Introduction
Lung cancer is one of the most fatal types of cancer
in developed countries, and remains the leading cause of
cancer-associated mortality (1),
with a higher mortality rate than prostate, breast and colon cancer
combined, and with >1 million deaths expected per year worldwide
(2). Although treatments for lung
cancer have improved in recent years, including surgery,
chemotherapy, radiation therapy, local treatment and targeted
therapy, >50% of patients with lung cancer present with advanced
disease at the first diagnosis and therefore, no longer qualify for
surgery (3).
Gene mutations serve an important role in
tumorigenesis. Since the discovery of epidermal growth factor
receptor (EGFR)-activating mutations and mutations of anaplastic
lymphoma kinase (ALK), the potential avenues for novel or
repurposed therapeutics have increased (4,5).
Compared with other cancers, lung cancer has one of the highest
rates of genetic alterations (6);
therefore, targeted therapeutics for specific genes/proteins may
lead to improvements in the outcomes of patients with lung cancer.
Therefore, targeted therapies hold great promise for patients with
lung cancer.
Mutant genes can be produced by chromosomal
rearrangements, including the translocation, insertion, inversion
and deletion of chromosomes (5).
EGFR mutations and ALK fusion genes are two well-studied and common
oncogenic drivers in patients with lung cancer, and small molecule
inhibitors of these target genes have been utilized in the clinic
to selectively inhibit the growth of gene-positive non-small cell
lung cancer (7,8). Furthermore, expression of the mutant
genes in cancer may additionally serve as identifying markers for
lung cancer and cancer typing.
The R-spondin (RSPO) family of proteins consists of
four members (RSPO1-4), all of which are secreted, and are widely
expressed in vertebrate embryos and in adults (9). The four genes that encode the RSPO
proteins share 40–60% pairwise amino acid sequence identity and are
predicted to possess substantial structural homology (10). Furthermore, the RSPO proteins are
characterized by two furin-like cysteine-rich domains, a
thrombospondin-type 1 repeat domain and a basic-amino acid-rich
C-terminal domain (9,11,12).
The Wnt/β-catenin signaling pathway has a vital role
in cell proliferation, development and disease pathogenesis
(13). RSPOs serve a regulatory role
in cell progress, which include substance transport and catabolism,
cell movement, cell growth and death, and cellular communication,
by activating and synergizing the Wnt/β-catenin signaling pathway
(14,15). For example, RSPO1 and the
leucine-rich repeat-containing G-protein coupled receptor 4/5
(LGR4/5) potentiate Wnt/β-catenin signaling, increased liver size
and improved liver regeneration (16). RSPO3 may promote vascular development
in Xenopus laevis by upregulating the expression of vascular
endothelial growth factor, through the activation of the Wnt/β
catenin signaling pathway (17).
Recently, the role of RSPOs in oncogenesis has also been
investigated. The RSPO genes may facilitate tumor development in
the colon by promoting Wnt signaling (18). In addition, a previous study
suggested that chromosomal rearrangements of the RSPO2 and RSPO3
genes may initiate hyperplasia and tumor development in vivo
(19). Furthermore, it has been
previously demonstrated that the RSPO family of proteins may serve
as tumor suppressors. RSPO1 and LGR5 co-localize and form complexes
with transforming growth factor β1 (TGF-β) receptor I and II,
enhancing TGF-β downstream signaling, and therefore, suppressing
metastasis of colon cancer (20).
Taken together, the aforementioned studies suggest
that the RSPO family of proteins may act as potential therapeutic
targets or prognostic biomarkers in certain types of cancer.
However, to the best of our knowledge, there are no studies
demonstrating the expression or role of RSPOs in lung cancer. In
the present study, the expression of the RSPO proteins in patients
with different pathological types of lung cancer and in lung cancer
cell lines was determined by utilizing publicly available databases
and their prognostic value was assessed.
