Introduction
Esophageal cancer is one of the most common
malignancies worldwide, and in 2012, the incidence of esophageal
cancer ranked 8th of all cancers (1). In 2015, cancer data from China revealed
that the incidence and mortality rate of esophageal cancer were
ranked 4th (2). Given that the early
symptoms of esophageal cancer are not typical, and that the methods
for early diagnosis are limited, 70–80% of patients cannot be
diagnosed until reaching the intermediate or late stages of
disease, when the opportunity for surgical treatment has been lost
(3). Clinically, radiotherapy,
chemotherapy and other comprehensive treatments are commonly used
(4). A limited number of molecular
and clinical studies have focused on the treatment of esophageal
cancer in regards to the genes involved in immunization (5), and the majority of these studies have
focused on adenocarcinoma at the junction of esophageal
adenocarcinoma and gastric esophageal cancer, which is more
frequently observed in Europe and the United States (6,7).
Isobaric tags for relative and absolute quantitation
(iTRAQ) is a high-throughput, high-sensitivity proteome
quantification technique that is widely used for screening
diagnostic and efficacy-related tumor markers (8,9). In the
present study, iTRAQ technology was first used to identify
differentially expressed serum proteins associated with
chemoradiotherapeutic efficacy in patients with esophageal squamous
cell carcinoma (ESCC); the patients were recruited from different
ethnic groups in the Xinjiang Uyghur Autonomous Region.
Concurrently, functional tests including MTT assay, clonogenic and
fluorescence assays, flow cytometry, Transwell assays and
wound-healing assays were performed to determine the effect of the
serum proteins with significantly differential expression, and RNA
interference (RNAi) technology was used to silence the expression
of ILK-associated gene; thus, their effects on the proliferation,
apoptosis, invasion and migration of ESCC cells was observed, the
results of which may be used as a reference for further research
into the efficacy and prognostic mechanisms of esophageal cancer
treatment.
Materials and methods
Patients
A total of 60 patients with esophageal cancer who
were treated at the First Affiliated Hospital of Xinjiang Medical
University between July 2008 and April 2017 were recruited into the
present study. the patients were recruited from different ethnic
groups (25 cases from Han, 15 cases from Kazakh and 20 cases from
Uygur). A control group containing 60 age- and sex-matched subjects
was also recruited; these subjects underwent physical examination
at the First Affiliated Hospital of Xinjiang Medical University.
The clinicopathological characteristics of the patients are shown
in Table I. The patients were
recruited according to the following inclusion criteria: i)
Pathologically confirmed advanced IIIB-IV esophageal cancer (using
the esophageal cancer Tumor-Node-Metastasis staging criteria
developed by the American Joint Committee on Cancer) (10) including those with postoperative
recurrence and re-grading, or who were inoperable; ii) measurable
lesions; iii) aged between 18–75 years of age (male or female) with
a physical performance score (WHO Performance Status) (11) ≤0-2; iv) had received and tolerated
radiotherapy or chemotherapy, and possessed an expected survival
period ≥6 months; iii) no major organ dysfunction, with normal
routine blood test results (white blood cell count, neutrophil
percentage, neutrophil count, hemoglobin and platelet count) and
normal liver, kidney and cardiac function; and iv) were able to
understand the aim of the study and give informed consent for
participation.
 | Table I.Clinicopathological characteristics
of the patients. |
Table I.
Clinicopathological characteristics
of the patients.
| Clinicopathological
characteristic | n | Proportion, % |
|---|
| Age |
|
|
|
≤60 | 32 | 53.33 |
|
>60 | 28 | 46.67 |
| Sex |
|
|
|
Male | 35 | 58.33 |
|
Female | 25 | 41.67 |
| Involved
region |
|
|
| Neck
section | 7 | 11.67 |
| Upper
thoracic | 11 | 18.33 |
| Middle
thoracic | 23 | 38.33 |
| Lower
thoracic | 19 | 31.67 |
| Clinical stage |
|
|
| Phase
III | 38 | 63.33 |
| Phase
IVA | 22 | 36.67 |
The exclusion criteria included: i) A history of
malignancy at other sites; ii) patients who were pregnant or
lactating; and iii) a history of uncontrollable psychosis,
diagnosed by an expert neurologist. The recruited patients received
platinum-based dual chemotherapy (docetaxel combined with cisplatin
or paclitaxel combined with cisplatin), of paclitaxel (135–150
mg/m2) or docetaxel (75/m2) plus carboplatin
(500 mg/time) or cis platinum (75 mg/m2), once every 3–4
weeks, for a total of 4–6 cycles. All patients underwent concurrent
chemoradiotherapy, followed immediately by 4–6 cycles of
chemotherapy. The radiotherapy program consisted of
three-dimensional conformal irradiation or intensity-modulated
radiation therapy, DT50-66 GY/25-33f (where the preventive and
radical irradiation doses were 50 and 60–66 GY, respectively).
Short-term efficacy was categorized as complete
remission (CR), partial remission (PR), stability disease (SD) and
progression disease (PD) according to the response evaluation
criteria in solid tumors (RECIST). The chemoradiotherapy-sensitive
group was classified as CR+PR+SD, and the
chemoradiotherapy-resistant group as PD.
Follow-up was conducted using the telephone,
outpatient consultation and medical records once every 3 months,
and the follow-up deadline was July 2017. During the follow-up
period, the date of death was recorded for the patients that had
died. For the patients who were lost to follow-up, the final
follow-up time was recorded.
Specimen collection and serum
preparation
A total of 2 ml of fasting peripheral venous blood
was collected from all participants. Following collection, the
samples were stored at 4°C for 2–3 h until the blood had
coagulated. The samples were then centrifuged at 3,000 × g for 10
min at 4°C to isolate the serum, and the supernatants were stored
at −80°C until required.
Screening and identification of target
proteins
The serum samples were divided into eight groups:
Normal control group; esophageal cancer patients prior to treatment
group; esophageal cancer patients following treatment group;
radiotherapy and chemotherapy sensitive group prior to treatment;
chemoradiotherapy antagonist group prior to treatment; total
survival period of 1 to 2 years; total survival period 2–3 years;
and total survival >3 years. Protein iTRAQ markers were used to
identify differentially expressed proteins. After removing
high-abundance proteins, such as albumin, immunoglobulin G and
antitrypsin, a Bradford protein concentration determination assay
was performed to determine the protein concentration. A total of 20
µg protein was loaded on a 12.5% gel and resolved using SDS-PAGE,
and proteolysis and peptide quantification were performed. A total
of 8 labeled peptide samples were re-dissolved in 100 µl of solvent
(98% H2O, 0.1% formic acid) and 8 µl was injected into a
Type C18 column size 100 mm × 75 µm, pore size 300A, particle size
5 µm. The samples were subjected to liquid phase separation using
an Ultra Nano LC system (Thermo Fisher Scientific, Inc.) with a
gradient of 5 to 80% buffer (98% acetonitrile, 2% H2O
and 0.1% formic acid) for 3 h at 300 nl/min. Peptides were eluted
using a gradient of 5–80% (v/v) acetonitrile in 0.1% formic acid
over 45 min at a flow rate of 300 nl min−1 combined with
a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.) The
eluates were directly entered the Q-Exactive mass spectrometer
(Thermo Fisher Scientific, Inc.), set in positive ion mode and
data-dependent manner with a full mass spectrometry scan from
350–2000 m/z, full scan resolution at 70,000 MS/MS scan resolution
at 17,500 MS/MS scan, with a minimum signal threshold of
1E+5, isolation width at 2 Da. The sample was repeatedly
identified 4 times. The raw mass spectrometry data was generated as
a RAW file. ProteomeDiscoveral 4 software (Thermo Fisher
Scientific, Inc.) was used to quantitatively analyze the peak
intensity of the reported peptide ions. The results were scanned
and filtered according to the misclassification rate ≤0.01. A ratio
>1.2 between the aforementioned groups was considered to
indicate significant upregulation, and a ratio <0.8 between
groups was considered to indicate significant downregulation;
P<0.05.
Integrin-linked kinase (ILK) siRNA
target sequence screening and lentiviral vector construction
The ESCC EC9706 (DMEM supplemented with 10% FBS,
37°C, 5% CO2), ECa109 (RPMI-1640 supplemented with 10%
FBS, cultured at 37°C, 5% CO2) and TE-1 (RPMI-1640
without Hepes supplemented with 10% FBS) cell lines were purchased
from Shanghai Jikai Gene Chemical Technology Co., Ltd. Using the
ILK gene as a template, multiple 19–21 nucleotide RNAi target
sequences were designed using Chromas target sequence analysis and
design software (Technelysium Pty, Ltd., version 2.1.3). siRNAs
were designed for the target gene, the target sequence was
5′-CGAAGCTCAACGAGAATCA-3′, and the n.
Following RNAi target design, single-stranded DNA
oligos containing interference sequences was synthesized and paired
by annealing (90°C water bath for 15 min) to produce
double-stranded DNA. These were subsequently ligated into the
lentiviral vector GV115 (Shanghai Jikai GeneChem Co., Ltd.).
