Introduction
Lung cancer has been the most common malignancy in
the world, with ~1,820,000 new cases and ~1,600,000 patients
succumbing to disease per year, for several decades (1). Small cell lung cancer (SCLC), a
malignant tumor type, accounts for almost 15% of all newly
diagnosed lung cancer cases (2).
SCLC most frequently results from smoking and has a particularly
aggressive behavior. Of patients with SCLC, it is reported that
60–70% have developed metastasis at the point of diagnosis
(3). Untreated patients with SCLC
rapidly succumb to the disease within 2–4 months (4). Patients with limited-stage SCLC are
usually treated with chemo- and radiotherapy and with consecutive
prophylactic cranial irradiation in the case of intracranial
metastasis (5,6), while chemotherapy is the primary choice
for patients with extensive-stage disease (7). Despite treatment, patients with SCLC
eventually relapse due to resistance. The 5-year survival rate is
<3% (8), making SCLC the lung
cancer subtype with the highest mortality rate.
In the 1980s, numerous researchers began to search
for prognostic markers and therapeutic targets for SCLC. A number
of serum components have been suggested as diagnostic biomarkers
and indicators of the clinical response to chemotherapy in patients
with SCLC (9), among which
neuron-specific enolase (NSE) is generally acknowledged as a
diagnostic and therapeutic marker of SCLC (10–12). NSE
is the r-subunit of the glycolytic enolase enzyme that catalyzes
the decomposition of glycerol in the glycolytic pathway and
consists of a, b and c subunits and aa, bb, cc, ac and bc isozymes
(13). The upregulation of NSE is
frequently reported in SCLC (14).
Molina et al (15) compared
the serum levels of squamous cell carcinoma antigen,
carcinoembryonic antigen, CYFRA21-1, several mucins and NSE in
patients with SCLC, and determined that the detection sensitivity
of NSE was 81.2% and was superior to that of other markers. It has
also been determined that the level of NSE has prognostic value.
Liu et al (16) reported that
a normal serum NSE level (hazard ratio, 0.447; P=0.017) at the time
of diagnosis was an independent positive prognostic factor for
patients with limited-stage SCLC, but not for extensive-stage SCLC.
Furthermore, NSE levels were reported to be a predictor of complete
response and survival following chemotherapy (17,18) and
to be associated with the tumor burden, for which it is generally
regarded as a reliable marker to evaluate the response to
chemotherapy (19).
As a useful predictive marker, NSE has been
investigated intensively in SCLC and other neuroendocrine
neoplasms, including neuroblastoma. However, the role of NSE in
regulating SCLC and the exact mechanisms remain to be elucidated.
In the present study, the influence of NSE knockdown on cell
proliferation and migratory ability, cell cycle and protein levels
of metastasis-associated genes in SCLC cell lines were evaluated
for the first time.
Materials and methods
Cell lines and culture
The NCI-H209, NCI-H345, NCI-H446 and NCI-H146 SCLC
cell lines were purchased from the American Type Culture Collection
and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; HyClone; GE Healthcare Life Sciences) and 100 mg/ml
penicillin and streptomycin in an incubator containing 5%
CO2 at 37°C.
RNA interference
Small interfering (si)RNAs against NSE were designed
and synthesized by GenePharma Co., Ltd. (Shanghai, China). The
sequences of the siRNAs targeting human NSE were siRNA-1:
5′-GGACAAAUACGGCAAGGAUTT-3′ and siRNA-2:
5′-GCCGGACAUAACUUCCGUATT-3′ and the sequence of the negative
control siRNA was: 5′-UUCUCCGAACGUGUCACGUTT-3′. For RNA
interference experiments, the siRNAs were transfected at a working
concentration of 50 nmol/l into NCI-H209 cells (1×105
cells/well) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The functional assays were performed 48 h following
transfection.
Western blot analysis
Western blot analysis was performed as previously
reported (20). Total protein was
extracted by radioimmunoprecipitation assay buffer (Sangon Biotech
Co., Ltd.), and the concentration was determined by the
bicinchoninic acid method. Protein (30 µg/lane) was separated by
10% SDS-PAGE and transferred to PVDF membranes (Merck KGaA),
followed by blocking in 5% skimmed milk at 25°C for 1.5 h.
