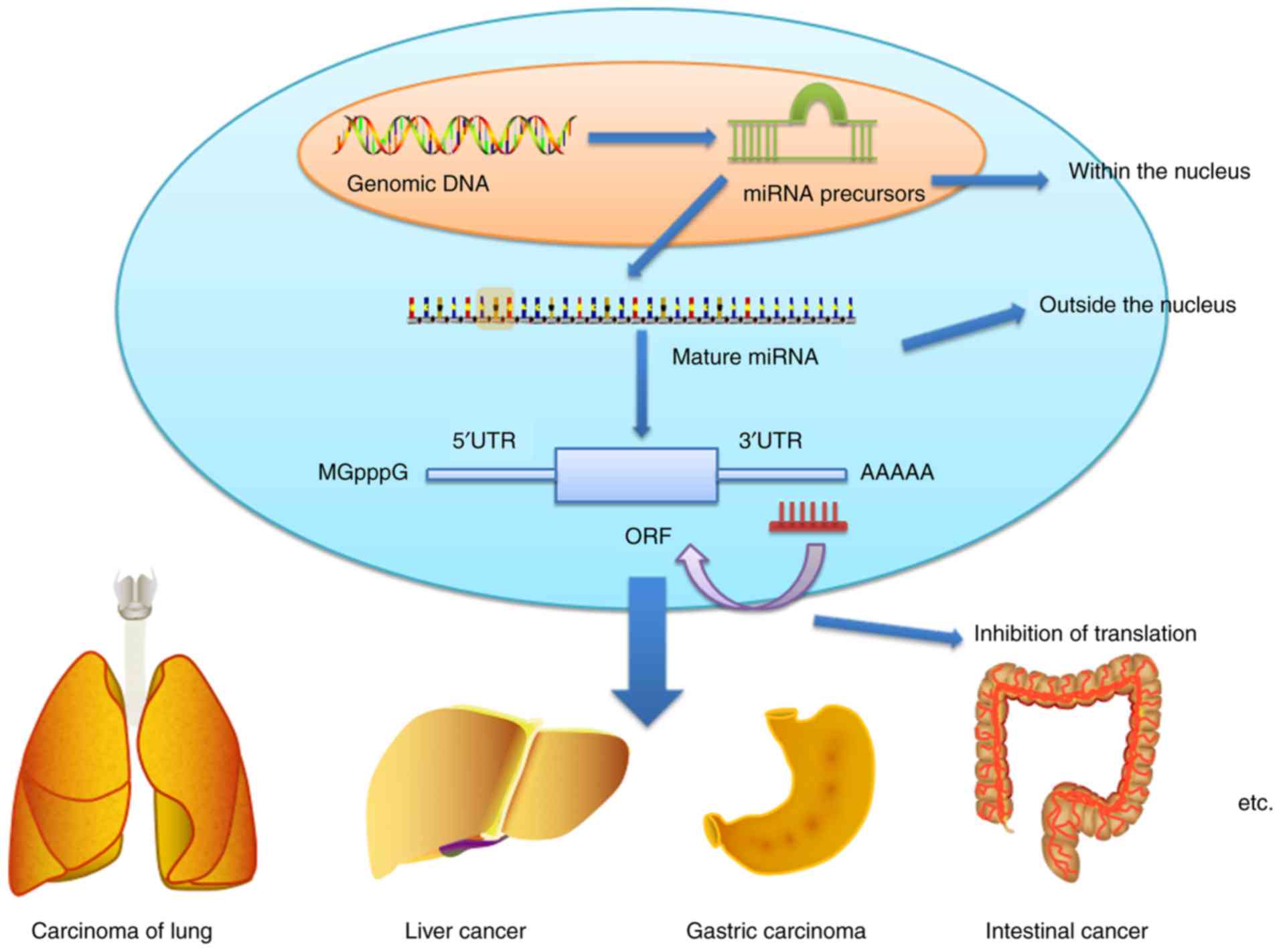
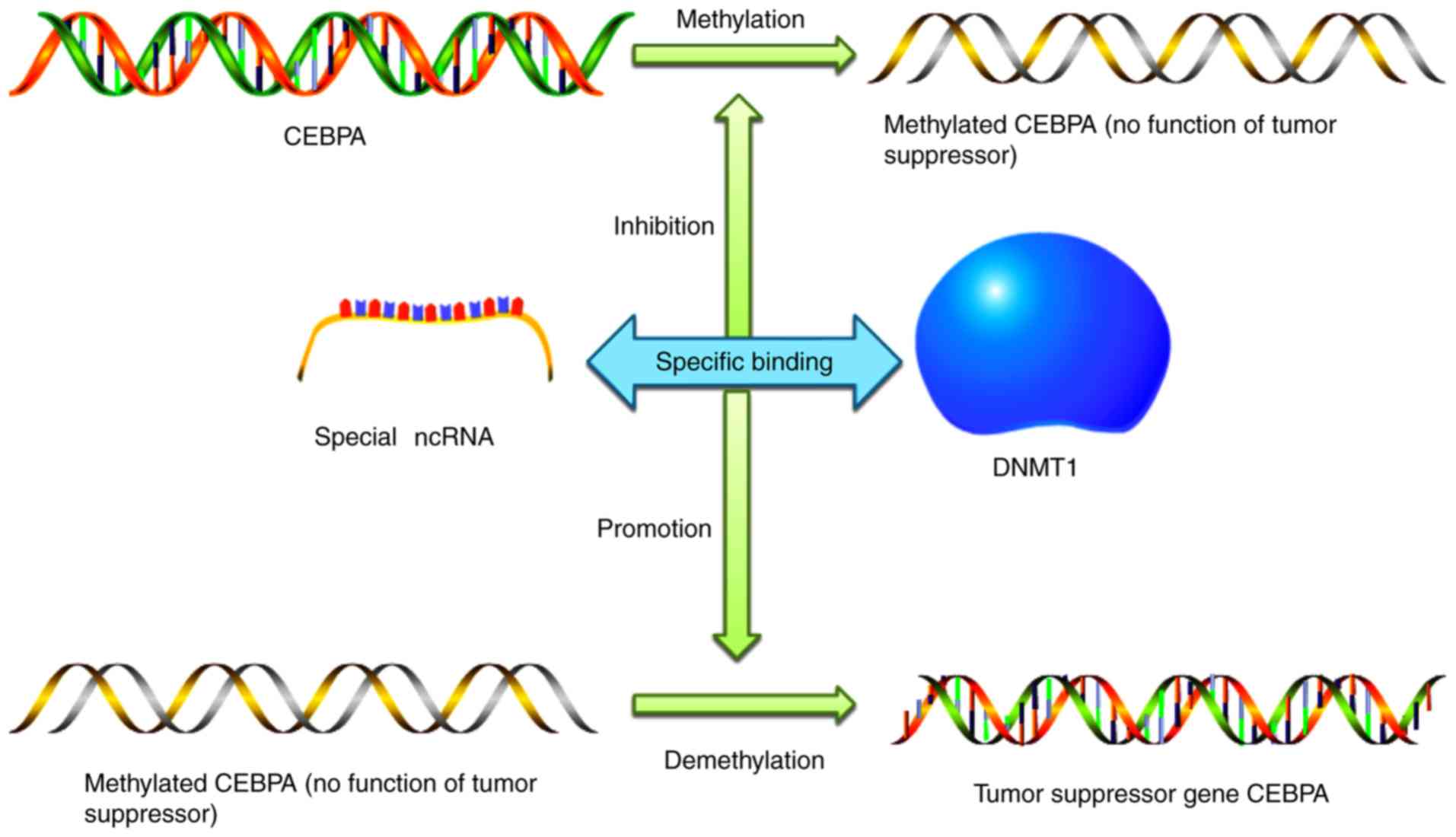
|
1
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peng JF, Zhuang YY, Huang FT and Zhang SN:
Noncoding RNAs and pancreatic cancer. World J Gastroenterol.
22:801–814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vorvis C, Hatziapostolou M, Mahurkar-Joshi
S, Koutsioumpa M, Williams J, Donahue TR, Poultsides GA, Eibl G and
Iliopoulos D: Transcriptomic and CRISPR/Cas9 technologies reveal
FOXA2 as a tumor suppressor gene in pancreatic cancer. Am J Physiol
Gastrointest Liver Physiol. 310:G1124–G1137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duguang L, Jin H, Xiaowei Q, Peng X,
Xiaodong W, Zhennan L, Jianjun Q and Jie Y: The involvement of
lncRNAs in the development and progression of pancreatic cancer.