Prepared TOP10 competent E. coli cells (GeneChem, Inc.) were
transformed using the ligation products, and PCR was used to
identify positive recombinant vectors. PCR and DNA sequencing were
performed on the recombinant positive clones. The upstream primer
of the positive clone was amplified using Taq Plus DNA Polymerase
(Vazyme BioTech Co. Ltd.). The sequences of the primers used were:
Forward, CCTATTTCCCATGATTCCTTCATA, and reverse
GTAATACGGTTATCCACGCG. The thermocycling conditions were: 94°C for 3
min; followed by 22 cycles of 94°C for 30 sec; 55°C for 30 sec;
72°C for 30 sec; and a final extension of 72°C for 5 min. Sanger
sequencing was used for verification. The sequencing results were
compared with the correct clones for plasmid extraction. Bacteria
containing the desired plasmids were transferred to 150 ml of LB
liquid medium containing ampicillin, and cultured overnight at 37°C
with shaking. The plasmids were extracted using an EndoFree Maxi
Plasmid kit according to the manufacturer's protocols and the
quality was determined. Plasmids of sufficient quality, as
determined using NanoDrop 2000 (Thermo Fisher Scientific, Inc.),
were used for virus packaging.
Following transfection, the TE-1 cells were cultured
at 37°C, in a humidified incubator with 5% CO2.
Lentiviral particles have different affinities for different cell
lines (12). First, the target cells
were pre-infected with a lentivirus, and different multiplicity of
infections (MOI) gradients were used to evaluate the MOI of the
target cells (Data not shown). A 48-well plate was used with a
total medium volume of 200 µl. For the fluorescence-tagged
lentivirus infection, the infection time point was determined in
the preliminary results. Green fluorescent protein expression was
observed under a fluorescence microscope (magnification, ×100). The
fluorescence rate was 70–80%, the cell confluence was approximately
80%, the cells appeared healthy, the infection-efficiency was
deemed sufficient, and cells were collected for further
experimentation.
Reverse transcription-quantitative
PCR
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) was used to analyze tumor
specimen. Total RNA was extracted using the TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturers protocol. cDNA was synthesized from 2 µg total RNA
using the M-MLV reverse transcriptase at 42°C for 1 h followed by
70°C for 10 min. GAPDH was used as the reference gene for RT-qPCR.
The sequences used in the present study were ILK forward,
5′-GACGACATTTTCACTCAGTGCC-3′ and reverse,
5′-ACGGTTCATTACATTGATCCGTG-3′; GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse
5′-CACCCTGTTGCTGTAGCCAAA-3′. Gene-specific amplification of the 12
µl PCR mixture was performed using a ABI 7500HT real-time PCR
system (Thermo Fisher Scientific, Inc.). The thermocycling
condition were: 94°C for 3 min; followed by 22 cycles of 94°C for
30 sec, 94°C for 60 sec and 72°C for 60 sec; followed by a final
extension step at 72°C for 5 min. DNA concentration was
approximately doubled following each cycle of denaturation,
annealing and elongation. The cycle quantification (Cq) of each
sample was calculated, and the relative expression of the gene was
calculated using the 2−ΔΔCq method (13).
Cell proliferation assay
Trypsinized TE-1 cells in the logarithmic growth
phase were resuspended in complete medium and plated in 96-well
culture plates at 2×103 cells/well. Cells were incubated
at 37°C in a humidified atmosphere with 5% CO2. The
Celigo® cytometer (Nexclom Bioscience) was used to
detect cell growth once a day for 5 days. By adjusting the input
parameters of the analysis settings using Cellometer Auto 2000 Cell
Viability Counter (Nexcelom Bioscience), the number of cells with
green fluorescence was calculated and a 5-day cell proliferation
curve was generated.
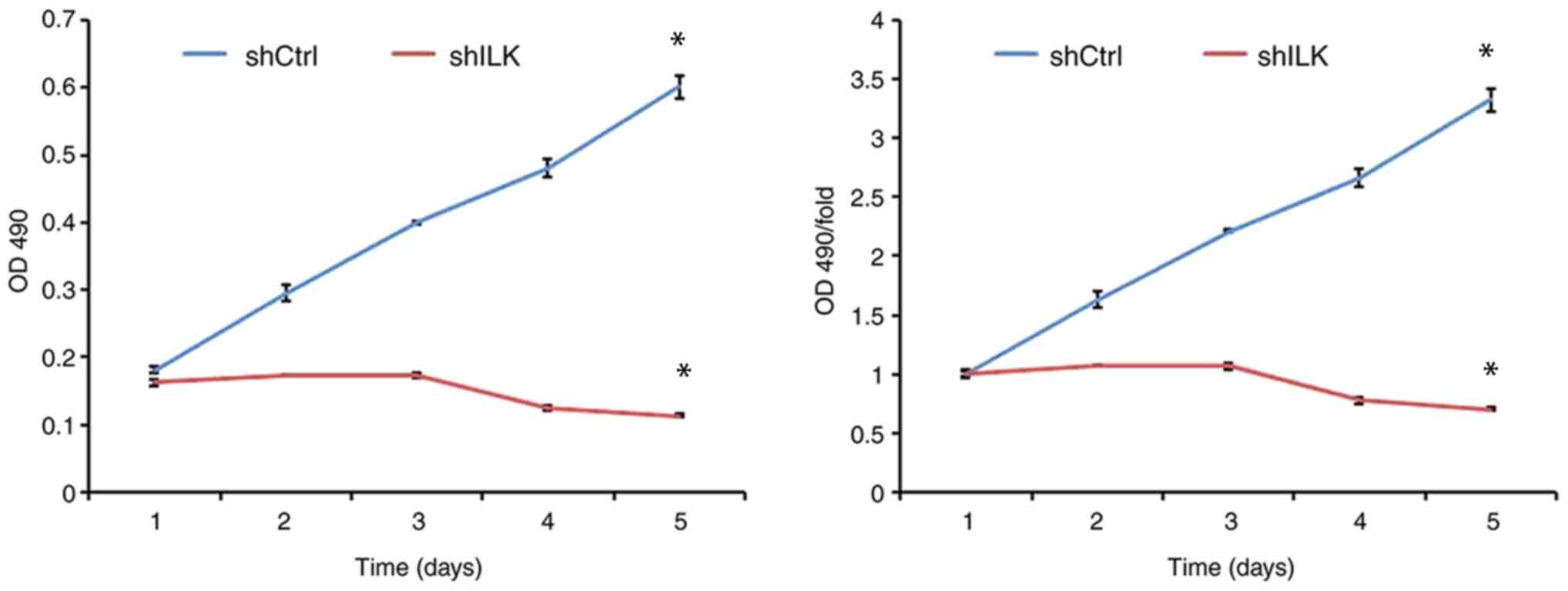
MTT assay
Cells in the logarithmic growth phase were
harvested, homogenized into a single-cell suspension, and
2×103 cells/well were plated in 96-well cell culture
plates. Cells were incubated at 37°C in a humidified incubator with
5% CO2. Subsequently, 20 µl MTT (5 mg/ml) was added to
each well, and the cells were incubated for a further 4 h. The
supernatant was discarded, 150 µl DMSO was added and the samples
were shaken for 2–5 min to fully dissolve the formazan crystals.
Absorbance was determined at 490 nm using a microplate reader. Time
was used as the abscissa and the optical density was used as the
ordinate for plotting the cell growth curve.
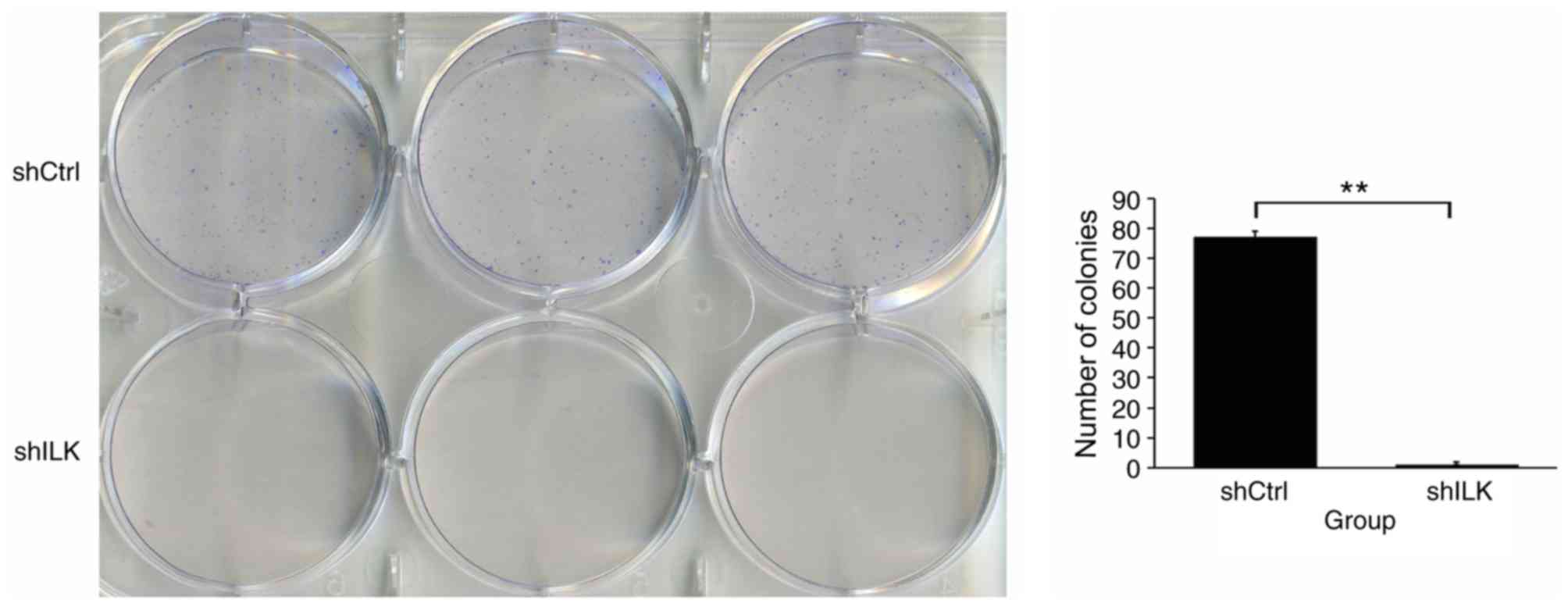
Cell colony formation assay
After transfection, TE-1 cells in the logarithmic
growth phase were trypsinized in each experimental group,
resuspended in complete medium and counted. The cells were seeded
into a 6-well culture plate (500 cells/well) and the cultured for
11 days with a media change once every 3 days. The cells were fixed
with 1 ml 4% paraformaldehyde at 37°C for 30–60 min and stained
with 500 µl Giemsa staining solution (Shanghai Dingguo
Biotechnology Co., Ltd.) at 37°C for 10–20 min and the colonies
were subsequently counted.
