms and
their use in miRNA functional studies
Despite the fact that Caenorhabditis elegans
do not develop cancer, they have been widely used as model
organisms to identify molecular functions and to describe pathways
involved in normal cellular processes that are severely impaired in
cancer, including cell proliferation, differentiation, metabolism
and death (19). Full sequencing of
C. elegans genomes showed that ~60% of miRNAs have a human
homologous gene. As the number of conserved miRNAs in the miRNA
family of C. elegans decreased, the study of miRNA function
in C. elegans eliminated redundancy as a barrier to be
overcome. Moreover, C. elegans are self-fertilized
hermaphrodites that can produce large numbers of genetically
identical offspring (19,20). The visual tracking of C.
elegans, with a well-organized transparent body, makes for an
excellent model system. Phenotypic and genetic screening, the
application of molecular techniques, and the development of
transgenic C. elegans have identified a number of key
miRNAs, and have elucidated their mechanisms of action. Research on
C. elegans has also contributed to the understanding of the
molecular basis of the miRNA biological genome. For example, Dicer
C. elegans ortholog gene, dcr-1, involved in RNA-mediated
silencing, has been identified as a key component necessary for the
processing of mature let-7 from its precursor molecule (19,21).
The zebrafish is a model system that plays an
important role in elucidating the functional role of miRNA in both
normal and cancer cells. Since let-7 was identified during in
silico prediction as a cross-species conserved miRNA, the other
miRNAs, initially found in zebrafish, have considerable homology
with the miRNAs encoded by humans and other vertebrates, in terms
of composition and function (20,21).
Thanks to a complete angiogenesis and immune system, as well as a
developed organ system, human cancer cells were successfully
transplanted into zebrafish embryos, enabling the tumor to grow in
the host microenvironment. Therefore, the zebrafish was also shown
to be a successful model system for assessing tumor response to
anticancer therapy in vivo. In summary, despite the fact
that the zebrafish is a simple model system, the striking
properties of D. rerio permit the use of this organism to
better understand the effects of aberrant changes in endogenous
levels of certain miRNAs (21,22). The
zebrafish has been proven to be a successful model system, leading
to the recognition of several miRNAs through basic biochemical and
molecular studies, and has great potential as a model to identify
clinically relevant miRNAs.
Previous studies used mir-520d-5p to confirm its
possible therapeutic effect on cancer cells (23,24). The
size and signal intensity of green fluorescent protein
(GFP)-expressing tumors was measured in mice weekly, for 4 weeks
after subcutaneous inoculation. It was found that, compared with
the control group, 520d treatment, which consisted of introducing
exogenous gene 520d into mice, inhibited tumor growth by >80%
per week, resulting in the disappearance of ~30% of the tumors. In
mice with tumor disappearance, the presence of human genomic
material at the injection site was quantitatively detected by
alu-PCR, confirming the coexistence of the two cell sources. GFP
was expressed in connective tissue at each site where the
immune-deficient mouse tumors disappeared, and ~0.1% of the
extracted DNA contained human genomic material.
miRNAs can be divided into two forms, namely those
with high and low expression in pancreatic cancer cells, similar to
proto-oncogenes and tumor suppressor genes. These two factors have
wide influence on biological pathways, and their abnormal
regulation is an important factor leading to the development of
pancreatic cancer (25).
Oncogenic miRNAs
Researchers detected 95 miRNAs with abnormal
expression in pancreatic cancer using RT-PCR. Among these,
mir-196a, mir-190, mir-186, mir-221, mir-222, mir-200b, mir-15b and
mir-95 were significantly upregulated, while let-7, mir-96, mir-15
and mir-132 were significantly downregulated (2). Studies reported that mir-217 and
mir-148a/b were downregulated in pancreatic cancer cells, mir-155
and mir-21 were found to be highly expressed in breast cancer and
they may also be used as landmarks of pancreatic cancer (10,18).
Previous studies analyzed tumors, including those of the lung,
stomach, prostate, rectum and pancreas, and found that mir-17-5p,
mir-20a, mir-21, mir-92, mir-106a and mir-155 were all highly
expressed in tumor cells (10,18). Of
these, mir-20a was highly sensitive and specific, and could be used
as a marker for differentiating various stages of pancreatic cancer
from paracarcinoma tissue and chronic pancreatitis. Previous
studies compared the plasma levels of 50 pancreatic cancer patients
and 10 healthy patients, and found that 37 types of miRNA were
decreased and 54 types of miRNA were increased (2,26,27).
Among these miRNAs, mir-21 is an oncogene. Therefore, it is thought
that the high expression of mir-21 can be used as an indicator of
poor survival in pancreatic cancer patients, as well as a
prognostic marker. Additionally, the miRNA expression profile,
including mir-99, mir-100 and mir-100-1/2, varied in pancreatic
cancer tissues, adjacent and chronic pancreatitis tissues (CP)
(27,28). mir-199a-1 and mir-199a-2 were
relatively highly expressed in pancreatic cancer and CP, possibly
indicating that they stimulate the growth of neoplasms. This could
be caused by the interference of inflammatory factors. The study
also found that mir-196a-2 was significantly correlated with
patient survival. Other studies found that miRNA precursors with
increased expression were mir-22-1, −424, −301, −100, −376a,
−125b-1, −021, −016-1, −181a, c, −092-1, −092-1, −212, −107,
−024-l, 2, 1et-7f-1, 1et-7d, mir-345, −142-p, −139 (14,29).
These results were similar to those of Volinia (30). Volinia (30) and Zhou et al (31) used endpoint immuno PCR to detect the
expression of mir-196a in normal pancreatic cells, pancreatic duct
epithelial neoplasms (PanIN, precancerous) cells and pancreatic
cancer cells. It was found that, in normal pancreatic cells,
mir-196a expression was not present; however, PanIN and all tumor
cells showed positive expression of miR-196a, suggesting that
mir-196a might indicate the carcinogenic transformation process of
pancreatic cells. Volinia (30) and
Yu et al (29) compared the
differences in expression of miRNAs in the peripheral blood of
ovarian cancer patients and normal individuals, and found that
mir-21, −92, −93, −126 and −29a were highly expressed in ovarian
cancer. Xiong et al (11) and
Yan et al (32) found that
mir-184 was highly expressed in the peripheral blood of tongue
squamous cell carcinoma. Yan et al (33) found that mir-92 was highly expressed
in the peripheral blood of rectal cancer patients.
Tumor suppressor gene-related
miRNAs
As opposed to proto-oncogenes, some miRNAs are less
well expressed in pancreatic tissue than in normal pancreatic
tissue; they inhibit the development of pancreatic cancer by
negatively regulating the cell cycle and proliferation. The
decreased levels of mature miRNAs may be caused by defects in any
step of miRNA biosynthesis, eventually leading to the expression of
inappropriate miRNA target proteins. Wang et al (34) found that the expression of two
miRNAs, mir-15a and mir-16a, was downregulated or missing in most
patients with chronic lymphocytic leukemia (CLL). Sun et al
(27) and Vassaux et al
(35) found that the expression of
mir-127 in some human primary tumors was downregulated or missing,
while it was normal in normal cells; therefore, mir-127 was
hypothesized to be a potential tumor suppressor gene. Recently, a
study reported that mir-615-5p was significantly downregulated in
pancreatic cancer compared with its expression in normal adjacent
tissues, and that it inhibited proliferation, migration and
invasion of pancreatic cancer cells by targeting AKT2 (36). The let-7 family miRNAs are also well
known among all types of miRNA. mir-1181 was found to have low
expression in the middle of pancreatic cancer, and it was shown to
directly inhibit the expression of sox2 and stat3 (37). mir193-b is a tumor suppressor gene
that demonstrates interaction inhibition with MillHG (38,39).
mir-193b negatively regulates mir31HG by directly targeting two
miRNA binding sites in the MillHG sequence and inhibits
proliferation of pancreatic cancer cells, while mir31HG can be used
as an endogenous ‘sponge’ to reduce its anticancer effect (10,40).