Cancer Biol Ther. 18:927–936. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao
X, Li W, Zheng S, Ye H, Wang L, et al: LncRNA HOTTIP modulates
cancer stem cell properties in human pancreatic cancer by
regulating HOXA9. Cancer Lett. 410:68–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y
and Zhang G: LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth
and invasion through regulating the miR-204/HMGB1 axis. Int J Biol
Macromol. 116:545–551. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chitkara D, Mittal A and Mahato RI: miRNAs
in pancreatic cancer: Therapeutic potential, delivery challenges
and strategies. Adv Drug Deliv Rev. 81:34–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen P, Wan D, Zheng D, Zheng Q, Wu F and
Zhi Q: Long non-coding RNA UCA1 promotes the tumorigenesis in
pancreatic cancer. Biomed Pharmacother. 83:1220–1226. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y and Sarkar FH: MicroRNA targeted
therapeutic approach for pancreatic cancer. Int J Biol Sci.
12:326–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiong G, Feng M, Yang G, Zheng S, Song X,
Cao Z, You L, Zheng L, Hu Y, Zhang T and Zhao Y: The underlying
mechanisms of non-coding RNAs in the chemoresistance of pancreatic
cancer. Cancer Lett. 397:94–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abreu FB, Liu X and Tsongalis GJ: miRNA
analysis in pancreatic cancer: The Dartmouth experience. Clin Chem
Lab Med. 55:755–762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Batchu RB, Gruzdyn OV, Qazi AM, Kaur J,
Mahmud EM, Weaver DW and Gruber SA: Enhanced phosphorylation of p53
by microRNA-26a leading to growth inhibition of pancreatic cancer.
Surgery. 158:981–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Czech B and Hannon GJ: One loop to rule
them all: The ping-pong cycle and piRNA-guided silencing. Trends
Biochem Sci. 41:324–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guil S and Esteller M: RNA-RNA
interactions in gene regulation: The coding and noncoding players.
Trends Biochem Sci. 40:248–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ferreira HJ and Esteller M: Non-coding
RNAs, epigenetics, and cancer: Tying it all together. Cancer
Metastasis Rev. 37:55–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai X, Wang M, McElyea SD, Sherman S,
House M and Korc M: A microRNA signature in circulating exosomes is
superior to exosomal glypican-1 levels for diagnosing pancreatic
cancer. Cancer Lett. 393:86–93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li A, Yu J, Kim H, Wolfgang CL, Canto MI,
Hruban RH and Goggins M: MicroRNA array analysis finds elevated
serum miR-1290 accurately distinguishes patients with low-stage
pancreatic cancer from healthy and disease controls. Clin Cancer
Res. 19:3600–3610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang J, Yao M, Li Y, Zhao D, Hu S, Cui X,
Liu G, Shi Q, Wang Y and Yang Y: MicroRNAs for osteosarcoma in the
mouse: A meta-analysis. Oncotarget. 7:85650–85674. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Delás MJ, Sabin LR, Dolzhenko E, Knott SR,
Munera Maravilla E, Jackson BT, Wild SA, Kovacevic T, Stork EM,
Zhou M, et al: lncRNA requirements for mouse acute myeloid leukemia
and normal differentiation. Elife. 6:e256072017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He L, Tian DA, Li PY and He XX: Mouse
models of liver cancer: Progress and recommendations. Oncotarget.
6:23306–23322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishihara Y, Tsuno S, Kuwamoto S, Yamashita
T, Endo Y, Miura K, Miura Y, Sato T, Hasegawa J and Miura N:
Tumor-suppressive effects of atelocollagen-conjugated
hsa-miR-520d-5p on un-differentiated cancer cells in a mouse
xenograft model. BMC Cancer. 16:4152016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei WF, Han LF, Liu D, Wu LF, Chen XJ, Yi
HY, Wu XG, Zhong M, Yu YH, Liang L and Wang W: Orthotopic xenograft
mouse model of cervical cancer for studying the role of MicroRNA-21
in promoting lymph node metastasis. Int J Gynecol Cancer.