Materials and methods
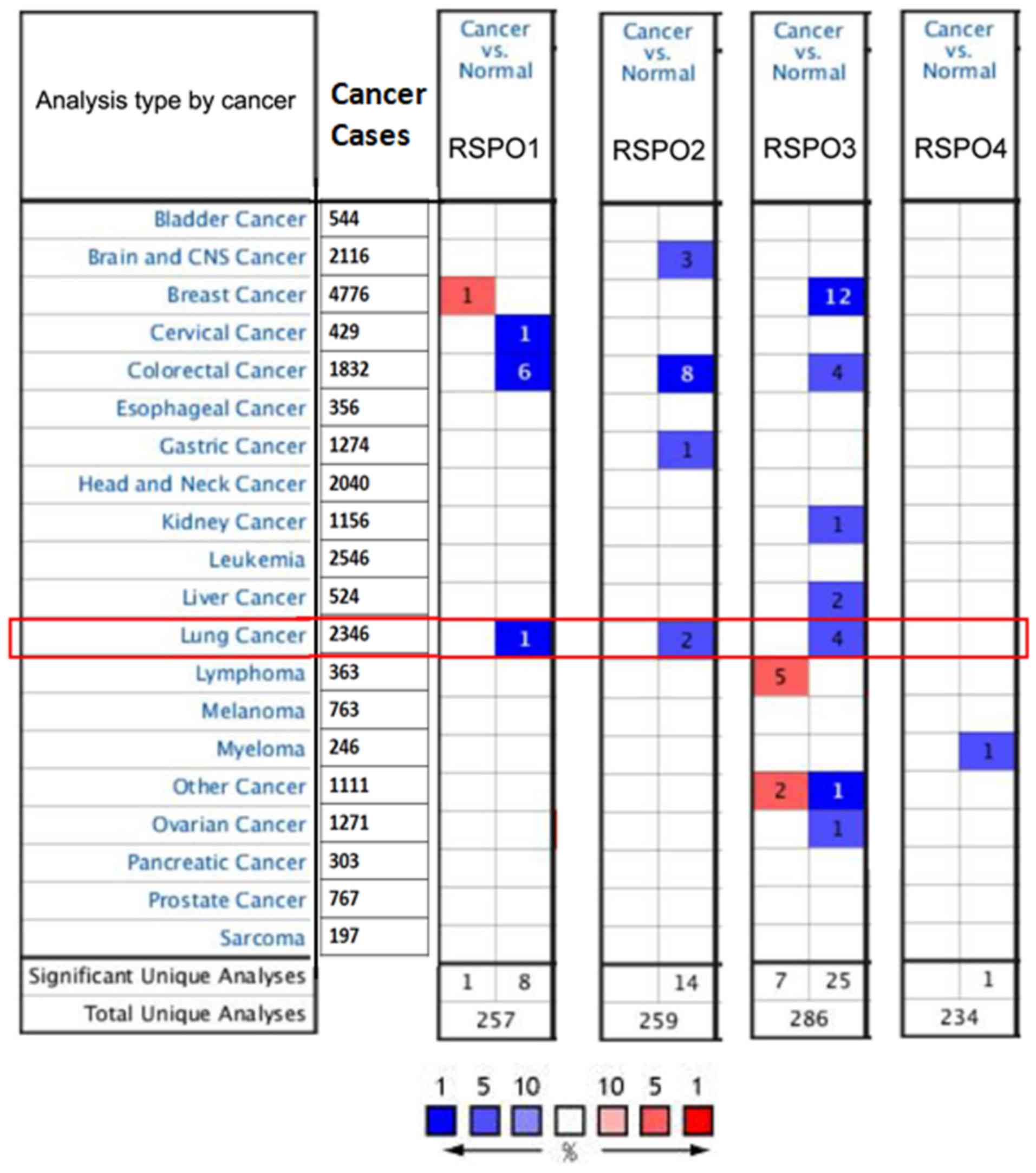
ONCOMINE database analysis
ONCOMINE is an online database of tumor-related gene
expression (www.oncomine.org). In the present
study, ONCOMINE was used to analyze the mRNA expression differences
of the RSPO family of proteins between tumor and normal tissues in
common types of human cancer, including lung cancer (2,346 cases),
liver cancer (524 cases) and leukemia (2,546 cases). In this
database, Student's t-test was used to compare tumor and normal
tissues. The thresholds of gene analysis were set as follows:
P<0.01; fold-change, 2; gene rank, top 10% [where the tumor
shown is within the top 10% of the tumor type of the RSPO gene,
which included bladder cancer, brain and CNS cancer, breast cancer
and lung cancer (Fig. 1)]; analysis
type, cancer vs. normal; and data type, mRNA. The above data were