Studies on miRNAs in the treatment of pancreatic
cancer have been ongoing. The Panpane-1, LPc111 and LPc006 cell
lines of pancreatic cancer showed increased tolerance to
gemcitabine when they were transfected with precursor mir-21;
apoptosis levels decreased or proliferation levels increased. By
contrast, mir-21 expression in the suit-2 pancreatic cancer cell
line was inhibited; apoptosis levels increased and proliferation
activity decreased (41). Li et
al (42) and Hancock and Skalsky
(43) showed that mir-21 can be used
as an indicator of resistance of pancreatic cancer cells in the
Hengtian region. Pancreatic cancer cell lines with low expression
of mir-21 are highly sensitive to fluorouracil. Li et al
(18) and Hancock and Skalsky
(43) screened and verified
pancreatic cancer tissues and two types of pancreatic cancer cell
lines that were resistant to gemcitabine using miRNA microarray. It
was found that cells with high expression of mir-142-5p and mir-204
had significantly increased tolerance to gemcitabine. It was
proposed that the expression levels of mir-142-5p can be thought of
as indicators of resistance of pancreatic cancer cells to
gemcitabine.
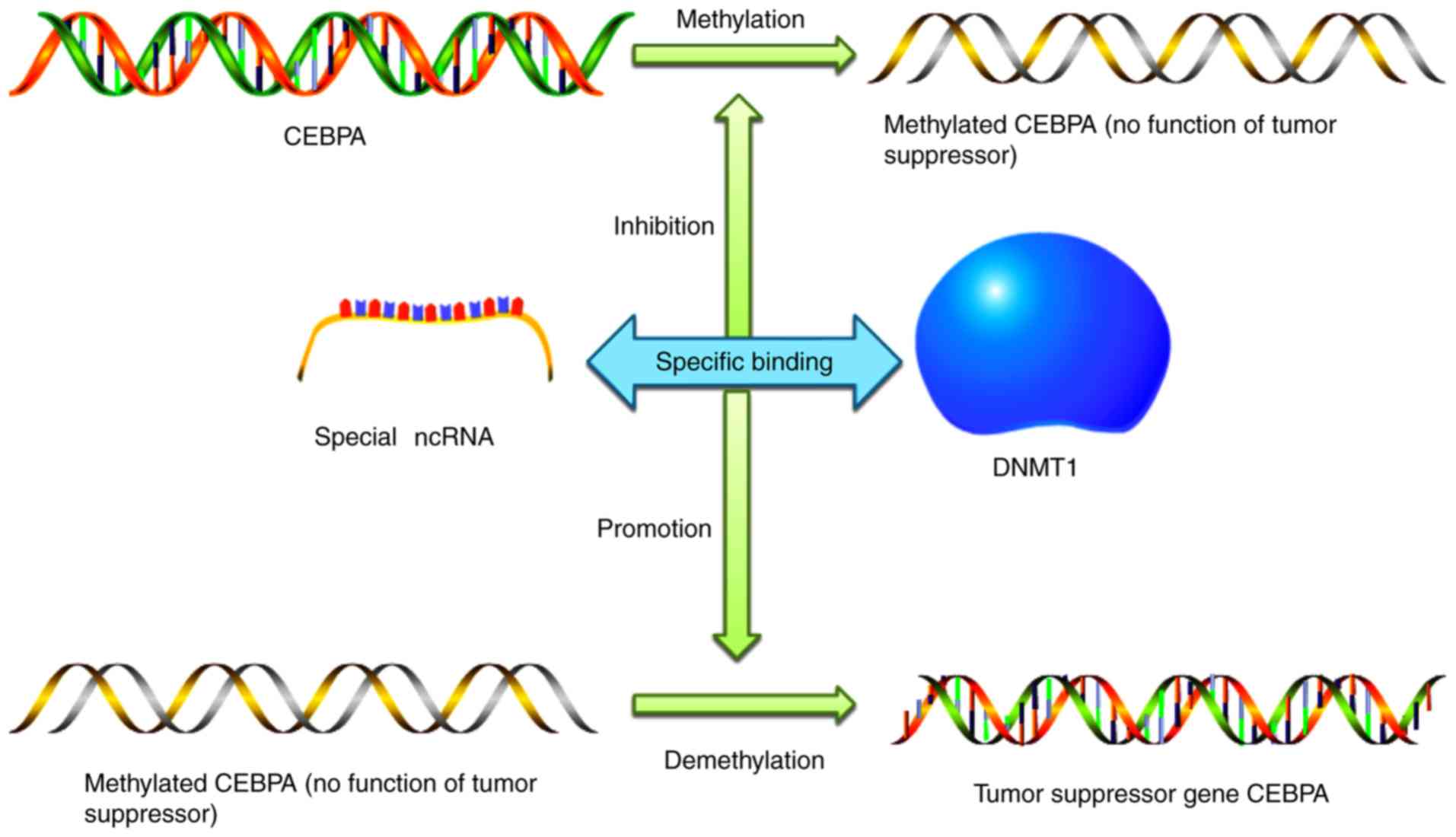
DNA methylation is a form of DNA chemical
modification that alters the genetic expression without changing
the DNA sequence, causing changes in chromatin structure, DNA
conformation, DNA stability and the way DNA interacts with
proteins, thereby controlling gene expression. DNA methylation has
a definite effect on cell carcinogenesis in various ways. A methyl
group attaches to the DNA region near the tumor suppressor gene,
inhibits the activity of the gene, and effectively ‘silences’ the
gene. Recently, researchers focused on a class of ncRNAs that
specifically affect CCAAT enhancer binding protein α (CEBPA), a
tumor suppressor gene (44,45). CEBPA silencing is associated with
acute myeloid leukemia, lung and pancreatic cancer, as well as
other types of cancer. ncRNA binds DNA methyltransferase 1 to
prevent methylation of CEBPA. This principle, which is likely to be
applied to thousands of other genes, holds promise for
‘reactivating’ downregulated cancer suppressors. Researchers
(46) found that the
newly-discovered ncRNA specifically inhibits DNA methylation, and
the introduction of RNA selectively demethylates certain genes,
thereby initiating the expression of tumor suppressors. This
mechanism is expected to be exploited in the treatment of a variety
of diseases, including cancer (Fig.