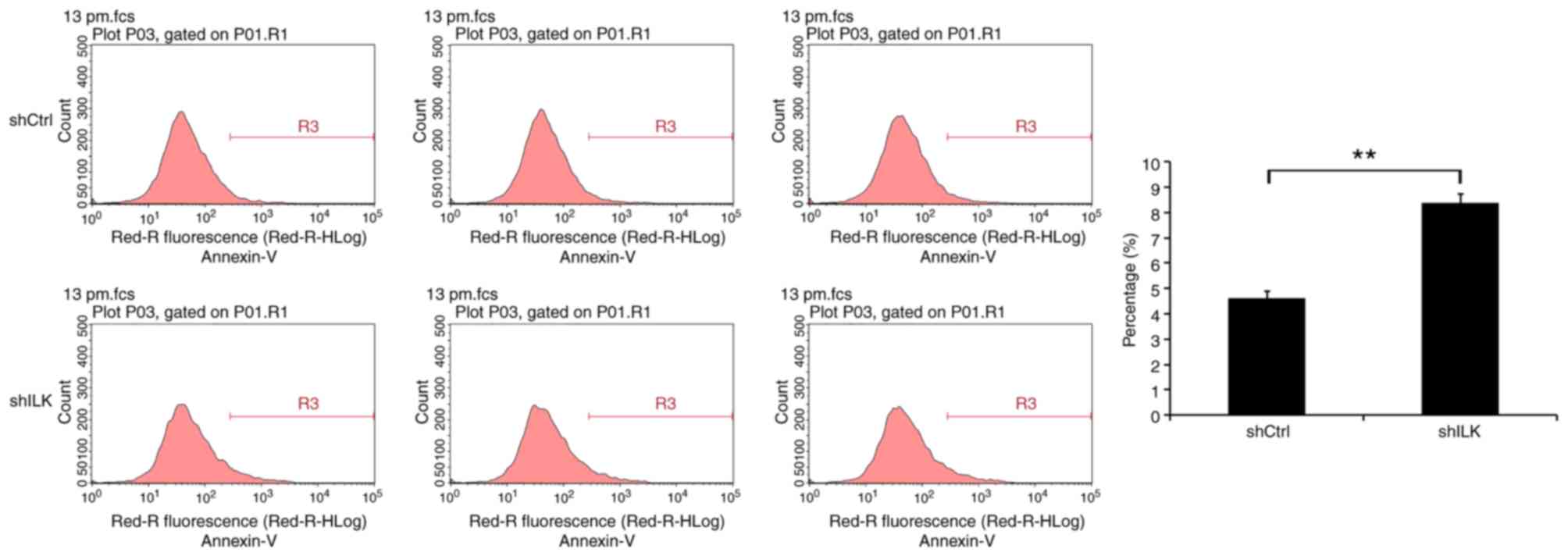
Apoptosis detection using flow
cytometry
Following transfection, cells in the logarithmic
growth from each experimental group were harvested and resuspended
in complete culture medium. The cells were collected by
centrifugation at 1,300 × g for 5 min at 4°C, the supernatant was
discarded, and the cell pellet was washed with pre-cooled D-Hanks
(Shanghai Research Extension Biotechnology Co., Ltd.) solution at
4°C. The cell pellet was washed with binding buffer (Shanghai Suo
Laibao Biotechnology Co., Ltd,) stained with 10 µl Annexin V-APC
and analyzed using a flow cytometer and Guava® InCyte
version 8HT (Luminex Corporation).
Transwell migration and invasion
assay
A total of 1×105 cells/well were seeded
on a Matrigel membrane in the upper chamber of a 24-well Transwell
plates for the invasion assay. For the migration assay cells were
seeded without Matrigel. In the bottom chamber, 600 µl medium
containing 30% FBS was added. Cells were incubated under standard
culture conditions for 20 h. Following invasion or migration, cells
which had not invaded or migrated were removed and the remaining
cells were stained with 800 µl of Giemsa staining solution onto the
lower surface of the membrane at 37°C for 3–5 min. The membranes
were visualized under a light microscope at ×200 and imaged to
determine differences in the migratory and invasive capacity. A
total of 9 random microscopic fields for each well were
counted.
Wound-healing assay
Transfected cells (~4×104) from each
group were added to 96-well plates, and cultured until ~90%
confluent. The following day, the concentration of serum in the
medium was reduced, and a wound was created in the monolayer. The
cells were gently rinsed 2–3 times with serum-free medium, and
incubated at 37°C in a humidified incubator with 5% CO2.
Images were captured at 0, 8 and 24 h using a fluorescence
microscope (magnification, ×200). Images were obtained after 0, 8
and 24 h using a light microscope (magnification, ×200). The width
of the scratched area in the picture at each time point of each
wound was obtained using the W value function of Adobe Photoshop
(Adobe Systems Europe, Ltd. version CC 2018). Wound closure was
measured relative to the starting wound width by dividing the wound
width at each time point by the width at the start.
Statistical analysis
Statistical data analysis was conducted using SPSS
version 19.0 software (IBM Corp.). A Student's t-test was used to
compare differences between two groups. Comparisons between shILK
and shCtrl were initially analyzed using an F-test (homogeneity of
variance test). If the F-test value is >0.05, a t-test value was
obtained using an equal variance two-sample test. If the F-test
value was <0.05, the heteroscedasticity was doubled. The sample
test yielded the t-test value. P<0.05 was considered to indicate
a statistically significant difference.
Results
Evaluation of clinical efficacy
According to RECIST, 36 cases were included in the
CR+PR+SD group by analyzing medical data from outpatient visits
records, which accounted for 60% of the total cases; 24 cases were
classified as PD, 40% of the total.
Differentially expressed serum
proteins in patients with ESCC
From the patients with ESCC, 46 differentially
expressed proteins were identified prior to chemoradiotherapy,
compared to those identified in the control subjects: 26 proteins
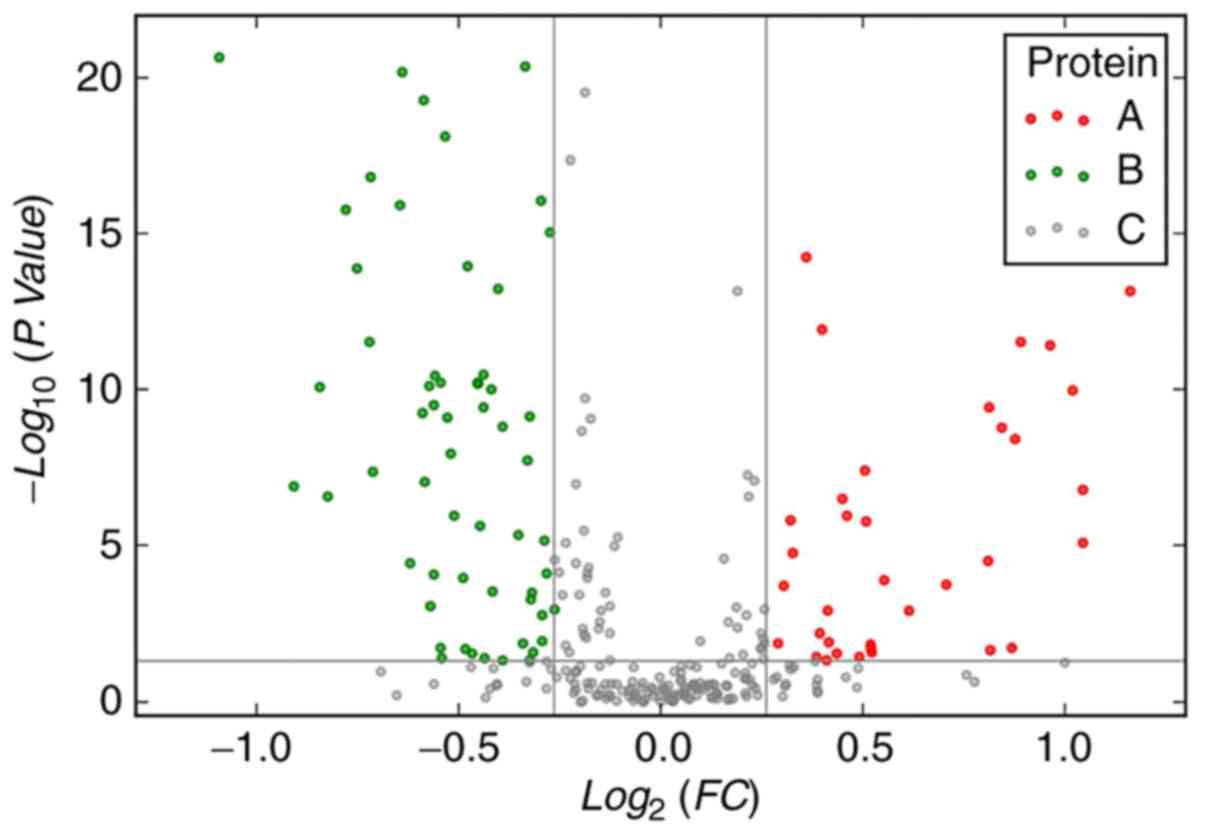
were upregulated and 20 were downregulated (Tables II and III). Volcano maps show that differential
expression of serum proteins between patients with ESCC (before
chemoradiotherapy) and those in the control group (Fig. 1).
 | Table II.Upregulated differentially expressed
proteins in the serum of Chinese patients with esophageal cancer,
before chemoradiotherapy. |
Table II.