obtained from studies that showed statistically differences in
expression. Furthermore, to analyze the mRNA expression differences
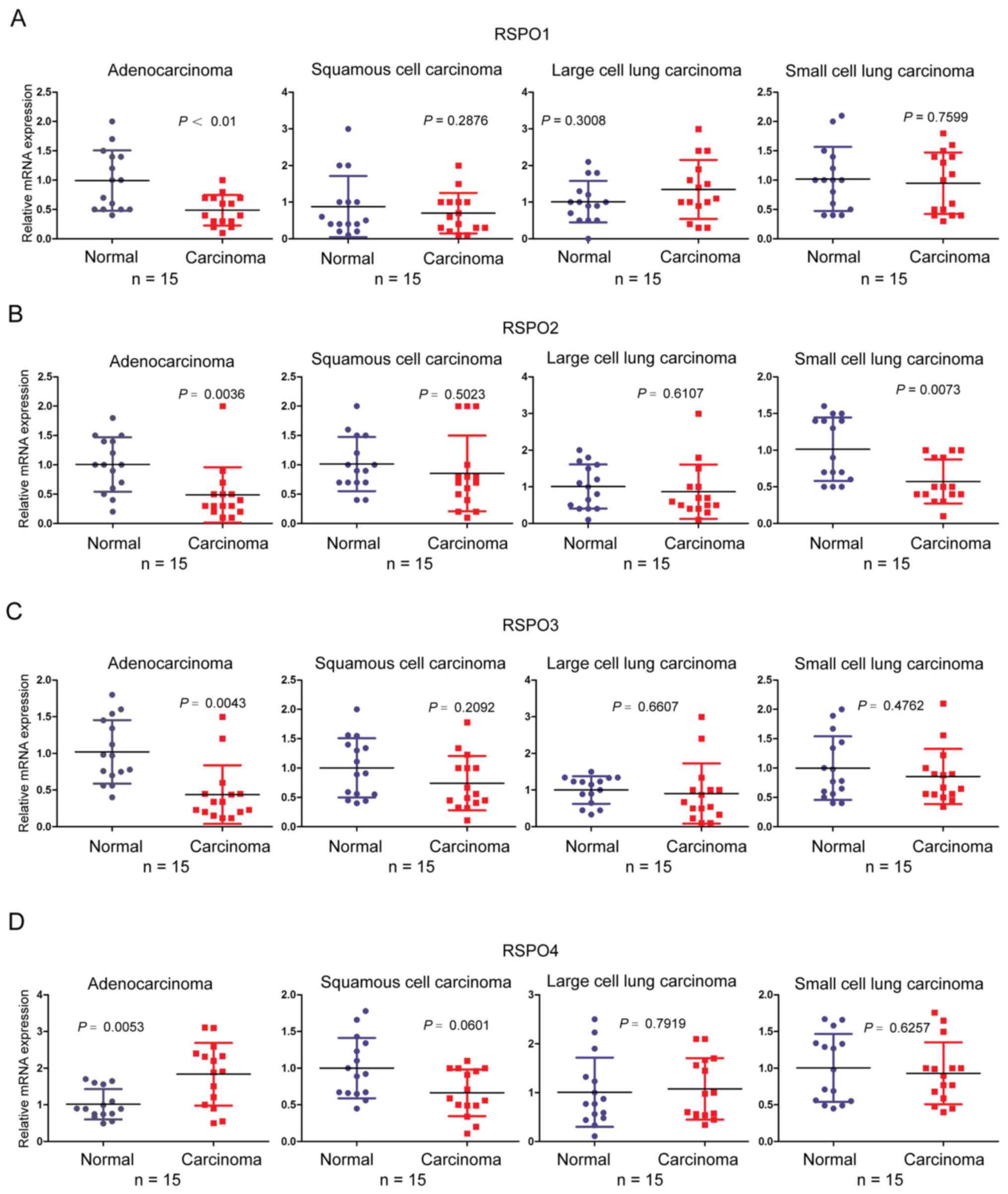
of the RSPO family of proteins between different pathological types
of lung cancer, the terms ‘adenocarcinoma’, ‘squamous cell
carcinoma’, ‘large cell lung cancer’ and ‘small cell lung cancer’
were used.
Cancer Cell Line Encyclopedia (CCLE)
database analysis
The CCLE database (portals.broadinstitute.org/ccle/home), is an online
encyclopedia of a compilation of gene expression, chromosomal copy
number and massively parallel sequencing data from 947 human cancer
cell lines (21). CCLE was used to
analyze the mRNA expression levels of RSPOs in a range of cell
lines for different types of cancer, including non-small lung
cancer, liver cancer and breast cancer, and marked as ‘lung_NSC’,
‘liver’ and ‘breast’, respectively in Fig. 2.