2) (1,8,46).
miRNA-301
mir-301 is highly expressed only in pancreatic
cancer, suggesting that mir-301 may be a molecular marker of
specificity in pancreatic cancer. Studies have shown that mir-301
is highly expressed in genes related to the invasion and metastasis
of pancreatic cancer, including cox-2 and mmp-2. mir-301 is closely
related to the invasion and metastasis of pancreatic cancer
(47). mir-301 inhibitors
effectively suppress the expression of the invasion- and
metastasis-related genes cox-2 and mmp-2, and significantly inhibit
the invasion and metastasis of pancreatic cancer cells, suggesting
that mir-301 is effective (10,12).
mir-301 may be a new molecular target for the therapy of pancreatic
cancer. The establishment of effective detection methods and
targeted treatment strategies for mir-301 will provide important
theoretical and experimental bases for the early diagnosis,
treatment option selection, efficacy evaluation and prognosis in
pancreatic cancer (47).
miRNA-20a
miRNA-20a was shown to be expressed at low levels in
pancreatic tissue and cell lines. The expression of the miRNA
inhibits the growth and invasion of pancreatic cancer cell lines,
and its role in pancreatic cancer is achieved through the negative
regulation of oncogenes Stat3 and cyclin D1 (48). Studies have reported that miRNA-20a
directly inhibits E2F TF1 (E2F1) and regulates the cell cycle. The
miRNA also plays the role of an oncogene as it is overexpressed in
a number of malignant tumors, including lung cancer and B-cell
lymphoma. The mechanism involved may be the downregulation of Stat3
and cyclin D1 expression by miRNA-20a (16,49).
mir-200 family
Peng et al (2)
and Qian et al (38) found
that the expression of the mir-200 family is downregulated or is
missing in the course of EMT. The results were related to EMT, and
it was reported that EMT is closely related to the relatively low
survival rate after surgical resection of pancreatic duct cell
carcinoma. Studies found that the gemcitabine resistance of
pancreatic cancer cells was associated with the lower expression of
mir-200 and let-7 in these cells. The epithelial phenotype was
restored by transfection of mir-200 with generate gemcitabine
resistant cells; therefore, the epithelial marker E-calcium mucins
was increased, and the leaf-cutting marker E-box zinc-finger
protein was combined with vimentin to inhibit EMT and invasion of
tumor cells (27,50).
Therapeutic application of miRNAs
miRNA can be involved in treatment in two ways,
namely the reduction and replacement of miRNA. Downregulation of
miRNA is targeted at the acquisition of functions, and oncogenic
miRNA can be inhibited in the following ways: i) Small molecule
inhibitors act on the regulation of miRNA expression at the
transcriptional level; ii) antisense oligonucleotides bind to
complementary miRNAs, inducing duplex formation or miRNA
degradation; iii) miRNA masking uses molecules complementary to the
3′UTR of the target miRNA to competitively inhibit downstream
target effects; and iv) miRNA sponge, constructed by
oligonucleotide with multiple complementary miRNA binding sites,
binds to target miRNA (51,52). The second strategy, miRNA
substitution, is targeted at functional loss, including the
restoration of functional loss by introducing systemic miRNA (miRNA
mimics) or inserting miRNA-coding genes into viral construction,
and reintroducing tumor suppressor miRNA (51,53).
Despite the promise of both approaches, two major problems have,
thus far, hampered the development and effectiveness of miRNA-based
therapies. The first is that it is necessary to obtain
tissue-specific delivery and develop more efficient cellular uptake
of synthetic oligonucleotides to achieve constant target
inhibition. The second important problem is the relatively low
efficiency of miRNAs in body fluids or tissues, seeking to restore
lost function or block tumor-enhanced miRNAs, which have been
partially overcome by the construction of locked nucleic acids
(LNA). The most successful use of the so-called ‘LNA anti-miRs’
resulted in the first miRNA-based clinical trial for the treatment
of hepatitis C virus infection, which used lna-anti-mir to target
mir-122 (54,55).
Chemotherapy is an important treatment strategy for
most pancreatic cancer patients. Chen Q and Hong L reported the
results of a phase III clinical trial that directly compared
gemcitabine with 5-FU and showed that gemcitabine significantly
improved the median survival time. Soon thereafter, gemcitabine
became the first-line and gold-standard chemotherapy for pancreatic
cancer (51,54). Nevertheless, drug resistance to
gemcitabine and other drugs may lead to treatment failure. Despite
the fact that research in drug resistance has been ongoing, the
mechanism of its occurrence has not been fully explained. The three
most common reasons for acquiring drug resistance are the
expression of energy-demanding transporters, insensitivity to
drug-induced apoptosis and drug detoxification-induced mechanisms
(53,55). In one study, oncogenic mir-155 levels
were shown to increase after gemcitabine was used for the treatment
of pancreatic cancer cells. mir-200a, mir-200b and mir-200c were
downregulated in gemcitabine-resistant pancreatic cancer cells
(55). A recent report showed that
mir-34 was involved in the self-renewal of pancreatic CSCs, and the
deletion of mir-34 in pancreatic cancer was related to the
enrichment of CSCs, which are not sensitive to chemotherapy
(52,56). There is also evidence that miRNAs may
regulate EMT by regulating cadherin-1 and other molecules,
mediating various cell resistance mechanisms (54,56).
When studying the expression levels of mir-200 and let-7 in
gemcitabine-resistant pancreatic cancer cells with the EMT
phenotype, the downregulated expression of mir-200 family
upregulated cadherin-1 and downregulated ZeB1 and vimentin (EMT
inducer). Downregulation of ZeB1 in mesenchymal Panc1 cells led to
upregulation of mir-200c (56).
Therefore, by targeting CSCs or EMT phenotype cells that lead to
tumor recurrence and metastasis, the re-expression of specific
miRNAs can be used as a new strategy for the treatment of
pancreatic cancer.
pIRNA
piRNA research began more recently. piRNAs are
ncRNAs of 24–30 nt in length. piRNAs rely on the piwi protein group
that inhibits the expression of transposable elements in two ways
by forming a complex with piwi. At present, there are two proposed
pathways for generating piRNAs, namely a primary processing pathway
and a ‘ping-pong’ amplification loop. Similar to miRNA, the 5′-end
also has a strong uracil bias (57,58). A
previous study has shown that piRNA exists in mammalian germ cells,
and regulates the gene silencing pathway by binding with the piwi
subfamily proteins to form piRNA complexes (2). The discovery of piRNA opens a new field
for the study of non-coding small molecular RNAs. To date, piRNA
research has achieved interim results. However, it is too early to
predict their use in biomedicine (2,58). At
present, piRNA regulates mRNA primarily via two mechanisms:
Degradation and shear. CAF1 mRNA in miRNA regulation has a very
important role in degradation. Studies in mouse CAF1 by Andersen
et al (57) demonstrated its
combination with piRNA/piwi complexes. This complex process
recognizes the target mRNA after removal of adenosine piRNA,
eventually causing mRNA degradation. Repoila et al (47) reported the piRNA/piwi-mediated shear
mechanism of mRNA. There remain numerous problems that require
further discussion (44,58). For example, the homologous
conservation of piRNA sequences in various species is not easy to
analyze. In piRNA panspermia, it remains unknown how the division
mechanisms of the 3′-end and the piRNA amplification cycle are
regulated. piRNA research not only enriches the research content of
small molecular RNA, but also helps further the understanding of
molecular regulation and mechanisms of biological gametes with
substantial theoretical value (57).