Subsequently, the membranes were probed with anti-NSE (cat. no.
8171S), anti-vascular endothelial growth factor (VEGF; cat. no.
9698S), anti-NM23 (cat. no. 3338S) and anti-E-cadherin (cat. no.
14472S) antibodies (1:1,000 dilution; Cell Signaling Technology,
Inc.) and anti-GAPDH antibody (1:1,000 dilution; cat. no. MB001;
Bioworld Technology, Inc.) at 4°C overnight, followed by incubation
with peroxidase-conjugated goat anti-mouse IgG (H+L; 1:2,000
dilution; cat. no. 33201ES60; Yeasen Biotechnology Co., Ltd.) as
the secondary antibody with ECL at 37°C for 2 h. Ultimately, the
intensity of the protein bands was visualized using an X-ray image
film processing machine (Kodak). The band intensity was quantified
by densitometric analysis using Image-Pro Plus version 4.5 software
(Media Cybernetics, Inc., Rockville, MD, USA).
MTT and clone formation assay
For the cell viability assays, the cells transfected
with NSE siRNA-1 or control siRNA for 24 h were seeded in 96-well
plates at 1.5×103 cells per well in a final volume of
150 µl. After 12 h of incubation, the cell viability was determined
using an MTT assay as described previously (21).
For the clone formation assay, the cells transfected
with siRNA or control were re-seeded in 6-well plates at 1,000
cells per well and cultured at 37°C for 2 weeks. The medium was
replaced every 3 days. At the end of the experiment, the cells were
washed twice with PBS, fixed with 4% paraformaldehyde at 37°C for
30 min and stained with 0.5% crystal violet. Following washing with
PBS, images of the plates were captured and the numbers of colonies
were counted under a light microscope (magnification, ×10; Olympus
Corporation).
Transwell assay
The Transwell assay was performed in modified Boyden
chambers (BD Biosciences) as described previously (22). In brief, 105 cells
suspended in serum-free DMEM were added to each of the upper
chambers present in the insert with 8-µm pore filters in a 24-well
culture plate. DMEM with FBS (10%) was added to the lower chamber.
After 8 h, the cells on the upper and lower sides were fixed with
4% paraformaldehyde at 37°C for 30 min and stained with 0.5%
crystal violet. The cells on the upper side, which had not
transgressed through the membrane, were gently removed with a
cotton swab and images of the cells located on the lower side of
the membrane were captured under a light microscope (magnification,
×100; Olympus Corporation), followed by counting of the migrated
cells.
Cell cycle analysis
Cell cycle analysis was performed as previously
described (20). In brief, the
NCI-H209 cells transfected with NSE siRNA-1 or control siRNA for 48
h were collected and fixed with 75% ice-cold ethanol, prior to
being stored at −20°C for 3 h. Subsequently, the cells were
centrifuged at 7,000 g at 37°C for 1 min, resuspended in 1 ml
lysis/propidium iodide (PI) buffer (0.1% Triton X-100, 0.05 mg/ml
PI and 1 mg/ml RNase A; Sigma-Aldrich; Merck KGaA), incubated at
37°C for 30 min and analyzed using a FACSCanto II flow cytometer
(BD Biosciences) with FACSDiva software (version 4.1; BD
Biosciences).
Statistical analysis
All statistical analyses were performed using SPSS
software vision 16.0 (SPSS, Inc.). A two-tailed Student's t-test
was performed to evaluate the differences between two groups.
Values are expressed as the mean ± standard deviation. P<0.05
was considered to indicate a statistically significant
difference.