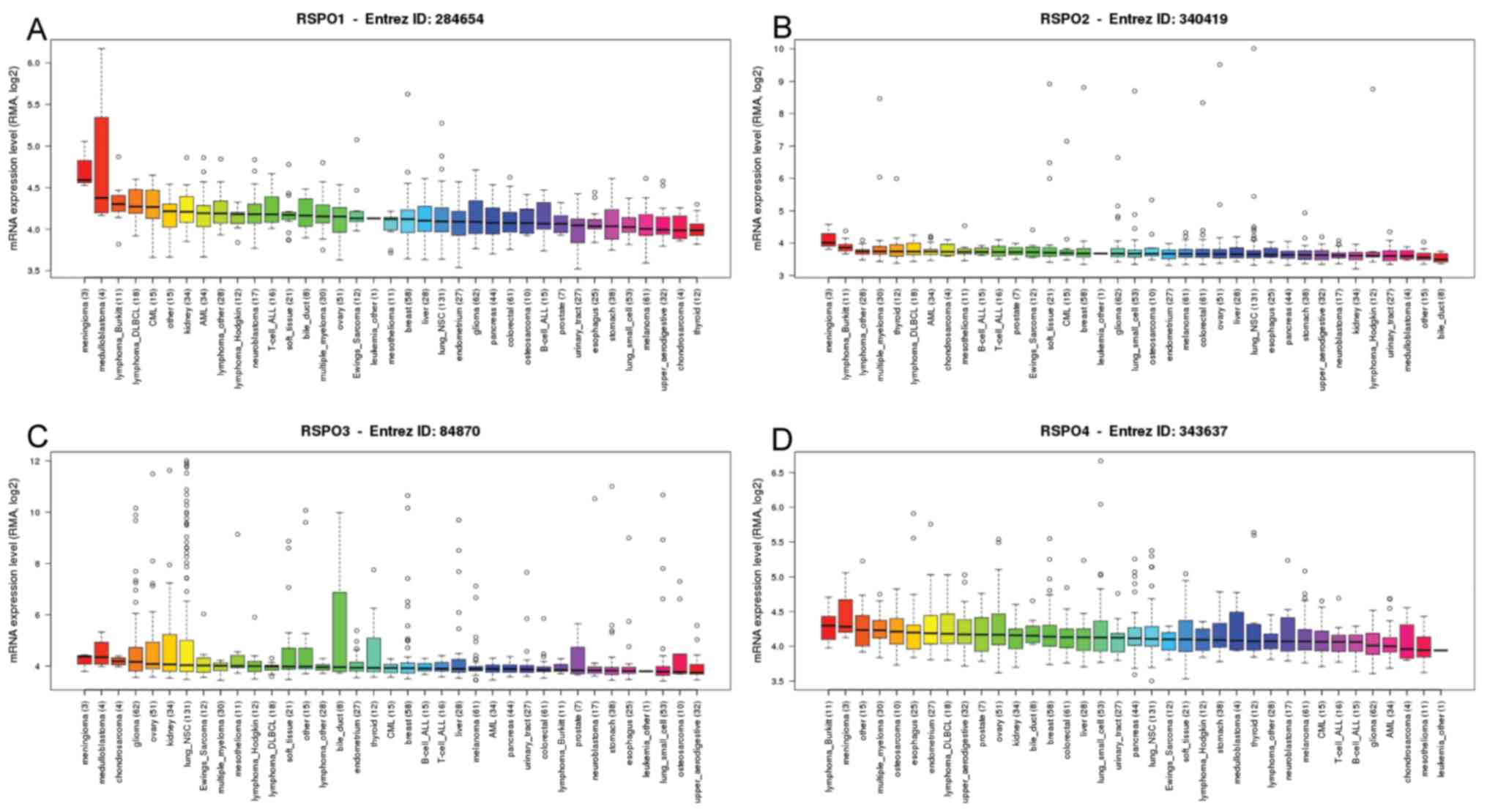 | Figure 2.RSPO protein family expression across
4,103 primary tumors from Cancer Cell Line Encyclopedia database.
Box-and-whisker plots showed the distribution of (A) RSPO1, (B)
RSPO2, (C) RSPO3 and (D) RSPO4 mRNA expression for each subtype,
ordered by the median RSPOs expression level (line), the
inter-quartile range (box) and up to 1.5× the inter-quartile range
(bars). RSPO, R-spondin; DLBCL, diffuse large B-cell lymphoma; CML,
chronic myeloid leukemia; AML, acute myeloid leukemia; ALL, acute
lymphoblastic leukemia; NSC, non-small cell. |
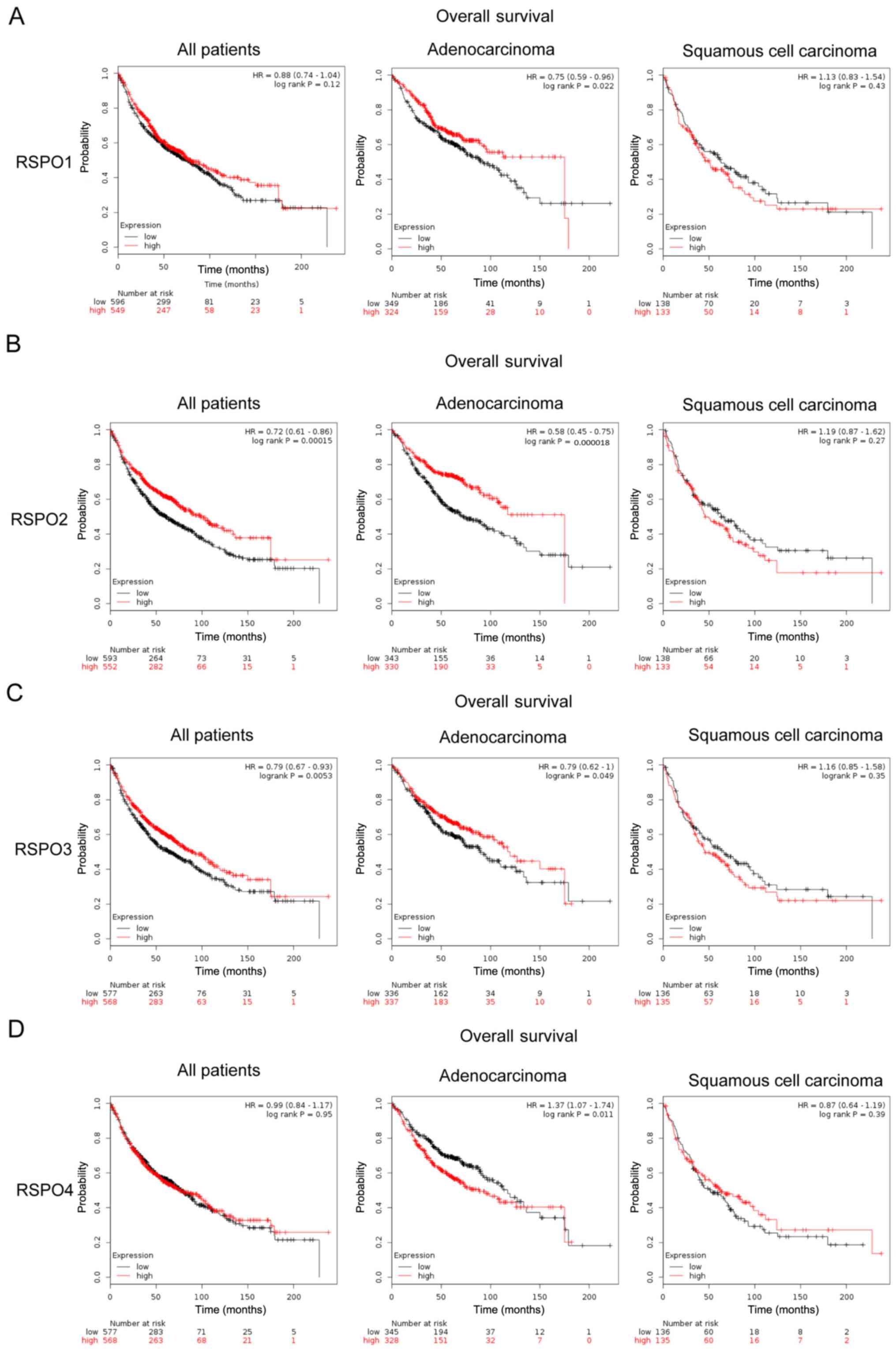
Kaplan-Meier (KM) plotter database
analysis
The KM plotter (kmplot.com/analysis/) is capable of assessing the
effect of 54,675 genes on survival using 10,461 cancer samples,
including 3,452 lung cancers samples, with a mean follow-up of
69.5/34.5/17 months for all patients with lung cancer,
adenocarcinoma and squamous cell carcinoma respectively. The cancer
patients were divided into high and low expression groups by the
median values of mRNA expression, where expression levels above
this value were classified as high expression and below as low
expression. The prognostic values of the members of the RSPO family
of proteins with increased expression in different pathological
types of lung cancer samples were further assessed to determine
overall survival (OS) using KM plotter. The log-rank test was used
to compare OS between high and low expression groups. P<0.05 was
used to indicate a statistically significant difference.
Results
Expression of RSPO1-3 is significantly
decreased in patients with lung cancer
The ONCOMINE database was used to determine whether
there was a difference in the mRNA expression levels of the members
of the RSPO family of proteins between cancer and normal tissues in
lung cancer. At present, four RSPO proteins have been identified in
different types of cancer, including hematological malignancies and
solid tumors (9). The database