There are three members of the piwi subfamily, namely MIWI, MILI
and MIWI2, which are specific proteins in germ cells and closely
related to the occurrence of sperm. Knockout of MIWI, MILI or MIWI2
resulted in sperm production disorders in male mice (58). MILI was expressed during early
spermatogenesis, from mitosis of spermatocytes to the pachytene
stage of spermatocytes, and the MILI deficient mice stopped at
pachytene stage. MIWI is expressed later, from pachytene to spheric
spermatogenesis. Knockout of MIWI causes spermatogenesis to stop at
spheric spermatogenesis. However, mice with knockout MIWI2 had
defects in meiosis progression at early meiosis stage and
significant loss of germ cells with age (57).
piRNA forms a complex with piwi protein that is
involved in the regulation of target sequences. Piwi is also
detected in cancer cells. There are four piwi proteins in humans:
PIWIL1/HIWI, PIWIL2/HILI, PIWIL3 and PIWIL4/HIWI2, and there are
specific types of piwi proteins in various cancers (57). Piwi proteins play four major roles in
cancer cells: The promotion of cell proliferation, the inhibition
of apoptosis, the enhancement cell migration and the maintenance of
genomic stability. Cancer cells have unlimited ability to
proliferate and migrate, and they also have the ability to escape
apoptosis and divide continuously (57,58). In
order to maintain this ability, the relative stability of their
genome structure must be ensured. Although piwi protein was found
in cancer cells, piRNA that binds to piwi protein was not found,
suggesting that a number of unknown functions of the piwi and piRNA
complex remain undiscovered.
lncRNA
lncRNA is a long-chain ncRNA of >200 nt. Studies
have shown that abnormal expression of lncRNA is closely related to
the development of various tumors and can show the function of
oncogenes and tumor suppressor genes (59,60).
According to the currently known lncRNAs, the following functions
can be summarized as follows: i) Some lncRNAs bind
epigenetic-related proteins after transcription, modifying specific
sequences, activating or inhibiting the activity of
chromatin-related regions, and regulating cell activities, such as
X-chromosome inactivation; ii) lncRNAs inhibit or promote gene
transcription via base pairing and recruitment of TFs; iii) lncRNA
are precursors to the formation of miRNAs and regulate the network
regulation of miRNA by mediating the generation of miRNAs; and iv)
lncRNAs participate in the formation of nuclear sub-structures by
regulating related genes (5,9). Furthermore, lncRNAs can be involved in
gene network regulation in three ways: i) In the transcriptional
level: When lncRNA itself is transcribed, wherein it interferes
with the transcription of adjacent genes, and where it can be used
as an endogenous competitive RNA to control myoblast
differentiation; ii) in the post-transcriptional level: By
interacting with miRNA, lncRNA affects post-transcriptional
translation of related mRNA, thereby promoting the malignant
transformation and proliferation of cells; and iii) in the
epigenetic level: lncRNA regulates histone modification to silence
expression of adjacent genes, also affecting chromosome
reconstruction to promote tumor invasion and metastasis (42). Nevertheless, the mechanism of action
of lncRNA on tumorigenesis and development remains in its infancy.
Further research and elaboration of the mechanism of action of
lncRNA will provide new approaches to tumor diagnosis, treatment
and prognosis.
In terms of structure, lncRNAs usually have the same
structure as mRNAs, including 5′-caps and 3′-polyadenylated tails,
and they are spliced to form the final product. Despite the fact
that lncRNAs are poorly sequentially conserved, some studies
suggest that their functions may be relatively conserved (3,61).
Furthermore, genes are usually located very close to lncRNA in the
genome, and according to the proximity of their protein-coding
genes, lncRNA can be divided into five categories: i) Antisense
lncRNA; ii) intronic transcript lncRNA; iii) large intergenic
lncRNA; iv) promoter-associated lncRNA; and v) UTR-associated
lncRNA (42,61). Similar to miRNA, lncRNA plays both
carcinogenic and anti-oncogene roles in terms of occurrence,
development and metastasis of various tumors. While similarities
between lncRNA and miRNA exist, they are usually highly diverse in
structure and function. HOTAIR was the first lncRNA shown to
regulate gene expression in the form of reverse transcription, as
identified by Vorvis et al (3) lncRNA HOTAIR showed higher expression in
pancreatic cancer with peripheral tissue infiltration and lymph
node metastasis (29,62). Survival analysis of pancreatic cancer
patients showed that patients with low expression of lncRNA HOTAIR
had a significantly longer survival time than those with high
expression, which is of great significance in predicting prognosis
(62).
Oncogenic lncRNA
To date, a partial oncogenic lncRNA has been
identified. Zhang et al (63)
used gene chip technology and found that ZEB1 antisense RNA 1
(zeb1-as1) was upregulated in liver cancer tissues, especially in
metastatic tumor tissues. zeb1-as1 overexpression was associated
with abnormal DNA methylation. Yan et al (33) and Volinia et al (64) found that colorectal cancer-associated
lncRNA showed high expression in colorectal cancer tissues. A
recent study showed that repulsive guidance molecule B as1
(rgmb-as1), upregulated in non-small cell lung cancer, was
associated with the degree of tumor differentiation, lymph node
metastasis and clinical stage, and was negatively correlated with
RGMB (25). As a pro-apoptotic
factor, GDFl5 inhibits the proliferation of cancer cells, and
HOTAIR binding with the GDFl5 gene region weakens its anticancer
effect in pancreatic cancer tissues.
Tumor suppressor gene-related
lncRNA
In contrast to oncogenic lncRNA, some lncRNAs have
been shown to act as tumor suppressors. Ferreira and Esteller
(16) and He et al (36) found that the expression of cell
adhesion molecule as1 (cadm1-as1) was downregulated in renal clear
cell carcinoma, and that interfering with the expression of
cadm1-as1 promoted the growth and migration of tumor cells, and
reduced the apoptosis rate. Some researchers found that the low
expression of taurine upregulated gene 1 (TUG1) was related to
high-level TNM stage, large tumor size and lower overall survival
in patients with non-small cell lung cancer. The inhibition of TUG1
expression significantly promoted the proliferation of tumor cells
and tissues (40,65). One study found that overexpression of
tslc1-as1 induced upregulation of TSLC1 and inhibited
proliferation, migration and invasion of tumor cells. The
expression of tslc1-as1 was positively correlated with other tumor
suppressor genes, including neurofibromatosis type 1, von
Hippel-Lindau tumor suppressor and phosphoinositide-3-kinase
regulatory subunit 1, while it was negatively correlated with the
oncogene B-Raf proto-oncogene serine/threonine kinase. In an in
vitro study, Li et al (42) and Yu et al (29) found that the overexpression of growth
arrest-specific 5 (GAS5) significantly inhibited the proliferation
and invasion of PANC1 and bxpc-3 cell lines. After inhibition of
GAS5 expression by RNA interference, GAS5 was arrested in the
G0/G1 stage. The proportion of cells in the
stage decreased, and more cells were in the S stage, suggesting
that GAS5 regulates the cell cycle of pancreatic cancer cells and
inhibits the invasion of cancer cells (66,67).