Results
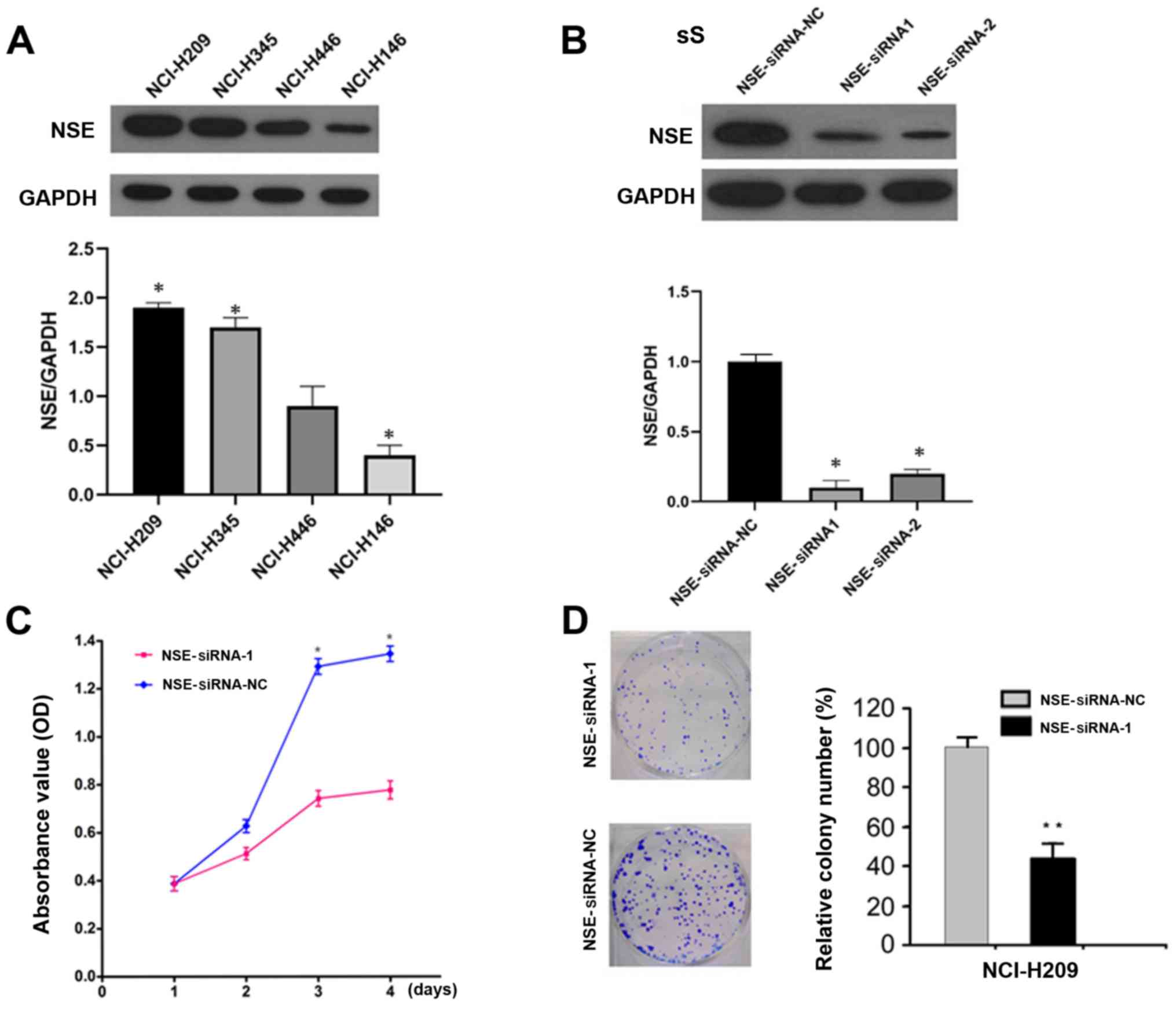
Silencing of NSE reduces the
proliferation and clone formation ability of SCLC cells
To determine the function of NSE in SCLC cells, the
protein levels of NSE were first examined in a panel of SCLC cell
lines, the NCI-H209 cell line exhibited the highest expression and
was therefore used for the subsequent knockdown experiments.
(Fig. 1A). The expression of NSE was
then knocked down in NCI-H209 cells using specific siRNAs against
NSE and the silencing efficiency was evaluated by western blot
analysis. The two siRNAs reduced the protein levels of NSE by
>80% (Fig. 1B). The knockdown of
NSE markedly decreased the proliferation of NCI-H209 cells, as
determined with an MTT assay (Fig.
1C). Furthermore, the silencing of NSE resulted in the
formation of smaller and a fewer colonies compared with
observations in the control group (Fig.