Upregulated differentially expressed
proteins in the serum of Chinese patients with esophageal cancer,
before chemoradiotherapy.
| Accession ID | Protein | Gene | Ratio | P-value |
|---|
| P23528 | Cofilin-1 | CFL1 | 1.532 |
1.16789×10−3 |
| P06753 | Tropomyosin α-3
chain | TPM3 | 1.633 |
1.76099×10−4 |
| P08185 |
Corticosteroid-binding globulin | SERPINA6 | 1.753 |
3.01686×10−5 |
| P67936 | Tropomyosin α-4
chain | TPM4 | 1.759 |
3.61117×10−10 |
| Q15746 | Myosin light chain
kinase, smooth muscle | MYLK | 1.76 |
2.32945×10−2 |
| P02749 | β-2-glycoprotein
1 | APOH | 1.794 |
1.68655×10−9 |
| P02750 | Leucine-rich
α-2-glycoprotein | LRG1 | 1.828 |
1.89684×10−2 |
| P0DJI8 | Serum amyloid A-1
protein | SAA1 | 1.836 |
3.64567×10−9 |
| P01019 |
Angiotensinogen | AGT | 1.855 |
2.96144×10−12 |
| P01009 |
α-1-antitrypsin | SERPINA1 | 1.944 |
3.41166×10−65 |
| P68871 | Hemoglobin subunit
β | HBB | 1.951 |
3.77869×10−12 |
| P05155 | Plasma protease C1
inhibitor | SERPING1 | 2.027 |
1.03898×10−10 |
| P05543 | Thyroxin-binding
globulin | SERPINA7 | 2.063 |
7.87217×10−6 |
| P19652 | α-1-acid
glycoprotein 2 | ORM2 | 2.065 |
1.57516×10−7 |
| P01023 |
α-2-macroglobulin | A2M | 2.209 |
2.24553×10−69 |
| P04217 |
α-1B-glycoprotein | A1BG | 2.241 |
7.01006×10−14 |
| P0DJI9 | Serum amyloid A-2
protein | SAA2 | 2.254 |
7.76843×10−16 |
| P01011 |
α-1-antichymotrypsin | SERPINA3 | 2.363 |
1.59357×10−28 |
| P02768 | Serum albumin | ALB | 2.477 |
1.89258×10−3 |
| P02741 | C-reactive
protein | CRP | 2.517 |
1.59305×10−91 |
| P01861 | Immunoglobulin
heavy constant γ4 | IGHG4 | 2.584 |
2.27127×10−3 |
| P02787 |
Serotransferrin | TF | 2.89 |
5.56713×10−82 |
| P02790 | Hemopexin | HPX | 2.999 |
5.72798×10−31 |
| P25311a |
Zinc-α-2-glycoprotein | AZGP1 | 3.122 |
5.90489×10−8 |
| Q13418a | Integrin-linked
protein kinase | ILK | 4.386 |
3.69519×10−31 |
| P02763a | α-1-acid
glycoprotein 1 | ORM1 | 5.234 |
5.50575×10−16 |
 | Table III.Downregulated differentially
expressed proteins in the serum of Chinese patients with esophageal
cancer, before chemoradiotherapy. |
Table III.
Downregulated differentially
expressed proteins in the serum of Chinese patients with esophageal
cancer, before chemoradiotherapy.
| Accession ID | Protein | Gene | Ratio | P-value |
|---|
| P02776a | Platelet factor
4 | PF4 | 0.469 |
2.12173×10−21 |
| Q08380a | Galectin-3-binding
protein | LGALS3BP | 0.483 |
4.59282×10−40 |
| P08697a |
α-2-antiplasmin | SERPINF2 | 0.495 |
2.5819×10−30 |
| P10720 | Platelet factor 4
variant | PF4V1 | 0.533 |
1.24972×10−7 |
| P02751 | Fibronectin | FN1 | 0.536 |
1.72558×10−11° |
| Q16610 | Extracellular
matrix protein 1 | ECM1 | 0.557 |
8.40852×10−11 |
| Q13103 | Secreted
phosphoprotein 24 | SPP2 | 0.565 |
2.66878×10−7 |
| P27918 | Properdin | CFP | 0.582 |
1.6419×10−16 |
| P00488 | Coagulation factor
XIII A chain | F13A1 | 0.594 |
1.3398×10−14 |
| Q5SYB0 | FERM and PDZ
domain-containing protein 1 | FRMPD1 | 0.607 |
2.98852×10−12 |
| Q15485 | Ficolin-2 | FCN2 | 0.608 |
1.51904×10−17 |
| O43852 | Calumenin | CALU | 0.61 |
4.42339×10−8 |
| P01860 | Immunoglobulin
heavy constant γ3 | IGHG3 | 0.639 |
1.20085×10−16 |
| P04114 | Apolipoprotein
B-100 | APOB | 0.641 |
4.42101×10−7° |
| O43866 | CD5
antigen-like | CD5L | 0.642 |
6.51383×10−21 |
| Q92954 | Proteoglycan 4 | PRG4 | 0.651 |
3.50318×10−5 |
| P04004 | Vitronectin | VTN | 0.658 |
4.9004×10−179 |
| P04180 |
Phosphatidylcholine-sterol
acyltransferase | LCAT | 0.665 |
5.75865×10−10 |
| P04196 | Histidine-rich
glycoprotein | HRG | 0.666 |
5.28207×10−20 |
| Q96IY4 | Carboxypeptidase
B2 | CPB2 | 0.667 |
8.99551×10−8 |
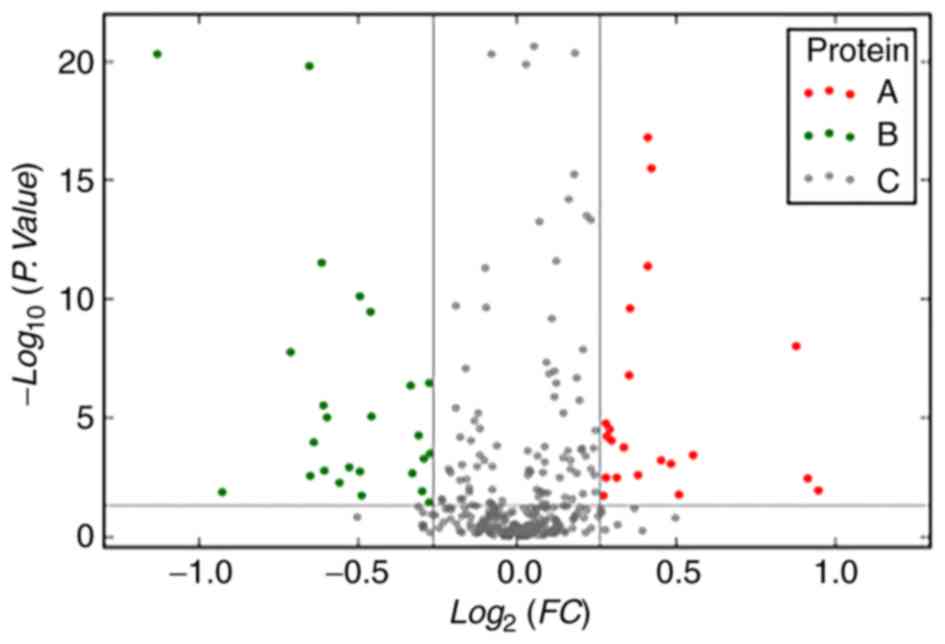
Significantly differentially expressed proteins were
compared between the chemoradiotherapy-sensitive and -resistant
groups and between the survival times of patients with ESCC,
classified as either 1–2, 2–3 and >3 years (Tables IV and V). The volcano plots show differential
expression of serum proteins between patients with esophageal
cancer of different severities group (Fig. 2).
 | Table IV.Differentially expressed proteins
between Chinese chemoradiotherapy-sensitive and
chemoradiotherapy-resistant patients with esophageal cancer. |
Table IV.