Bc-819 is a plasmid vector capable of expressing
diphtheria toxin A chain under the regulation of H19, which has
antitumor effects. Bc-819 was initially used in the treatment of
bladder cancer. In human trials, after ingestion, it expresses a
large amount of diphtheria toxin in cells, causing tumor volume
reduction. With the extensive development of clinical trials, the
effective concentration and dose of bc-819 have been further
determined (33,64). On this basis, Zhang et al
(63) and Zhou et al
(68) used gemcitabine, a first-line
chemotherapy drug for pancreatic cancer, in combination with bc-819
in an animal model of pancreatic cancer, and achieved good
efficacy. Pvt1 oncogene (PVT1) is directly related to the
sensitivity of pancreatic cancer to gemcitabine. In the human
pancreatic cancer aspc-1 cell line, decreased PVT1 activity
improved the sensitivity to gemcitabine. When Kim et al
(69) conducted high-throughput
sequencing for pancreatic cancer, it was found that homeobox A
distal transcript antisense RNA (HOTTIP) increased the expression
in pancreatic cancer and was involved in the development of
resistance to gemcitabine through HOXAl3. Tseng et al
(50) found that PVT1 in the aspc-1
cell line regulated the sensitivity of cells to gemcitabine, and
showed that the overexpression of full-length PVT1 positive-sense
cDNA increased the sensitivity of cells to gemcitabine. Meller
et al (40) confirmed that
malat-1 reduces the sensitivity of aspc-1 and cfpac-1 cell lines to
gemcitabine chemotherapy.
lncRNA mir31HG
lncRNA mir31HG is one of the lncRNA molecules with
the most significant differential expression, primarily located in
the cytoplasm. Recent studies have shown that mir-31 is located in
the intron region of mir31HG, and the transcription of mir-31 is
regulated by mir31HG. Both miR-31 and mir31HG were found to be
significantly positively correlated in pancreatic cancer tissues,
while the expression of mir31HG was not changed with the decrease
of mir-31 level, suggesting that both may exert their respective
roles independently (33,64). mir-31 expression did not change at
low concentrations of mir31HG, and the inhibition or expression of
mir-31 did not affect the expression of mir31HG. At the
transcriptional level, mir31HG and mir-31 did not regulate the
expression of each other, and their biological functions were
independent. The pancreatic cancer gene function of mir31HG acted
by promoting the proliferation, migration and invasion of
pancreatic cancer cells (34,64).
Another study suggested that the low expression of mir-193b in
pancreatic cancer has a significant negative correlation with the
expression of mir31HG (70).
mir-193b was found to negatively regulate the expression and
function of mir31HG by directly targeting mir31HG.
Maternally expressed 3 (MEG3)
MEG3 is an imprinted gene expressed by the mother,
located in the positive q32 region of chromosome 14, and its
transcription product MEG3 is a 1,595-bp long RNA molecule. Another
study found that tumor protein p53 and growth differentiation
factor 15 play important roles in the regulation of MEG3 (71). There are two differentially
methylated regions (DMRs) upstream of the MEG3 gene: IG-DMR and
meg3-dmr. Previous studies found that these two DMRs may be the
control centers of gene expression for the entire dlk1-meg3 unit
(61,71). Zhang et al (63) found that the meg3-dmr was fully
methylated in a neuroblastoma cell line. The ultimate effect was an
increase in the methylation level of the MEG3 gene promoter, which
was also the direct cause of MEG3 inactivation. Zhang et al
(63) and Peng and Jiang (61) found that the expression of MEG3 in
pancreatic cancer was negatively correlated with the
clinicopathological features of the tumor. MEG3 overexpression
inhibited pancreatic cancer activity by regulating the
PI3K/AKT/bcl-2/BAX/cyclin D1/p53 and PI3K/AKT/mmp-2/mmp-9 signaling
pathways. AKT, bcl1-2 cell cycle protein and D1 are found in normal
tissues and inhibit pyruvate dehydrogenase kinase during cell
division to promote the expression of p53 and BAX. They inhibit the
invasion and migration of the pancreatic cancer PANC1 cell line by
inhibiting the expression of mmp-2 and mmp-9 (39,72).
H19 imprinted maternally expressed
transcript (H19)
H19 plays an oncogenic role by antagonizing
hmga2-mediated EMT through antagonizing the anti-oncogene let-7,
which is primarily located in the cytoplasm and has a high
expression level during embryonic development, while it is strongly
suppressed after birth (70). As the
first exon of H19 transcription, mir-675 is negatively correlated
with the expression level of H19. As a key TF, E2F-1 is positively
correlated with the expression levels of H19, and E2F-1 is a direct
target of miR675. mir-675 reduces H19 activation by affecting the
expression of E2F-1 protein by binding to the 3′UTR on E2F-1 mRNA
(73). Furthermore, some studies
have shown that H19 has the dual function of oncogene and tumor
suppressor. H19 was the earliest lncRNA used for pancreatic cancer
treatment. DNA treatment controlled by the H19 gene sequence,
whether used alone or combined with gemcitabine, significantly
improves the therapeutic effect in pancreatic cancer (36,74,75).
PVT1
PVT1 has the potential to cause cancer in numerous
malignant tumors. However, only recently, studies revealed the role
of PVT1 in pancreatic cancer. Tseng et al (50) found that the expression levels of
PVT1 were positively correlated with clinical stage and total
survival time in pancreatic cancer. PVT1 is a regulator of the
sensitivity of pancreatic cancer cells to gemcitabine. Other
studies showed that curcumin inhibited the prc2-pvtl-c-myc axis by
inhibiting the prc2-subunit enhancer of zeste 2 polycomb repressive
complex 2 subunit, targeting CSCs and enhancing the sensitivity of
cancer cells to chemical therapeutic agents (2,42). Tutar
(39) and Tseng et al
(50) found that PVT1 promoted the
proliferation and metastasis of pancreatic cancer cells by acting
as a ‘molecular sponge’ to inhibit mir-448 activation, thus
inhibiting the effect of SERPINE1 mRNA binding protein 1 on target
cells. Nevertheless, studies on the detailed mechanism of PVT1 in
pancreatic cancer and other tumor types are rare. Furthermore,
other genes play a crucial role in pancreatic cancer, and they are
located at various sites (11)
(Table II).
 | Table II.Function and location of typical
genes involved in pancreatic cancer. |
Table II.
Function and location of typical
genes involved in pancreatic cancer.
| First author
(year) | Gene name | Genomic
localization | Involvement in
pancreatic cancer | (Refs.) |
|---|
| Wang (2017), Liu
(2016) | H19 | 11p15.5 | Cell proliferation,
invasion, targeted therapy | (70,73) |
| Martens-Uzunova
(2014) | PVT 1 | 8q24.21 | Drug
resistance | (65) |
| Vallot (2017), Xie
(2017) | HULC | 6p24.3 | Cell
proliferation | (66,67) |
| Meller (2015) | AF 339813 | 13q31.3 | Cell proliferation
and apoptosis | (40) |
| Pang (2015) | Gas5 | 1q25.1 | Cell proliferation,
the cell cycle | (72) |
| Kim (2013) | HOTAIR | 12q12.13 | Proliferation,
apoptosis, drug resistance, chemotherapy sensitivity | (69) |
| Fuschi (2019) | UCA1 | 19pl13.12 | Proliferation,
migration, invasion | (75) |
| He (2018),
Martens-Uzunova (2014) | ROR | 1p31.3 | Proliferation,
migration, invasion, drug resistance (autophagy) | (36,65) |
| Li (2017) | TUG1 | 22q12.2 | Proliferation,
migration (EMT) | (42) |
| Meller (2015),
Hancock (2018) |
ENST00000480739 | 12q13 | Unspecified | (40,43) |
Drug resistance and sensitivity in
drug application
The clinical efficacy of some pancreatic
cancer-targeted drugs, such as gemcitabine, remains controversial.