1D).
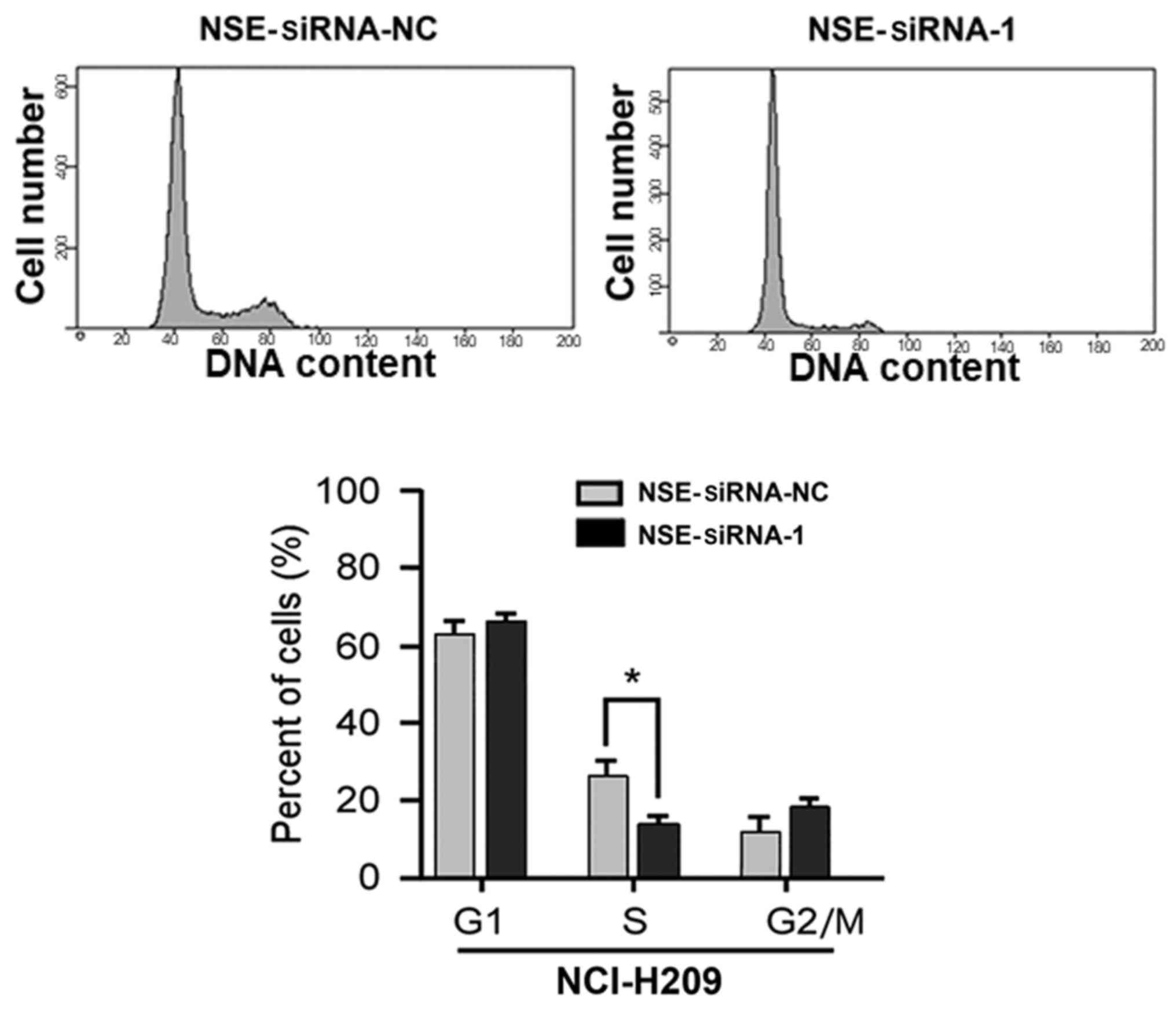
Silencing of NSE inhibits the cell
cycle of SCLC cells
Based on the observation that NSE affected the
proliferation of NCI-H209 cells, experiments were then performed to
examine whether the silencing of NSE affects the cell cycle of SCLC
cells. As presented in Fig. 2, the
proportion of cells in the S-phase was significantly decreased
following the knockdown of NSE (P<0.05). The proportions of
cells in the G1-phase and G2/M-phases were increased, although
these changes were not significant (P>0.05). Taken together,
these results indicate that the depletion of NSE suppressed the
proliferative and colony formation ability of the cells through
repressing cell cycle progression of the NCI-H209 cells.