contained a total of 257,259,286 and 234 unique analyses for RSPO1,
RSPO2, RSPO3 and RSPO4, respectively. ONCOMINE analysis revealed
that RSPO1, 2 and 3 mRNA expression levels were significantly lower
in lung cancer tissue compared with normal samples in a wide
variety of datasets, which differed from many other types of cancer
(Fig. 1). RSPO1 had a 2-fold
increase in normal tissues compared with that in lung cancer
tissue, while RSPO2 and RPSO3 had a 4- and 8-fold increase,
respectively. In addition, analysis of the CCLE was consistent with
that of ONCOMINE analysis, demonstrating that RSPO 1–3 were
distinctively downregulated in lung cancer cell lines (Fig. 2). The mRNA expression of RSPO1, 2 and
3 was lower in lung adenocarcinoma compared with normal tissues
(P<0.01). However, except for the expression level of RSPO2 in
small cell lung cancer, which was decreased (P=0.0073), the RSPO1
and 3 expression levels were not significantly different between
cancer tissue and normal tissue in other types of lung cancer,
including squamous cell carcinoma, large cell lung carcinoma and
small cell lung carcinoma (all P>0.05; Fig. 3A-C). However, there was no
significant difference in the mRNA expression level of RSPO4
between squamous cell carcinoma, large cell lung carcinoma and
small cell lung carcinoma samples and normal controls. As shown in
Fig. 1, there was no statistical
significant difference in the expression level of RSPO4 mRNA in
lung cancer. Furthermore, except for the high expression of RSPO4
in lung adenocarcinoma (P=0.0053), there was no statistical
significant difference of RSPO4 mRNA expression level between tumor
tissues and normal tissues in other types of lung cancer, including
squamous cell carcinoma, large cell carcinoma and small cell
carcinoma (P=0.0601, P=0.7919, P=0.6257, respectively) (Fig. 3D). These results suggest that the
function of RSPO1, RSPO2 and RSPO3 may differ from the function of
RSPO4, and may have unique roles in tumorigenesis in lung
cancer.