Differentially expressed proteins
between Chinese chemoradiotherapy-sensitive and
chemoradiotherapy-resistant patients with esophageal cancer.
| Accession ID | Protein | Gene | Ratio | P-value | Regulation |
|---|
| P02763 | α-1-acid
glycoprotein 1 | ORM1 | 0.348 |
1.00117×10−13 | Down |
| P00738 | Haptoglobin | HP | 0.456 |
4.58182×10−21 | Down |
| P01833a | Polymeric
immunoglobulin receptor | PIGR | 0.526 |
1.30577×10−2 | Down |
| P04217 |
α-1B-glycoprotein | A1BG | 0.653 |
1.50285×10−20 | Down |
| P25311 |
Zinc-α-2-glycoprotein | AZGP1 | 0.656 |
2.97886×10−6 | Down |
| P08571 | Monocyte
differentiation antigen CD14 | CD14 | 0.657 |
1.63716×10−3 | Down |
| P11597 | Cholesteryl ester
transfer protein | CETP | 0.661 |
9.93464×10−6 | Down |
| Q13418 | Integrin-linked
kinase | ILK | 0.587 |
9.41968×10−4 | Down |
| A0M8Q6a | Ig λ-7 chain C
region | IGLC7 | 1.886 |
3.64692×10−3 | Up |
| Q15848a | Adiponectin | ADIPOQ | 1.836 |
9.54065×10−9 | Up |
| P59665a | Neutrophil defensin
1 | DEFA1 | 1.927 |
1.16427×10−2 | Up |
 | Table V.Differentially expressed proteins
between Chinese patients with esophageal cancer with >3 years
and 1–2 years survival rates. |
Table V.
Differentially expressed proteins
between Chinese patients with esophageal cancer with >3 years
and 1–2 years survival rates.
| Accession ID | Description | Gene | Ratio | P-value | Regulation |
|---|
| P05154a | Plasma serine
protease inhibitor | SERPINA5 | 0.421 |
4.45068×10−18 | Down |
| Q92496 | Complement factor
H-related protein 4 | CFHR4 | 0.558 |
5.80371×10−11 | Down |
| P59665 | Neutrophil defensin
1 | DEFA1 | 0.58 |
5.35236×10−3 | Down |
| P62805a | Histone H4 | HIST1H4A | 0.584 |
1.79658×10−6 | Down |
| P68431a | Histone H3.1 | HIST1H3A | 0.596 |
1.23326×10−3 | Down |
| P20160 | Azurocidin | AZU1 | 0.606 |
2.56589×10−4 | Down |
| Q13418a | Integrin-linked
kinase | ILK | 0.633 |
1.45967×10−2 | Down |
| P02776 | Platelet factor
4 | PF4 | 0.664 |
5.87470×10−17 | Down |
| P49720 | Proteasome subunit
β type-3 | PSMB3 | 1.552 |
1.44523×10−2 | Up |
| P0DJI9a | Serum amyloid A-2
protein OS=Homo | SAA2 | 1.68 |
1.57258×10−13 | Up |
Among the differentially-expressed proteins, ILK
appeared most frequently at all outcomes in patients with
esophageal cancer. This protein was upregulated in patients with
ESCC before chemoradiotherapy, compared with the control group
[ratio (r)=4.386; P<0.05], and downregulated in the
chemoradiotherapy-sensitive group compared with the
chemoradiotherapy-resistant group (r=0.587, P<0.05; survival
period >3 years compared with a survival period of 1–2 years;
r=0.633, P<0.05; Table VI).
 | Table VI.Expression of ILK in Chinese patients
with esophageal cancer of different severities. |
Table VI.
Expression of ILK in Chinese patients
with esophageal cancer of different severities.
| Group | Accession | Description | Gene | Ratio | P-value | Regulation |
|---|
| Esophageal cancer
patients prior to treatment vs. control group | Q13418 | Integrin-linked
kinase | ILK | 4.386 |
3.69519×10−31 | Up |
|
Chemoradiotherapy-sensitive group vs.
chemoradiotherapy-resistant group | Q13418 | Integrin-linked
kinase | ILK | 0.587 |
9.41968×10−4 | Down |
| Survival group
>3 years vs. survival group 1–2 years | Q13418 | Integrin-linked
kinase | ILK | 0.633 |
1.45967×10−2 | Down |
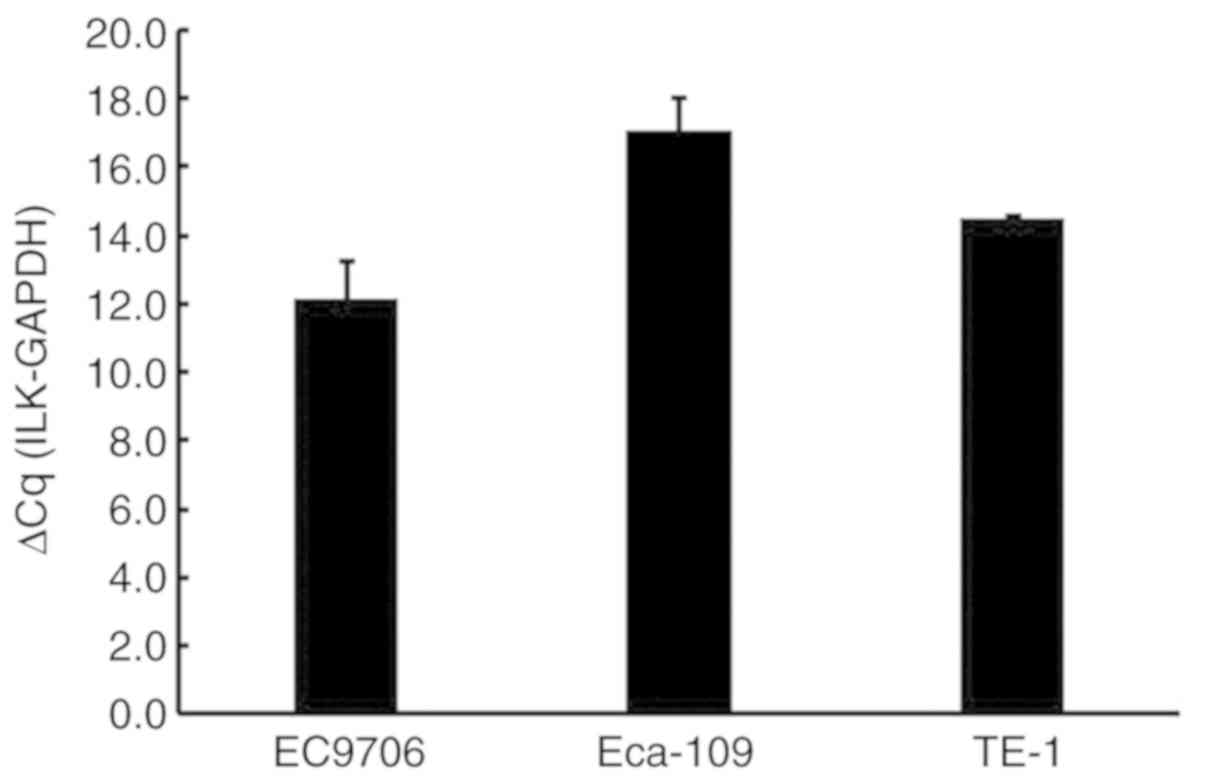
The effect of ILK expression on cell
proliferation, apoptosis, invasion and migration in ESCC
The ILK mRNA expression levels were determined in
the TE-1 ESCC cell line (Fig. 3).
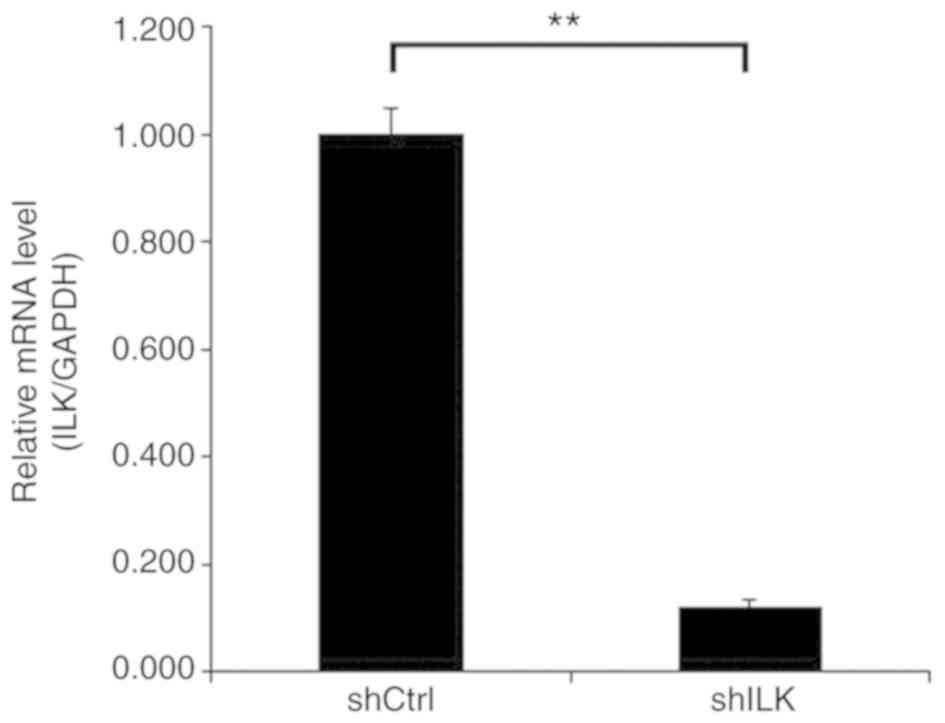
Lentivirus was used for target cell transfection, and the results
confirmed a transfection efficiency of >80%. Following
transfection, there were no noticeable changes in the morphology
and the cells appeared healthy. After siRNA lentivirus infection,
the mRNA expression level of ILK was determined using RT-qPCR. The
results showed that the expression of ILK in TE-1 cells of the
shILK group was successfully inhibited compared with the shCtrl
group (P<0.05), and that the knockdown efficiency was 88%
(Fig. 4).