27:1587–1595. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu J, Zhu W and Wu W: MicroRNAs change
the landscape of cancer resistance. Methods Mol Biol. 1699:83–89.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Taucher V, Mangge H and Haybaeck J:
Non-coding RNAs in pancreatic cancer: Challenges and opportunities
for clinical application. Cell Oncol (Dordr). 39:295–318. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moriya C, Taniguchi H, Miyata K, Nishiyama
N, Kataoka K and Imai K: Inhibition of PRDM14 expression in
pancreatic cancer suppresses cancer stem-like properties and liver
metastasis in mice. Carcinogenesis. 38:638–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun XJ, Liu BY, Yan S, Jiang TH, Cheng HQ,
Jiang HS, Cao Y and Mao AW: MicroRNA-29a promotes pancreatic cancer
growth by inhibiting tristetraprolin. Cell Physiol Biochem.
37:707–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang P, Zhang J, Zhang L, Zhu Z, Fan J,
Chen L, Zhuang L, Luo J, Chen H, Liu L, et al: MicroRNA 23b
regulates autophagy associated with radioresistance of pancreatic
cancer cells. Gastroenterology. 145:1133–1143.e12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu X, Zheng H, Chan MT and Wu WK: HULC: An
oncogenic long non-coding RNA in human cancer. J Cell Mol Med.
21:410–417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Volinia S: Unexpected findings of
variability in microRNAs suggest roles in human genetics. Genome
Med. 4:692012. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou B, Sun C, Hu X, Zhan H, Zou H, Feng
Y, Qiu F, Zhang S, Wu L and Zhang B: MicroRNA-195 suppresses the
progression of pancreatic cancer by targeting DCLK1. Cell Physiol
Biochem. 44:1867–1881. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan H, Li Q, Wu J, Hu W, Jiang J, Shi L,
Yang X, Zhu D, Ji M and Wu C: MiR-629 promotes human pancreatic
cancer progression by targeting FOXO3. Cell Death Dis. 8:e31542017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan L, Zhang J, Guo D, Ma J, Shui SF and
Han XW: IL-21R functions as an oncogenic factor and is regulated by
the lncRNA MALAT1/miR-125a-3p axis in gastric cancer. Int J Oncol.
54:7–16. 2019.PubMed/NCBI
|
|
34
|
Wang W, Zhao L, Wei X, Wang L, Liu S, Yang
Y, Wang F, Sun G, Zhang J, Ma Y, et al: MicroRNA-320a promotes 5-FU
resistance in human pancreatic cancer cells. Sci Rep. 6:276412016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vassaux G, Angelova A, Baril P, Midoux P,
Rommelaere J and Cordelier P: The promise of gene therapy for
pancreatic cancer. Hum Gene Ther. 27:127–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P,
Liu D, Tian L, Yin J, Jiang K and Miao Y: ALKBH5 inhibits
pancreatic cancer motility by decreasing long non-coding RNA
KCNK15-AS1 methylation. Cell Physiol Biochem. 48:838–846. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Wang J, Huang S, Zhu X, Liu J,
Yang N, Song D, Wu R, Deng W, Skogerbø G, et al: Systematic
identification and characterization of chicken (Gallus gallus)
ncRNAs. Nucleic Acids Res. 37:6562–6574. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qian Y, Feng L, Wu W, Weng T, Hu C, Hong
B, Wang FXC, Shen L, Wang Q, Jin X and Yao H: MicroRNA expression
profiling of pancreatic cancer cell line L3.6p1 following B7-H4
knockdown. Cell Physiol Biochem. 44:494–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tutar Y: miRNA and cancer; computational
and experimental approaches. Curr Pharm Biotechnol. 15:4292014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meller VH, Joshi SS and Deshpande N:
Modulation of chromatin by noncoding RNA. Annu Rev Genet.