High RSPO 1–3 mRNA expression is
associated with improved OS in patients with lung cancer
The prognostic effect of RSPO1, RSPO2 and RSPO3 in
patients with lung cancer was subsequently assessed. High mRNA
expression levels of RSPO2 and RSPO3 were associated with improved
OS in all patients with lung cancer [RSPO2, hazard ratio (HR)=0.72,
P=0.00015; RSPO3, HR=0.79, P=0.0053; Fig. 4]. In particular, analysis of lung
cancer pathological sub-types revealed that high mRNA expression of
RSPO1, 2 and 3 was significantly associated with improved OS in
patients with lung adenocarcinoma (RSPO1, HR=0.75, P=0.022; RSPO2,
HR=0.58, P=0.000018; RSPO3, HR=0.79, P=0.049); however, there was
no significant association between expression of RSPO1, 2 and 3 and
OS in patients with squamous cell carcinoma, which suggested that
RSPO1, RSPO2 and RSPO3 may have prognostic value for patients with
lung adenocarcinoma (Fig. 4).
Contrary to RSPO1, 2 and 3, RSPO4 expression was associated with
worse OS in patients with lung adenocarcinoma (HR=1.37; P=0.011).
However, high RSPO4 mRNA expression level was independent of OS in
all patients with lung cancer (HR=0.99; P=0.95; Fig. 4).
Discussion
Lung cancer presents with the highest prevalence and
mortality rates among all types of cancer, with a 5-year overall
survival rate of only 15%, even with conventional chemotherapy,
radiotherapy and surgical interventions (22). The low overall survival rates are
primarily due to resistance to chemotherapy, radiotherapy and
targeted therapy, in addition to the frequent presence of distant
metastases and recurrent or unresectable tumors (3). Therefore, the development of effective
anticancer therapies against lung cancer is required.
The RSPO family of secreted proteins has a
regulatory role in cell progress and has been demonstrated to be
involved in tumorigenesis (17,19). The
RSPO protein family also had lower expression in normal tissues
compared with that in colorectal cancer tissue. Recently, research
showed that the RSPO family fusion gene was found in superficially
serrated adenoma, which is a recently proposed subtype of
colorectal serrated lesion, suggesting that the deletion of RSPO
protein family is closely related to the development of colorectal
cancer (23). This suggests that
RSPO protein family also plays an important role in the development
of lung cancer. In the present study, the mRNA expression levels of
the RSPO proteins in various types of cancers, and lung cancer in
particular, were systematically examined using the ONCOMINE
database and CCLE. In addition, the prognostic values of the four
RSPO proteins were examined in patients with different subtypes of
lung cancer through the KM plotter. The present study suggested
that, among the four RSPO proteins, RSPO1, RSPO2 and RSPO3
expression was significantly lower in lung cancer compared to
normal controls, suggesting a potential protective role in lung
cancer.
Wnt signaling is a highly conserved signaling
pathway that controls cell proliferation and homeostatic processes
(24). Wnt signaling primarily
consists of three distinct signaling pathways: Wnt/Ca2+
pathway; Wnt/planar cell polarity pathway; and Wnt/β-catenin
pathway (24). The role of
Wnt/β-catenin signaling in inflammatory responses and cell
development has been extensively studied (10,12,14). The
four RSPO proteins act through their cognate LGR4, LGR5 and LGR6
receptors to amplify Wnt/β-catenin signaling (10,19,20).
It has been demonstrated that RSPO1 interferes with
Dickkopf-related protein 1/Kremen-mediated internalization of LRP6,
therefore increasing cell surface retention of LRP6 and
subsequently regulating the cellular responsiveness to Wnt ligands
(25). Furthermore, studies have
shown that RSPO1/LGR5 directly activates TGF-β signaling, and when
RSPO1 activated LGR5, LGR5 formed complexes with TGF-β receptors,
and was therefore able to suppress colon cancer progression
(20). In Fig. 1, there was low expression of RSPO1-3
in colon cancer, which may be related to the progression of colon
cancer. Survival analysis of RSPO1 in the present study
demonstrated similar results, high expression of RSPO1 was
associated with improved OS in patients with lung cancer,
particularly in lung adenocarcinoma, suggesting RSPO1
tumor-suppressive role.