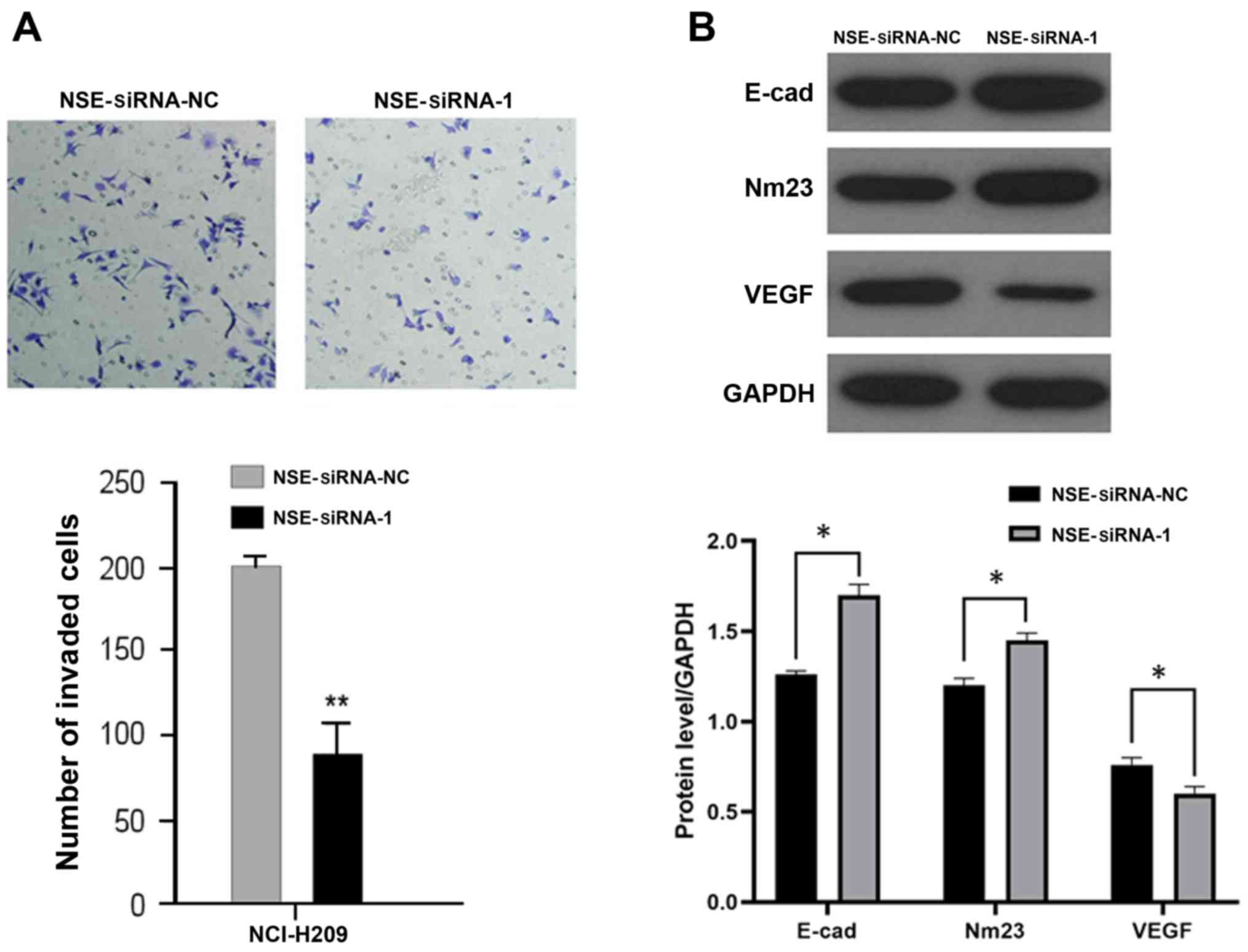
Knockdown of NSE represses the
migration of NCI-H209 cells
As elevated NSE is reported to be positively
associated with the metastatic status of patients with SCLC
(12), the present study
investigated whether NSE regulates the migration of NCI-H209 cells.