To date, some lncRNAs have been found to be associated with
pancreatic cancer therapy. Sethi et al (76) and Taucher et al (25) used a high-throughput microarray to
measure the expression profiles of lncRNAs in pancreatic cancer
tissue and found that quantitative PCR upregulated the expression
of HOTTIP in pancreatic cancer. Moreover, the study also reported
that HOTTIP promoted gemcitabine resistance through HOXA13,
suggesting that HOTTIP and HOXA13 may be new therapeutic targets
for pancreatic cancer. Unfortunately, the study failed to identify
the downstream signaling pathways that HOTTIP regulates (77). Tang et al (78) found that the PVT1 gene regulated the
sensitivity of gemcitabine in the human aspc-1 pancreatic cancer
cell line. It was found that that the sensitivity of gemcitabine
increased when the full-length PVT1 cDNA was overexpressed from the
antisense strand, and decreased when the full-length PVT1 cDNA was
overexpressed from the positive-sense strand. Nevertheless, the
underlying mechanism by which PVT1 regulates gemcitabine in
pancreatic tumorigenesis remains unknown. Xiong et al
(11) found that malat-1 reduced the
chemical sensitivity of gemcitabine in aspc-1 and cfpac-1
cells.
circRNA
Circular RNAs (circRNAs) are a class of ncRNAs
involved in the regulation of transcription and
post-transcriptional gene expression; they are also a class of
covalently bonded single-stranded ncRNAs. circRNA has the following
biological characteristics: i) As opposed to linear RNA, circRNA
forms a closed covalently circular structure that it is more stable
than linear RNA; ii) compared with linear mRNA, the species of
circRNA are more abundant and varied; iii) non-circRNA is primarily
derived from exons, and a large number of circRNAs exist in the
cytoplasm of eukaryotic cells; nevertheless, a small number of
circRNAs, derived from introns or intron fragments, exist in the
nucleus with spatiotemporal specificity of expression in various
tissues at various stages of development; iv) most circRNAs are
ncRNAs that are evolutionarily conserved; and v) most circRNAs are
regulated at the transcriptional or post-transcriptional level
(79,80). Knupp (81) and Li et al (82) compared microarray expression profiles
of circRNA in pancreatic ductal adenocarcinoma and normal
first-line tissues, and found that the expression of circRNA was
different in two different tissues, and the most typical of which
were ci-7 and ci-sirt7. circRNA alters the occurrence of pancreatic
cancer, and is expressed in normal and tumor tissues with
specificity, being more stable than linear RNA, and at higher
concentration in serum. These characteristics make circRNA an ideal
biomarker of pancreatic cancer and a substantial potential
therapeutic target (83–85). Nevertheless, research on the
correlation between circRNA and pancreatic cancer remains in the
preliminary stage, and the study of the association between circRNA
and pancreatic cancer is a hotspot in the current field of
pancreatic cancer research.
Conclusions
ncRNAs have become hotspots of modern medical
research; they promote the proliferation and metastasis of
pancreatic cancer through EMT, competitive endogenous RNA (ceRNA),
the cell cycle, apoptosis and other pathways. These various aspects
are isolated; nevertheless, they are interwoven with one another.
Several lncRNAs have been identified as oncogenes or tumor
suppressors involved in tumorigenesis. Despite the fact that
protein biomarkers are widely used in cancer detection and in
follow-up studies of diseases, lncRNAs have their own advantages.
lncRNA itself is an effector molecule that reflects the actual
level of a tumor. The correlation between lncRNA levels and
specific types of tumors makes it an accurate tool for tumor
diagnosis and classification (2,85). The
expression of lncRNA is related to tumor therapeutic response and
can be used as an indicator of therapeutic effect. Combined with
other prognostic methods, lncRNAs can be used as new tumor markers,
facilitating the identification of patients with poor prognosis and
prolonging their survival. Furthermore, lncRNA has extensive
application prospects in terms of tumor treatment (37,85).
Currently, lncRNA has been used in the treatment of various tumors.
However, most of the current studies on lncRNA focus on the
differences in the expression levels, and relatively few studies
have identified molecular functions. The discovery and
understanding of carcinogenic effects in pancreatic cancer are also
incomplete, and the details of these mechanisms remain to be
elucidated. With the continuous innovation and development of
research methods, lncRNAs may become potential biomarkers and
therapeutic targets for pancreatic cancer (2,35).
Acknowledgements
Not applicable.
Funding
The study was supported by grants from The Youth
Fund of The First Affiliated Hospital of Zhengzhou University.
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
YL and SH contributed to the design, as well as the
writing of the review. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peng JF, Zhuang YY, Huang FT and Zhang SN:
Noncoding RNAs and pancreatic cancer. World J Gastroenterol.
22:801–814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vorvis C, Hatziapostolou M, Mahurkar-Joshi
S, Koutsioumpa M, Williams J, Donahue TR, Poultsides GA, Eibl G and
Iliopoulos D: Transcriptomic and CRISPR/Cas9 technologies reveal
FOXA2 as a tumor suppressor gene in pancreatic cancer. Am J Physiol
Gastrointest Liver Physiol. 310:G1124–G1137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duguang L, Jin H, Xiaowei Q, Peng X,
Xiaodong W, Zhennan L, Jianjun Q and Jie Y: The involvement of
lncRNAs in the development and progression of pancreatic cancer.
Cancer Biol Ther. 18:927–936. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao
X, Li W, Zheng S, Ye H, Wang L, et al: LncRNA HOTTIP modulates
cancer stem cell properties in human pancreatic cancer by
regulating HOXA9. Cancer Lett. 410:68–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y
and Zhang G: LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth
and invasion through regulating the miR-204/HMGB1 axis. Int J Biol
Macromol. 116:545–551. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chitkara D, Mittal A and Mahato RI: miRNAs
in pancreatic cancer: Therapeutic potential, delivery challenges
and strategies. Adv Drug Deliv Rev. 81:34–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen P, Wan D, Zheng D, Zheng Q, Wu F and
Zhi Q: Long non-coding RNA UCA1 promotes the tumorigenesis in
pancreatic cancer. Biomed Pharmacother. 83:1220–1226. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y and Sarkar FH: MicroRNA targeted
therapeutic approach for pancreatic cancer. Int J Biol Sci.
12:326–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiong G, Feng M, Yang G, Zheng S, Song X,
Cao Z, You L, Zheng L, Hu Y, Zhang T and Zhao Y: The underlying
mechanisms of non-coding RNAs in the chemoresistance of pancreatic
cancer. Cancer Lett. 397:94–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abreu FB, Liu X and Tsongalis GJ: miRNA
analysis in pancreatic cancer: The Dartmouth experience. Clin Chem
Lab Med. 55:755–762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Batchu RB, Gruzdyn OV, Qazi AM, Kaur J,
Mahmud EM, Weaver DW and Gruber SA: Enhanced phosphorylation of p53
by microRNA-26a leading to growth inhibition of pancreatic cancer.
Surgery. 158:981–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Czech B and Hannon GJ: One loop to rule
them all: The ping-pong cycle and piRNA-guided silencing. Trends
Biochem Sci. 41:324–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guil S and Esteller M: RNA-RNA
interactions in gene regulation: The coding and noncoding players.