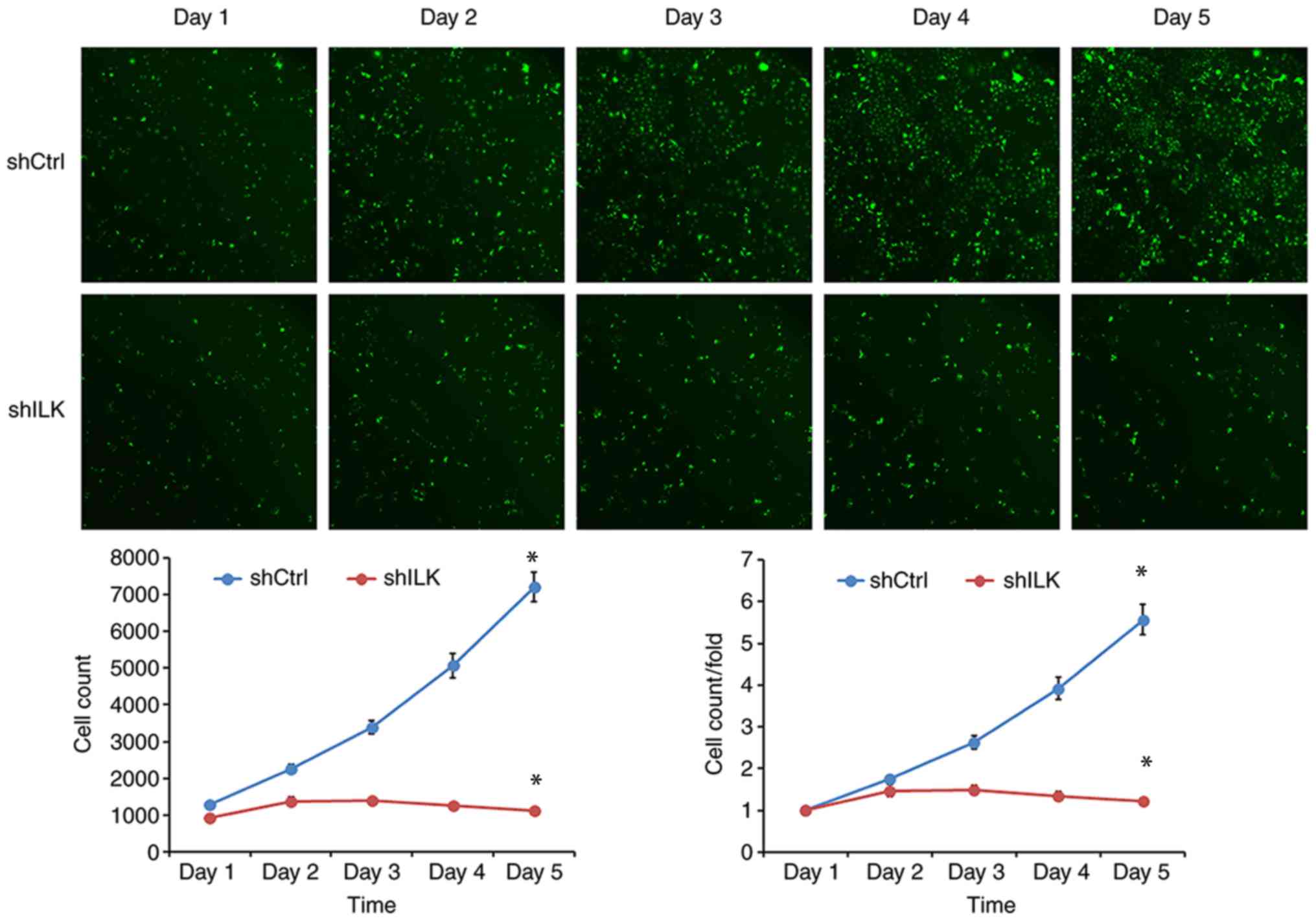
To examine the effect of ILK gene ablation on cell
proliferation, cell number was determined using fluorescence and
MTT assays three days post-transfection with shRNA lentivirus.
After five days, the proliferation rate was decreased in the shILK
group compared with the shCtrl group (P<0.05) in both assays
(Figs. 5 and 6). ILK gene-silencing was shown to inhibit
the proliferation of TE-1 cells.
Flow cytometry was used to detect the impact of ILK
gene inhibition on apoptosis, five days following shRNA lentivirus
transfection. The degree of apoptosis in TE-1 cells of the shILK
group increased significantly (P<0.01), indicating that the ILK
gene is significantly associated with apoptosis in these cells
(Fig. 7).
In addition, the impact of ILK gene ablation on cell
colony formation was also detected. The number of TE-1 cell
colonies in the shILK group was significantly lower than that of
the shCtrl group (P<0.01), suggesting that the ILK gene is
significantly associated with the colony forming ability of TE-1
cells (Fig. 8).
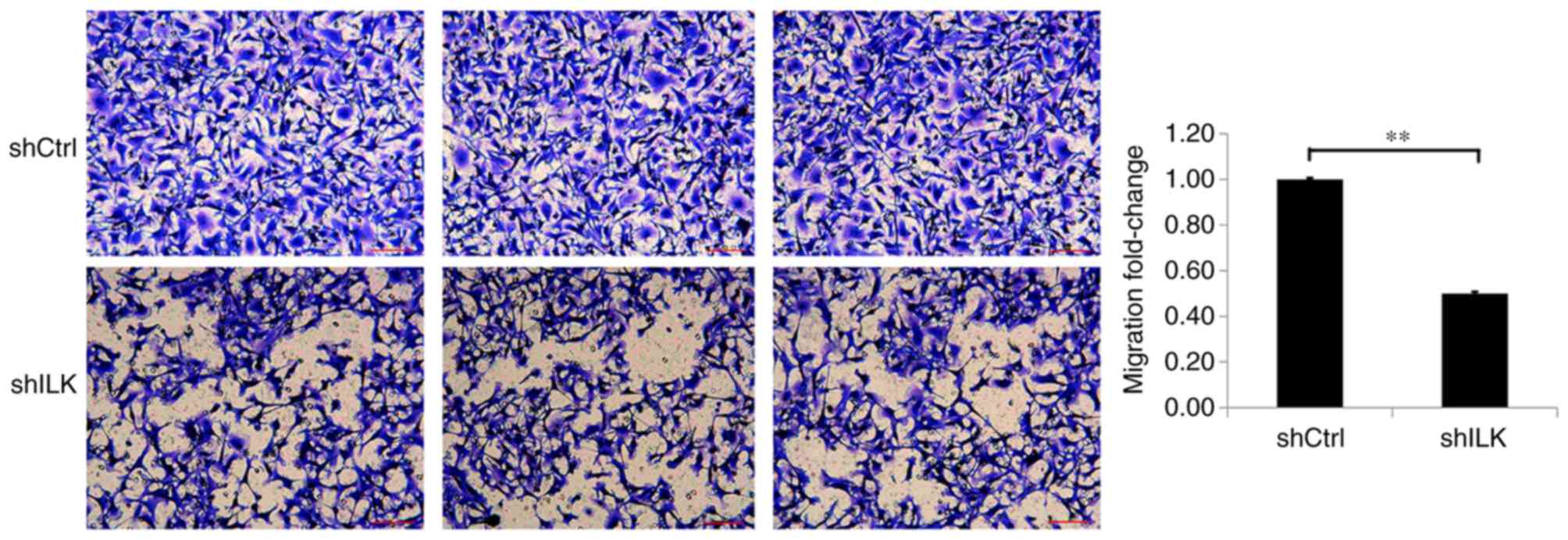
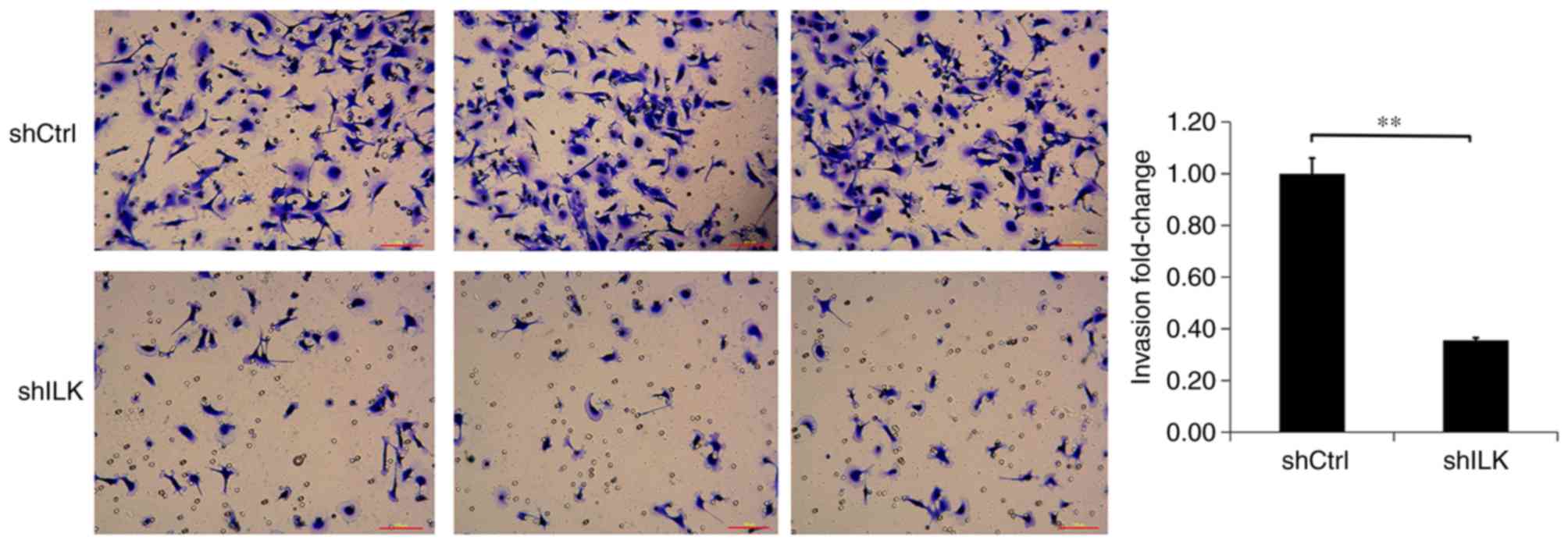
Migration and invasion assays were used to detect
the effect of ILK-knockdown on migration and invasion capacities of
TE-1 cells, three days post-transfection with shRNA lentivirus. The
migration and invasion abilities of TE-1 cells in the shILK group
were significantly inhibited compared with those of the shCtrl
group (P<0.01), suggesting that ILK gene-silencing inhibits the
migration and invasion abilities of TE-1 cells (Figs. 9 and 10).