Recently, it was demonstrated that RSPO2 acts as a
direct antagonistic ligand to E3 ubiquitin-protein ligase RNF43 and
E3 ubiquitin-protein ligase ZNRF3, which together, constitute a
master switch that governs limb development independent of LGR4
(26). A previous study has also
indicated that induced mucosal RSPO2 expression in susceptible mice
lead to the generation of a poorly differentiated epithelium and to
fatal colitis, and implicated RSPO2-mediated Wnt signaling
activation in intestinal morphogenesis, proliferation and
carcinogenesis (27). Furthermore,
in tumorigenesis, RSPO2 enhances Wnt signaling conferring
stemness-associated traits to susceptible pancreatic cancer cells
(28). However, its role in lung
cancer progression remains largely unknown. To the best of our
knowledge, the present study is the first to demonstrate that high
RSPO2 expression predicted better survival in patients with lung
cancer, particularly in lung adenocarcinoma, although there was no
significant effect on survival in patients with squamous cell
carcinoma.
RSPO3 is also associated with Wnt/β-catenin
signaling. Vidal et al (29)
suggested that deletion of RSPO3 resulted in a loss of
Sonic-hedgehog signaling and therefore impaired organ growth. A
previous study demonstrated that targeting protein tyrosine
phosphatase receptor type K-RSPO3-fusion inhibits colorectal tumor
growth and promotes differentiation (30). In the present study it was
demonstrated that similar to RSPO1 and RSPO2, RSPO3 expression was
significantly lower in samples from patients with lung
adenocarcinoma. Furthermore, high levels of RSPO3 predicted
improved OS in patients with lung cancer, particularly in lung
adenocarcinoma. Taken together, RSPO1, 2 and 3 expression may be
favorable prognostic predictors in patients with lung cancer,
particularly lung adenocarcinoma.
There are considerably fewer studies investigating
the function of RSPO4. In a recent study, it was demonstrated that
an RSPO4-mutant may have been associated with isolated congenital
anonychia/hyponychia (31). In the
present study, RSPO4 was not detected in almost all types of cancer
tissues and normal tissues. Survival analysis showed that the
expression level of RSPO4 was not directly associated with OS of
patients with lung cancer, suggesting a difference in the function
of RSPO4 in lung cancer compared with the other family members.
In conclusion, high expression levels of RSPO1,
RSPO2 and RSPO3 in lung cancer predicted improved OS, particularly
in patients with lung adenocarcinoma. RSPO1, RSPO2 and RSPO3 may
serve important protective roles in lung cancer, and may represent
novel biomarkers and prognostic factors for patients with lung
cancer. One of the limitations of this study is that validation of
the expression of RSPO family proteins in lung cancer cells lines
and tissues was not performed, and therefore further clinical and
mechanism analysis are required to verify the results of the
current study.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Science
Technology Project of Suzhou (grant no. SS201862).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the author on reasonable request.
Authors' contributions
WZ and JQ conceived and designed the project. LW, WZ
and JW performed data mining processes and prepared the figures.
LW, LJ and CL analyzed and interpreted the data. LJ and CL wrote
the manuscript. LW and WZ participated in the revision of the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moya-Horno I, Viteri S, Karachaliou N and
Rosell R: Combination of immunotherapy with targeted therapies in
advanced non-small cell lung cancer (NSCLC). Ther Adv Med Oncol.
10:17588340177450122018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó
L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Puig de la Bellacasa R, Karachaliou N,
Estrada-Tejedor R, Teixidó J, Costa C and Borrell JI: ALK and ROS1
as a joint target for the treatment of lung cancer: A review.
Transl Lung Cancer Res. 2:72–86. 2013.PubMed/NCBI
|
|
8
|
Wen M, Wang X, Sun Y, Xia J, Fan L, Xing
H, Zhang Z and Li X: Detection of EML4-ALK fusion gene and features
associated with EGFR mutations in Chinese patients with
non-small-cell lung cancer. Onco Targets Ther. 9:1989–1995. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim KA, Zhao J, Andarmani S, Kakitani M,
Oshima T, Binnerts ME, Abo A, Tomizuka K and Funk WD: R-Spondin
proteins: A novel link to beta-catenin activation. Cell Cycle.