Trends Biochem Sci. 40:248–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ferreira HJ and Esteller M: Non-coding
RNAs, epigenetics, and cancer: Tying it all together. Cancer
Metastasis Rev. 37:55–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai X, Wang M, McElyea SD, Sherman S,
House M and Korc M: A microRNA signature in circulating exosomes is
superior to exosomal glypican-1 levels for diagnosing pancreatic
cancer. Cancer Lett. 393:86–93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li A, Yu J, Kim H, Wolfgang CL, Canto MI,
Hruban RH and Goggins M: MicroRNA array analysis finds elevated
serum miR-1290 accurately distinguishes patients with low-stage
pancreatic cancer from healthy and disease controls. Clin Cancer
Res. 19:3600–3610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang J, Yao M, Li Y, Zhao D, Hu S, Cui X,
Liu G, Shi Q, Wang Y and Yang Y: MicroRNAs for osteosarcoma in the
mouse: A meta-analysis. Oncotarget. 7:85650–85674. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Delás MJ, Sabin LR, Dolzhenko E, Knott SR,
Munera Maravilla E, Jackson BT, Wild SA, Kovacevic T, Stork EM,
Zhou M, et al: lncRNA requirements for mouse acute myeloid leukemia
and normal differentiation. Elife. 6:e256072017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He L, Tian DA, Li PY and He XX: Mouse
models of liver cancer: Progress and recommendations. Oncotarget.
6:23306–23322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishihara Y, Tsuno S, Kuwamoto S, Yamashita
T, Endo Y, Miura K, Miura Y, Sato T, Hasegawa J and Miura N:
Tumor-suppressive effects of atelocollagen-conjugated
hsa-miR-520d-5p on un-differentiated cancer cells in a mouse
xenograft model. BMC Cancer. 16:4152016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei WF, Han LF, Liu D, Wu LF, Chen XJ, Yi
HY, Wu XG, Zhong M, Yu YH, Liang L and Wang W: Orthotopic xenograft
mouse model of cervical cancer for studying the role of MicroRNA-21
in promoting lymph node metastasis. Int J Gynecol Cancer.
27:1587–1595. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu J, Zhu W and Wu W: MicroRNAs change
the landscape of cancer resistance. Methods Mol Biol. 1699:83–89.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Taucher V, Mangge H and Haybaeck J:
Non-coding RNAs in pancreatic cancer: Challenges and opportunities
for clinical application. Cell Oncol (Dordr). 39:295–318. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moriya C, Taniguchi H, Miyata K, Nishiyama
N, Kataoka K and Imai K: Inhibition of PRDM14 expression in
pancreatic cancer suppresses cancer stem-like properties and liver
metastasis in mice. Carcinogenesis. 38:638–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun XJ, Liu BY, Yan S, Jiang TH, Cheng HQ,
Jiang HS, Cao Y and Mao AW: MicroRNA-29a promotes pancreatic cancer
growth by inhibiting tristetraprolin. Cell Physiol Biochem.
37:707–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang P, Zhang J, Zhang L, Zhu Z, Fan J,
Chen L, Zhuang L, Luo J, Chen H, Liu L, et al: MicroRNA 23b
regulates autophagy associated with radioresistance of pancreatic
cancer cells. Gastroenterology. 145:1133–1143.e12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu X, Zheng H, Chan MT and Wu WK: HULC: An
oncogenic long non-coding RNA in human cancer. J Cell Mol Med.
21:410–417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Volinia S: Unexpected findings of
variability in microRNAs suggest roles in human genetics. Genome
Med. 4:692012. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou B, Sun C, Hu X, Zhan H, Zou H, Feng
Y, Qiu F, Zhang S, Wu L and Zhang B: MicroRNA-195 suppresses the
progression of pancreatic cancer by targeting DCLK1. Cell Physiol
Biochem. 44:1867–1881. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan H, Li Q, Wu J, Hu W, Jiang J, Shi L,
Yang X, Zhu D, Ji M and Wu C: MiR-629 promotes human pancreatic
cancer progression by targeting FOXO3. Cell Death Dis. 8:e31542017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan L, Zhang J, Guo D, Ma J, Shui SF and
Han XW: IL-21R functions as an oncogenic factor and is regulated by
the lncRNA MALAT1/miR-125a-3p axis in gastric cancer. Int J Oncol.
54:7–16. 2019.PubMed/NCBI
|
|
34
|
Wang W, Zhao L, Wei X, Wang L, Liu S, Yang
Y, Wang F, Sun G, Zhang J, Ma Y, et al: MicroRNA-320a promotes 5-FU
resistance in human pancreatic cancer cells. Sci Rep. 6:276412016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vassaux G, Angelova A, Baril P, Midoux P,
Rommelaere J and Cordelier P: The promise of gene therapy for
pancreatic cancer. Hum Gene Ther. 27:127–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P,
Liu D, Tian L, Yin J, Jiang K and Miao Y: ALKBH5 inhibits
pancreatic cancer motility by decreasing long non-coding RNA
KCNK15-AS1 methylation. Cell Physiol Biochem. 48:838–846. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Wang J, Huang S, Zhu X, Liu J,
Yang N, Song D, Wu R, Deng W, Skogerbø G, et al: Systematic
identification and characterization of chicken (Gallus gallus)
ncRNAs. Nucleic Acids Res. 37:6562–6574. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qian Y, Feng L, Wu W, Weng T, Hu C, Hong
B, Wang FXC, Shen L, Wang Q, Jin X and Yao H: MicroRNA expression
profiling of pancreatic cancer cell line L3.6p1 following B7-H4
knockdown. Cell Physiol Biochem. 44:494–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tutar Y: miRNA and cancer; computational
and experimental approaches. Curr Pharm Biotechnol. 15:4292014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meller VH, Joshi SS and Deshpande N:
Modulation of chromatin by noncoding RNA. Annu Rev Genet.
49:673–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li M, Radvanyi L, Yin B, Li J, Chivukula
R, Lin K, Lu Y, Shen J, Chang DZ, Li D, et al: Downregulation of
human endogenous retrovirus type K (HERV-K) viral env RNA in
pancreatic cancer cells decreases cell proliferation and tumor
growth. Clin Cancer Res. 23:5892–5911. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hancock MH and Skalsky RL: Roles of
non-coding RNAs during herpesvirus infection. Curr Top Microbiol
Immunol. 419:243–280. 2018.PubMed/NCBI
|
|
44
|
Farshidfar F, Zheng S, Gingras MC, Newton
Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J,
et al: Integrative genomic analysis of cholangiocarcinoma
identifies distinct IDH-mutant molecular profiles. Cell Rep.
18:2780–2794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kamerkar S, LeBleu VS, Sugimoto H, Yang S,
Ruivo CF, Melo SA, Lee JJ and Kalluri R: Exosomes facilitate
therapeutic targeting of oncogenic KRAS in pancreatic cancer.