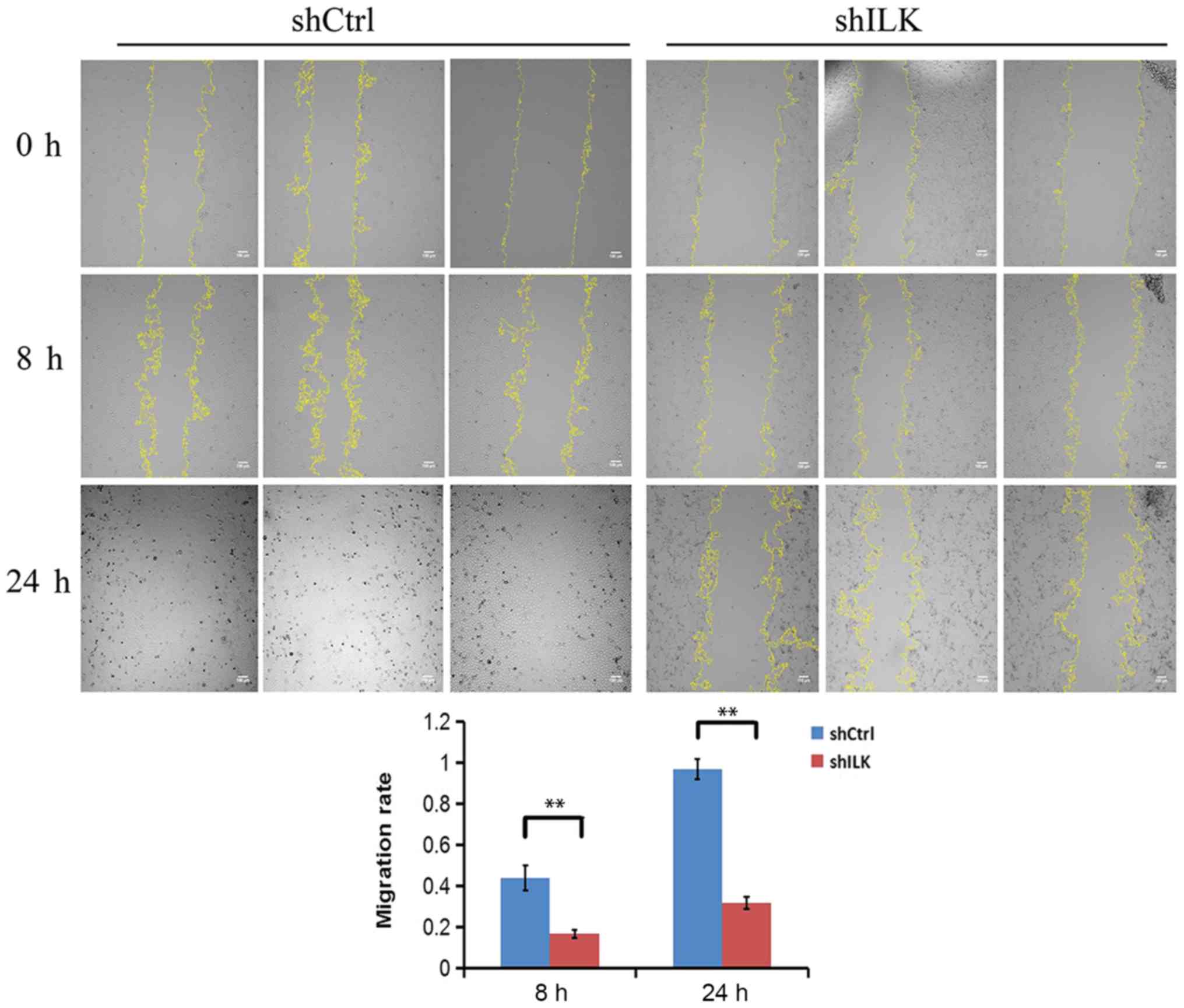
The effect of ILK gene-silencing on TE-1 cell
migration was also detected using a wound-healing assay three days
post-transfection with shRNA lentivirus. The migratory ability at 8
and 24 h in the shILK group compared with the shCtrl group at the
same time point was significantly reduced (P<0.01), suggesting
that ILK is significantly associated with the migratory ability of
TE-1 cells (Fig. 11).
Discussion
According to China's esophageal cancer and the USA
National Cancer Comprehensive Network guidelines, advanced ESCC is
primarily treated with chemoradiotherapy-based comprehensive
treatment (14). However, resistance
to chemoradiotherapy is the primary reason for its limited effect,
which results in poor overall efficacy (15). Studies in China and worldwide have
suggested that the abnormal expression of molecular markers, such
as cell division cycle 25C (16),
suppressor of cytokine signaling 1 (17), single-minded homolog 2 (18) and
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(19) in esophageal cancer tissues
has significance when predicting the efficacy of esophageal cancer
chemotherapy. However, to date, there are no established molecular
markers for predicting the efficacy of esophageal cancer
chemotherapy.
In the present study, iTRAQ technology was used to
screen serum differentially-expressed proteins associated with the
efficacy of chemoradiotherapy in ESCC. The results revealed that
ILK expression levels in patients with esophageal cancer (before
chemoradiotherapy) were higher compared with those in the control
subjects, and that ILK expression in chemoradiotherapy-sensitive
patients was reduced compared with that in the
chemoradiotherapy-resistant group. These findings indicated that
the abnormal expression of ILK may be associated with the
occurrence and development of ESCC, and that ILK may be involved in
the regulation of chemoradiotherapy sensitivity, and thus closely
associated with sensitivity to chemoradiotherapy in patients with
esophageal cancer. Based on these results, the effect of ILK
expression on ESCC cells characteristics was investigated. To the
best of our knowledge, the present study shows for the first time
that ILK gene-silencing significantly inhibits the proliferation,
invasion and migration, and promotes apoptosis in ESCC TE-1
cells.
ILK is a serine/methionine protein kinase that was
discovered by Hannigan et al (20) in a double yeast hybridization
screening experiment in human cDNA libraries, using the β1 integrin
cytoplasmic domain. As a key kinase in integrin and growth factor
receptor signaling pathways, ILK regulates cell growth,
differentiation, adhesion and apoptosis, and is closely associated
with tumor formation, invasion, and metastasis (21). Furthermore, ILK upregulation has been
reported to occur in bladder (22)
and colon cancer (23). Regarding
ILK research in esophageal cancer, Driver and Veale (24) used RT-PCR to study the expression and
subtypes of ILK, epidermal growth factor and transforming growth
factor-β1 in five human ESCC cell lines of South African origin.
The effect of ILK expression suggests that epidermal growth factor
and transforming growth factor-β1 may regulate ILK expression in
ESCC.
The present study suggested that cell proliferation
was reduced when ILK expression was inhibited in TE-1 cells, as
shown by fluorescence detection, MTT and colony formation assays.
These findings suggest that ILK may promote the proliferation of
TE-1 cells. Abnormal expression of the ILK gene may be an important
factor to increase the malignancy of ESCC. Apoptosis is closely
associated with the occurrence, development and prognosis of
various diseases, particularly tumors (25). The effect of ILK gene inhibition on
apoptosis in TE-1 cells was assessed using flow cytometry. The
apoptotic rate of TE-1 cells was increased relative to the control
group following ILK knockdown. Therefore, ILK inhibits apoptosis
and may be an important factor for the proliferation of TE-1 cells.