49:673–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li M, Radvanyi L, Yin B, Li J, Chivukula
R, Lin K, Lu Y, Shen J, Chang DZ, Li D, et al: Downregulation of
human endogenous retrovirus type K (HERV-K) viral env RNA in
pancreatic cancer cells decreases cell proliferation and tumor
growth. Clin Cancer Res. 23:5892–5911. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hancock MH and Skalsky RL: Roles of
non-coding RNAs during herpesvirus infection. Curr Top Microbiol
Immunol. 419:243–280. 2018.PubMed/NCBI
|
|
44
|
Farshidfar F, Zheng S, Gingras MC, Newton
Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J,
et al: Integrative genomic analysis of cholangiocarcinoma
identifies distinct IDH-mutant molecular profiles. Cell Rep.
18:2780–2794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kamerkar S, LeBleu VS, Sugimoto H, Yang S,
Ruivo CF, Melo SA, Lee JJ and Kalluri R: Exosomes facilitate
therapeutic targeting of oncogenic KRAS in pancreatic cancer.
Nature. 546:498–503. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Binenbaum Y, Na'ara S and Gil Z:
Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug
Resist Updat. 23:55–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Repoila F and Darfeuille F: Small
regulatory non-coding RNAs in bacteria: Physiology and mechanistic
aspects. Biol Cell. 101:117–131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY,
Fang C, Huang Q and Tian L: microRNA-7 impairs autophagy-derived
pools of glucose to suppress pancreatic cancer progression. Cancer
Lett. 400:69–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Karamitopoulou E, Haemmig S, Baumgartner
U, Schlup C and Wartenberg M: MicroRNA dysregulation in the tumor
microenvironment influences the phenotype of pancreatic cancer. Mod
Pathol. 30:1116–1125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen Q, Wang P, Fu Y, Liu X, Xu W, Wei J,
Gao W, Jiang K, Wu J and Miao Y: MicroRNA-217 inhibits cell
proliferation, invasion and migration by targeting Tpd52l2 in human
pancreatic adenocarcinoma. Oncol Rep. 38:3567–3573. 2017.PubMed/NCBI
|
|
52
|
Gayral M, Jo S, Hanoun N, Vignolle-Vidoni
A, Lulka H, Delpu Y, Meulle A, Dufresne M, Humeau M, Chalret du
Rieu M, et al: MicroRNAs as emerging biomarkers and therapeutic
targets for pancreatic cancer. World J Gastroenterol.
20:11199–11209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hao J, Zhang S, Zhou Y, Liu C, Hu X and
Shao C: MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer.
Biochem Biophys Res Commun. 406:552–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hong L, Yang Z, Ma J and Fan D: Function
of miRNA in controlling drug resistance of human cancers. Curr Drug
Targets. 14:1118–1127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Humeau M, Torrisani J and Cordelier P:
miRNA in clinical practice: Pancreatic cancer. Clin Biochem.
46:933–936. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kokuryo T, Hibino S, Suzuki K, Watanabe K,
Yokoyama Y, Nagino M, Senga T and Hamaguchi M: Nek2 siRNA therapy
using a portal venous port-catheter system for liver metastasis in
pancreatic cancer. Cancer Sci. 107:1315–1320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Andersen PR, Tirian L, Vunjak M and
Brennecke J: A heterochromatin-dependent transcription machinery
drives piRNA expression. Nature. 549:54–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Charlotte S: piRNAs power sperm
development in the adult. Biol Reprod. 94:2016.
|
|
59
|
Gao ZQ, Wang JF, Chen DH, Ma XS, Yang W,
Zhe T and Dang XW: Long non-coding RNA GAS5 antagonizes the
chemoresistance of pancreatic cancer cells through down-regulation
of miR-181c-5p. Biomed Pharmacother. 97:809–817. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gu L, Zhang J, Shi M, Zhan Q, Shen B and
Peng C: lncRNA MEG3 had anti-cancer effects to suppress pancreatic
cancer activity. Biomed Pharmacother. 89:1269–1276. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Peng W and Jiang A: Long noncoding RNA
CCDC26 as a potential predictor biomarker contributes to
tumorigenesis in pancreatic cancer. Biomed Pharmacother.
83:712–717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM
and Chen Y: The long non-coding RNA HOTAIR enhances pancreatic
cancer resistance to TNF-related apoptosis-inducing ligand. J Biol
Chem. 292:10390–10397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang L, Yang Z, Trottier J, Barbier O and
Wang L: Long noncoding RNA MEG3 induces cholestatic liver injury by
interaction with PTBP1 to facilitate shp mRNA decay. Hepatology.