5:23–26. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kazanskaya O, Glinka A, del Barco
Barrantes I, Stannek P, Niehrs C and Wu W: R-Spondin2 is a secreted
activator of Wnt/beta-catenin signaling and is required for Xenopus
myogenesis. Dev Cell. 7:525–534. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Lau WB, Snel B and Clevers HC: The
R-spondin protein family. Genome Biol. 13:2422012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rong X, Chen C, Zhou P, Zhou Y, Li Y, Lu
L, Liu Y, Zhou J and Duan C: R-spondin 3 regulates dorsoventral and
anteroposterior patterning by antagonizing Wnt/β-catenin signaling
in zebrafish embryos. PLoS One. 9:e995142014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sher I, Yeh BK, Mohammadi M, Adir N and
Ron D: Structure-based mutational analyses in FGF7 identify new
residues involved in specific interaction with FGFR2IIIb. FEBS
Lett. 552:150–154. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zebisch M, Xu Y, Krastev C, MacDonald BT,
Chen M, Gilbert RJ, He X and Jones EY: Structural and molecular
basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by
the Wnt agonist R-spondin. Nat Commun. 4:27872013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim KA, Wagle M, Tran K, Zhan X, Dixon MA,
Liu S, Gros D, Korver W, Yonkovich S, Tomasevic N, et al: R-Spondin
family members regulate the Wnt pathway by a common mechanism. Mol
Biol Cell. 19:2588–2596. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Planas-Paz L, Orsini V, Boulter L,
Calabrese D, Pikiolek M, Nigsch F, Xie Y, Roma G, Donovan A, Marti
P, et al: The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver
zonation and size. Nat Cell Biol. 18:467–479. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kazanskaya O, Ohkawara B, Heroult M, Wu W,
Maltry N, Augustin HG and Niehrs C: The Wnt signaling regulator
R-spondin 3 promotes angioblast and vascular development.
Development. 135:3655–3664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seshagiri S, Stawiski EW, Durinck S,
Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman
V, Jaiswal BS, et al: Recurrent R-spondin fusions in colon cancer.
Nature. 488:660–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han T, Schatoff EM, Murphy C, Zafra MP,
Wilkinson JE, Elemento O and Dow LE: R-spondin chromosome
rearrangements drive Wnt-dependent tumour initiation and
maintenance in the intestine. Nat Commun. 8:159452017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Geng L, Wang D, Yi H, Talmon G and
Wang J: R-spondin1/LGR5 activates TGFβ signaling and suppresses
colon cancer metastasis. Cancer Res. 77:6589–6602. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cancer facts and figures 2008American
Cancer Society; Atlanta, GA, USA: pp. 9–14. 2008
|
|
23
|
Hashimoto T, Ogawa R, Yoshida H, Taniguchi
H, Kojima M, Saito Y and Sekine S: EIF3E-RSPO2 and PIEZO1-RSPO2
fusions in colorectal traditional serrated adenoma. Histopathology.
75:266–273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xing Y, Clements WK, Kimelman D and Xu W:
Crystal structure of a beta-catenin/axin complex suggests a
mechanism for the beta-catenin destruction complex. Genes Dev.
17:2753–2764. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Binnerts ME, Kim KA, Bright JM, Patel SM,
Tran K, Zhou M, Leung JM, Liu Y, Lomas WE III, Dixon M, et al:
R-Spondin1 regulates Wnt signaling by inhibiting internalization of
LRP6. Proc Natl Acad Sci USA. 104:14700–14705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szenker-Ravi E, Altunoglu U, Leushacke M,
Bosso-Lefèvre C, Khatoo M, Thi Tran H, Naert T, Noelanders R,
Hajamohideen A, Beneteau C, et al: RSPO2 inhibition of RNF43 and
ZNRF3 governs limb development independently of LGR4/5/6. Nature.
557:564–569. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Papapietro O, Teatero S, Thanabalasuriar
A, Yuki KE, Diez E, Zhu L, Kang E, Dhillon S, Muise AM, Durocher Y,
et al: R-spondin 2 signalling mediates susceptibility to fatal
infectious diarrhoea. Nat Commun. 4:18982013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ilmer M, Boiles AR, Regel I, Yokoi K,
Michalski CW, Wistuba II, Rodriguez J, Alt E and Vykoukal J: RSPO2
enhances canonical Wnt signaling to confer stemness-associated
traits to susceptible pancreatic cancer cells. Cancer Res.
75:1883–1896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vidal V, Sacco S, Rocha AS, da Silva F,
Panzolini C, Dumontet T, Doan TM, Shan J, Rak-Raszewska A, Bird T,
et al: The adrenal capsule is a signaling center controlling cell
renewal and zonation through Rspo3. Genes Dev. 30:1389–1394. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Storm EE, Durinck S, de Sousa e Melo F,
Tremayne J, Kljavin N, Tan C, Ye X, Chiu C, Pham T, Hongo JA, et
al: Targeting PTPRK-RSPO3 colon tumours promotes differentiation
and loss of stem-cell function. Nature. 529:97–100. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hsu CK, Romano MT, Nanda A, Rashidghamat
E, Lee JYW, Huang HY, Songsantiphap C, Lee JY, Al-Ajmi H, Betz RC,
et al: Congenital anonychia and uncombable hair syndrome:
Coinheritance of homozygous mutations in RSPO4 and PADI3. J Invest
Dermatol. 137:1176–1179. 2017. View Article : Google Scholar : PubMed/NCBI
|