Furthermore, wound-healing assays revealed that ILK inhibited the
migratory ability of TE-1 cells compared with that of the control
group. It was hypothesized that ILK may promote the migration of
TE-1 cells, leading to enhanced biological characteristics and
increasing the risk of distant metastasis in malignant tumors. To
simulate the invasive tumor cell environment, a Transwell chamber
was utilized. The results showed that ILK silencing inhibited the
invasive properties of TE-1 cells, and therefore, ILK may promote
the invasion of ESCC cells.
In addition, studies have reported that abnormal
expression of ILK affects the chemoradiotherapeutic sensitivity of
tumor cells. Duxbury et al (26) found that overexpression of ILK
increased the chemotherapeutic resistance of pancreatic cancer
cells to gemcitabine, and that ILK knockout increased the
sensitivity of caspase-3-mediated apoptosis to gemcitabine. A
previous study by Lanvin et al (27) showed that ILK regulated mitochondrial
death caused by radiotherapy via the hypoxia inducible factor-1α
and survivin pathways, thereby altering the radiosensitivity of
glioma cells. These studies suggest that ILK increases tumor cell
resistance to radiotherapy. However, there is no experimental
indication that ILK influences the chemoradiotherapeutic
sensitivity of esophageal cancer.
In the present study, iTRAQ technology was used to
identify the molecular marker ILK, which is predictive of the
efficacy of esophageal cancer chemotherapy. It was revealed for the
first time that the inhibition of ILK expression promoted the
biological characteristics of ESCC, and increased the degree of
tumor malignancy. However, the present study has several
limitations and was not able to determine the mechanism by which
ILK promotes the characteristics of ESCC; therefore, further
research is required. Moreover, proteins associated with metastasis
were not investigated, and the lack of animal studies and the
investigation of proteins associated with epithelial-mesenchymal
transition are also limitations of the present study. In addition,
2-dimensional migration and invasion cannot fully recapitulate the
metastatic phenotype. At the same time, ILK is a potential mediator
of tumor chemoradiotherapy resistance. Therefore, in the future, an
in-depth understanding of the molecular mechanisms of resistance to
esophageal cancer chemoradiotherapy, and the exploration of new
strategies for targeting chemoradiotherapy resistance, will have
important clinical implications for improving treatment efficacy
and patient survival.
Acknowledgements
Not applicable.
Funding
The present study was supported by funding from the
National Science Foundation of China (grant no. 81860421).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XLM, HY and LZ performed the experiments, analyzed
the data and wrote the manuscript. XW, YW, LYC and QZ performed the
experiments and analyzed the data. XLM and LZ contributed to the
design of the study and the experiments, and wrote the manuscript.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
All patients agreed to participate in the present
study, and written consent was obtained from each patient. The
research protocol was approved by the institutional review
committee of the First Affiliated Hospital of Xinjiang Medical
University.
Patient consent for the publication
All patients provided written informed consent for
the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
iTRAQ
|
isobaric tags for relative and
absolute quantitation
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
ILK
|
integrin-linked kinase
|
|
CR
|
complete remission
|
|
PR
|
partial remission
|
|
SD
|
stability disease
|
|
PD
|
progression disease
|
|
RECIST
|
response evaluation criteria in solid
tumors
|
References
|
1
|
Soerjomataram I, Lortet-Tieulent J, Parkin
DM, Ferlay J, Mathers C, Forman D and Bray F: Global burden of
cancer in 2008: A systematic analysis of disability-adjusted
life-years in 12 world regions. Lancet. 380:1840–1850. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A Review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Clegg LX, Ward E, Ries LA, Wu X,
Jamison PM, Wingo PA, Howe HL, Anderson RN and Edwards BK: Annual
report to the nation on the status of cancer, 1975–2001, with a
special feature regarding survival. Cancer. 101:3–27. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Doi T, Piha-Paul SA, Jalal SI, Saraf S,
Lunceford J, Koshiji M and Bennouna J: Safety and antitumor
activity of the anti-programmed death-1 antibody pembrolizumab in
patients with advanced esophageal carcinoma. J Clin Oncol.
36:61–67. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Giri S, Pathak R, Aryal MR, Karmacharya P,
Bhatt VR and Martin MG: Incidence trend of esophageal squamous cell
carcinoma: An analysis of Surveillance Epidemiology and End Results
(SEER) database. Cancer Causes Control. 26:159–161. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang J, Zhi C, Zhen F, Yuan X, Jiao C,
Zhu H, Zhu H and Feng Y: iTRAQ-based quantitative proteomic
analyses of high grade esophageal squamous intraepithelial
neoplasia. Proteomics Clin Appl. Dec 11–2017.(Epub ahead of print).
doi: 10.1002/prca.201600167. View Article : Google Scholar
|
|
9
|
Hsu CH, Hsu CW, Hsueh C, Wang CL, Wu YC,
Wu CC, Liu CC, Yu JS, Chang YS and Yu CJ: Identification and
characterization of potential biomarkers by quantitative tissue
proteomics of primary lung adenocarcinoma. Mol Cell Proteomics.
15:2396–2410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rice TW: Esophageal cancer staging. Korean
J Thorac Cardiovasc Surg. 48:157–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Young J, Badgery-Parker T, Dobbins T,
Jorgensen M, Gibbs P, Faragher I, Jones I and Currow D: Comparison
of ECOG/WHO performance status and ASA score as a measure of
functional status. J Pain Symptom Manage. 49:258–264. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bandaranayake AD, Correnti C, Ryu BY,
Brault M, Strong RK and Rawlings DJ: Daedalus: A robust, turnkey
platform for rapid production of decigram quantities of active
recombinant proteins in human cell lines using novel lentiviral
vectors. Nucleic Acids Res. 39:e1432011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta DeltaC (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herskovic A, Russell W, Liptay M, Fidler
MJ and Al-Sarraf M: Esophageal carcinoma advances intreatment
results for locally advanced disease: Review. Ann Oncol.
23:1095–1103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li BZ, Chen ZL, Shi SS, Feng XL, Tan XG,
Zhou F and He J: Overexpression of Cdc25C predicts response to
radiotherapy and survival in esophageal squamous cell carcinoma
patients treated with radiotherapy followed by surgery. Chin J
Cancer. 32:403–409. 2013.PubMed/NCBI
|
|
17
|
Sugase T, Takahashi T, Serada S, Fujimoto
M, Hiramatsu K, Ohkawara T, Tanaka K, Miyazaki Y, Makino T,
Kurokawa Y, et al: SOCS1 gene therapy improves radiosensitivity and
enhances irradiation-induced DNA damage in esophageal squamous cell
carcinoma. Cancer Res. 77:6975–6986. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tamaoki M, Komatsuzaki R, Komatsu M,
Minashi K, Aoyagi K, Nishimura T, Chiwaki F, Hiroki T, Daiko H,
Morishita K, et al: Multiple roles of single-minded 2 in esophageal
squamous cell carcinoma and its clinical implications. Cancer Sci.
109:1121–1134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shigaki H, Baba Y, Watanabe M, Murata A,
Ishimoto T, Iwatsuki M, Iwagami S, Nosho K and Baba H: PIK3CA
mutation is associated with a favorable prognosis among patients
with curatively resected esophageal squamous cell carcinoma. Clin
Cancer Res. 19:2451–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hannigan GE, Leung-Hagesteijn C,
Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC and Dedhar
S: Regulation of cell adhesion and anchorage-dependent growth by a
new beta 1-integrin-linked protein kinase. Nature. 379:91–96. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong RP, Ng P, Dedhar S and Li G: The role
of integrin-linked kinase in melanoma cell migration, invasion and
tumor growth. Mol Cancer Ther. 6:1692–1700. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhuang X, Lv M, Zhong Z, Zhang L, Jiang R
and Chen J: Interplay between intergrin-linked kinase and
ribonuclease inhibitor affects growth and metastasis of bladder
cancer through signaling ILK pathways. J Exp Clin Cancer Res.
35:1302016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen H, Ma JL, Zhang Y, Deng GL, Qu YL, Wu
XL, He JX, Zhang S and Zeng S: Integrin-linked kinase
overexpression promotes epithelial-mesenchymal transition via
nuclear factor-κB signaling in colorectal cancer cells. World J
Gastroenterol. 22:3969–3977. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Driver GA and Veale RB: Modulation of
integrin-linked kinase (ILK) expression in human oesophageal
squamous cell carcinoma cell lines by the EGF and TGFbeta1 growth
factors. Cancer Cell Int. 6:122006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mohamed MS, Bishr MK, Almutairi FM and Ali
AG: Inhibitors of apoptosis: Clinical implications in cancer.
Apoptosis. 22:1487–1509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duxbury MS, Ito H, Benoit E, Waseem T,
Ashley SW and Whang EE: RNA interference demonstrates a novel role
for integrin-linked kinase as a determinant of pancreatic
adenocarcinoma cell gemcitabine chemoresistance. Clin Cancer Res.
11:3433–839. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lanvin O, Monferran S, Delmas C, Couderc
B, Toulas C and Cohen-Jonathan-Moyal E: Radiation-induced mitotic
cell death and glioblastoma radioresistance: A new regulating
pathway controlled by integrin-linked kinase, hypoxia-inducible
factor 1 alpha and survivin in U87 cells. Eur J Cancer.
49:2884–2891. 2013. View Article : Google Scholar : PubMed/NCBI
|