65:604–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Volinia S, Visone R, Galasso M, Rossi E
and Croce CM: Identification of microRNA activity by targets'
reverse EXpression. Bioinformatics. 26:91–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Vallot C, Patrat C, Collier AJ, Huret C,
Casanova M, Liyakat Ali TM, Tosolini M, Frydman N, Heard E,
Rugg-Gunn PJ and Rougeulle C: XACT noncoding RNA competes with XIST
in the control of X chromosome activity during human early
development. Cell Stem Cell. 20:102–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xie VK, Li Z, Yan Y, Jia Z, Zuo X, Ju Z,
Wang J, Du J, Xie D, Xie K and Wei D: DNA-Methyltransferase 1
induces dedifferentiation of pancreatic cancer cells through
silencing of Kruppel-Like factor 4 expression. Clin Cancer Res.
23:5585–5597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhou DD, Liu XF, Lu CW, Pant OP and Liu
XD: Long non-coding RNA PVT1: Emerging biomarker in digestive
system cancer. Cell Prolif. Oct 12–2017.(Epub ahead of print). doi:
10.1111/cpr.12398. View Article : Google Scholar
|
|
69
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z,
Wang R, Feng J and Luo Y: Long noncoding RNA H19 promotes
neuroinflammation in ischemic stroke by driving histone deacetylase
1-dependent M1 microglial polarization. Stroke. 48:2211–2221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang G, Pan J, Zhang L, Wei Y and Wang C:
Long non-coding RNA CRNDE sponges miR-384 to promote proliferation
and metastasis of pancreatic cancer cells through upregulating
IRS1. Cell Prolif. Sep 21–2017.(Epub ahead of print). doi:
10.1111/cpr.12389. View Article : Google Scholar
|
|
72
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu L, An X, Li Z, Song Y, Li L, Zuo S,
Liu N, Yang G, Wang H, Cheng X, et al: The H19 long noncoding RNA
is a novel negative regulator of cardiomyocyte hypertrophy.
Cardiovasc Res. 111:56–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Heiler S, Wang Z and Zöller M: Pancreatic
cancer stem cell markers and exosomes-the incentive push. World J
Gastroenterol. 22:5971–6007. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fuschi P, Maimone B, Gaetano C and
Martelli F: Noncoding RNAs in the vascular system response to
oxidative stress. Antioxid Redox Signal. 30:992–1010. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sethi S, Sethi S and Bluth MH: Clinical
implication of MicroRNAs in molecular pathology: An update for
2018. Clin Lab Med. 38:237–251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Vicentini C, Fassan M, D'Angelo E, Corbo
V, Silvestris N, Nuovo GJ and Scarpa A: Clinical application of
microRNA testing in neuroendocrine tumors of the gastrointestinal
tract. Molecules. 19:2458–2468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tang YT, Xu XH, Yang XD, Hao J, Cao H, Zhu
W, Zhang SY and Cao JP: Role of non-coding RNAs in pancreatic
cancer: The bane of the microworld. World J Gastroenterol.
20:9405–9417. 2014.PubMed/NCBI
|
|
79
|
Zhang S, Zhu D, Li H, Li H, Feng C and
Zhang W: Characterization of circRNA-associated-ceRNA networks in a
senescence- accelerated mouse prone 8 brain. Mol Ther.
25:2053–2061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kishikawa T, Otsuka M, Ohno M, Yoshikawa
T, Takata A and Koike K: Circulating RNAs as new biomarkers for
detecting pancreatic cancer. World J Gastroenterol. 21:8527–8540.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Knupp D and Miura P: CircRNA accumulation:
A new hallmark of aging? Mech Ageing Dev. 173:71–79. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Vassella E, Chaudhary AK, Mondal G, Kumar
V, Kattel K and Mahato RI: Chemosensitization and inhibition of
pancreatic cancer stem cell proliferation by overexpression of
microRNA-205. Cancer Lett. 402:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yoshida K, Toden S, Ravindranathan P, Han
H and Goel A: Curcumin sensitizes pancreatic cancer cells to
gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1
expression. Carcinogenesis. 38:1036–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as Heracles in autophagy.
Autophagy. 13:1004–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|