Nature. 546:498–503. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Binenbaum Y, Na'ara S and Gil Z:
Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug
Resist Updat. 23:55–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Repoila F and Darfeuille F: Small
regulatory non-coding RNAs in bacteria: Physiology and mechanistic
aspects. Biol Cell. 101:117–131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY,
Fang C, Huang Q and Tian L: microRNA-7 impairs autophagy-derived
pools of glucose to suppress pancreatic cancer progression. Cancer
Lett. 400:69–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Karamitopoulou E, Haemmig S, Baumgartner
U, Schlup C and Wartenberg M: MicroRNA dysregulation in the tumor
microenvironment influences the phenotype of pancreatic cancer. Mod
Pathol. 30:1116–1125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen Q, Wang P, Fu Y, Liu X, Xu W, Wei J,
Gao W, Jiang K, Wu J and Miao Y: MicroRNA-217 inhibits cell
proliferation, invasion and migration by targeting Tpd52l2 in human
pancreatic adenocarcinoma. Oncol Rep. 38:3567–3573. 2017.PubMed/NCBI
|
|
52
|
Gayral M, Jo S, Hanoun N, Vignolle-Vidoni
A, Lulka H, Delpu Y, Meulle A, Dufresne M, Humeau M, Chalret du
Rieu M, et al: MicroRNAs as emerging biomarkers and therapeutic
targets for pancreatic cancer. World J Gastroenterol.
20:11199–11209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hao J, Zhang S, Zhou Y, Liu C, Hu X and
Shao C: MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer.
Biochem Biophys Res Commun. 406:552–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hong L, Yang Z, Ma J and Fan D: Function
of miRNA in controlling drug resistance of human cancers. Curr Drug
Targets. 14:1118–1127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Humeau M, Torrisani J and Cordelier P:
miRNA in clinical practice: Pancreatic cancer. Clin Biochem.
46:933–936. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kokuryo T, Hibino S, Suzuki K, Watanabe K,
Yokoyama Y, Nagino M, Senga T and Hamaguchi M: Nek2 siRNA therapy
using a portal venous port-catheter system for liver metastasis in
pancreatic cancer. Cancer Sci. 107:1315–1320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Andersen PR, Tirian L, Vunjak M and
Brennecke J: A heterochromatin-dependent transcription machinery
drives piRNA expression. Nature. 549:54–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Charlotte S: piRNAs power sperm
development in the adult. Biol Reprod. 94:2016.
|
|
59
|
Gao ZQ, Wang JF, Chen DH, Ma XS, Yang W,
Zhe T and Dang XW: Long non-coding RNA GAS5 antagonizes the
chemoresistance of pancreatic cancer cells through down-regulation
of miR-181c-5p. Biomed Pharmacother. 97:809–817. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gu L, Zhang J, Shi M, Zhan Q, Shen B and
Peng C: lncRNA MEG3 had anti-cancer effects to suppress pancreatic
cancer activity. Biomed Pharmacother. 89:1269–1276. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Peng W and Jiang A: Long noncoding RNA
CCDC26 as a potential predictor biomarker contributes to
tumorigenesis in pancreatic cancer. Biomed Pharmacother.
83:712–717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM
and Chen Y: The long non-coding RNA HOTAIR enhances pancreatic
cancer resistance to TNF-related apoptosis-inducing ligand. J Biol
Chem. 292:10390–10397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang L, Yang Z, Trottier J, Barbier O and
Wang L: Long noncoding RNA MEG3 induces cholestatic liver injury by
interaction with PTBP1 to facilitate shp mRNA decay. Hepatology.
65:604–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Volinia S, Visone R, Galasso M, Rossi E
and Croce CM: Identification of microRNA activity by targets'
reverse EXpression. Bioinformatics. 26:91–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Vallot C, Patrat C, Collier AJ, Huret C,
Casanova M, Liyakat Ali TM, Tosolini M, Frydman N, Heard E,
Rugg-Gunn PJ and Rougeulle C: XACT noncoding RNA competes with XIST
in the control of X chromosome activity during human early
development. Cell Stem Cell. 20:102–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xie VK, Li Z, Yan Y, Jia Z, Zuo X, Ju Z,
Wang J, Du J, Xie D, Xie K and Wei D: DNA-Methyltransferase 1
induces dedifferentiation of pancreatic cancer cells through
silencing of Kruppel-Like factor 4 expression. Clin Cancer Res.
23:5585–5597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhou DD, Liu XF, Lu CW, Pant OP and Liu
XD: Long non-coding RNA PVT1: Emerging biomarker in digestive
system cancer. Cell Prolif. Oct 12–2017.(Epub ahead of print). doi:
10.1111/cpr.12398. View Article : Google Scholar
|
|
69
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z,
Wang R, Feng J and Luo Y: Long noncoding RNA H19 promotes
neuroinflammation in ischemic stroke by driving histone deacetylase
1-dependent M1 microglial polarization. Stroke. 48:2211–2221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang G, Pan J, Zhang L, Wei Y and Wang C:
Long non-coding RNA CRNDE sponges miR-384 to promote proliferation
and metastasis of pancreatic cancer cells through upregulating
IRS1. Cell Prolif. Sep 21–2017.(Epub ahead of print). doi:
10.1111/cpr.12389. View Article : Google Scholar
|
|
72
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu L, An X, Li Z, Song Y, Li L, Zuo S,
Liu N, Yang G, Wang H, Cheng X, et al: The H19 long noncoding RNA
is a novel negative regulator of cardiomyocyte hypertrophy.
Cardiovasc Res. 111:56–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Heiler S, Wang Z and Zöller M: Pancreatic
cancer stem cell markers and exosomes-the incentive push. World J
Gastroenterol. 22:5971–6007. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fuschi P, Maimone B, Gaetano C and
Martelli F: Noncoding RNAs in the vascular system response to
oxidative stress. Antioxid Redox Signal. 30:992–1010. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sethi S, Sethi S and Bluth MH: Clinical
implication of MicroRNAs in molecular pathology: An update for
2018. Clin Lab Med. 38:237–251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Vicentini C, Fassan M, D'Angelo E, Corbo
V, Silvestris N, Nuovo GJ and Scarpa A: Clinical application of
microRNA testing in neuroendocrine tumors of the gastrointestinal
tract. Molecules. 19:2458–2468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tang YT, Xu XH, Yang XD, Hao J, Cao H, Zhu
W, Zhang SY and Cao JP: Role of non-coding RNAs in pancreatic
cancer: The bane of the microworld. World J Gastroenterol.
20:9405–9417. 2014.PubMed/NCBI
|
|
79
|
Zhang S, Zhu D, Li H, Li H, Feng C and
Zhang W: Characterization of circRNA-associated-ceRNA networks in a
senescence- accelerated mouse prone 8 brain. Mol Ther.
25:2053–2061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kishikawa T, Otsuka M, Ohno M, Yoshikawa
T, Takata A and Koike K: Circulating RNAs as new biomarkers for
detecting pancreatic cancer. World J Gastroenterol. 21:8527–8540.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Knupp D and Miura P: CircRNA accumulation:
A new hallmark of aging? Mech Ageing Dev. 173:71–79. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Vassella E, Chaudhary AK, Mondal G, Kumar
V, Kattel K and Mahato RI: Chemosensitization and inhibition of
pancreatic cancer stem cell proliferation by overexpression of
microRNA-205. Cancer Lett. 402:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yoshida K, Toden S, Ravindranathan P, Han
H and Goel A: Curcumin sensitizes pancreatic cancer cells to
gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1
expression. Carcinogenesis. 38:1036–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as Heracles in autophagy.
Autophagy. 13:1004–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|