The NCI-H209 cells were transfected with NSE siRNA-1 or negative
control for 48 h and their migratory ability was examined using a
Transwell assay. As presented in Fig.
3A, the silencing of NSE markedly reduced the number of cells
that transgressed through the filter compared with that in the
negative control group. The expression levels of several
metastasis-associated proteins, including E-cadherin, NM23 and
VEGF, were then examined following the silencing of NSE in NCI-H209
cells. As presented in Fig. 3B, the
knockdown of NSE increased the protein levels of suppressors of
metastasis E-cadherin and NM23 but decreased the protein level of
pro-metastatic gene VEGF.
Taken together, these results indicated that the
knockdown of NSE suppressed the migratory ability of NCI-H209 cells
by regulating the expression of a panel of metastasis-associated
genes.
Discussion
NSE is generally regarded as a useful tumor marker
for patients with SCLC (23). In the
present study, loss-of-function experiments were performed to
examine the function of NSE in regulating the progression of SCLC
and the underlying mechanisms. The silencing of NSE suppressed the
proliferation, clone formation and migratory ability of the
NCI-H209 SCLC cells. Mechanistically, NSE silencing led to a
reduction in the S-phase population of NCI-H209 cells, which meant
that the cells in the G1-phase no longer progressed to the S-phase.
Furthermore, it was indicated that the depletion of NSE markedly
repressed the migration of SCLC cells by regulating the expression
of a panel of metastasis-associated genes. Collectively, the
results of the present study are the first, to the best of our
knowledge, to demonstrate the cancer-promoting function of NSE in
SCLC cells.
NSE, the r-subunit of the glycolytic enolase enzyme,
is specifically expressed in neuroendocrine cells and neurogenic
tumors (24). It is almost
undetectable in the serum of healthy individuals. In patients with
SCLC, necrotic SCLC cells excrete a large quantity of NSE, which
leads to elevated protein levels of NSE in the serum of patients
with SCLC. Therefore, NSE has been widely used as a serum biomarker
for SCLC. Elevated NSE has also been detected in SCLC tissues and
pleural fluid (25,26). In view of previous studies reporting
that NSE was elevated in patients with SCLC, the present study
aimed to determine the functional contributions of NSE in the
progression of SCLC. The downregulation of NSE by siRNAs reduced
the proliferation and colony formation of the NCI-H209 cells,
indicating that NSE may function as an oncogene by promoting the
proliferation and malignant transformation of SCLC cells.
Uncontrolled cell-cycle progression has been considered as one of
the important contributors in the carcinogenesis of cancer cells.
In the present study, the cell cycle profile of NCI-H209 cells
following the silencing of NSE was analyzed. The results indicated
that the knockdown of NSE led to downregulation of the S-phase
population of NCI-H209 cells. Taken together, the results of the
present study support the hypothesis that NSE promotes the cell
cycle progression of NCI-H209 cells and increases the proliferation
and tumorigenesis of these cells.
Metastasis is the major cause of mortality in
patients with SCLC. The correlation between serum levels of NSE and
the metastasis status of patients with SCLC have also been
investigated. Gronowitz et al (27) reported that NSE levels were markedly
higher in patients with metastasis compared with those with
early-stage disease. Therefore, the present study examined whether
NSE regulates the metastasis of SCLC cells.
Tumor angiogenesis and metastasis are the most
valuable prognostic factors for patients with SCLC, as they are
associated with treatment failure and poor prognosis (28). Angiogenesis serves a significant role
in tumor growth and metastasis (29); tumor blood vessels not only provide
nutrients to tumor tissues but also export a large number of tumor
cells to the tumor host, leading to tumor spread and metastasis.
VEGF is considered to be one of the most important regulators of
angiogenesis. It has been confirmed that VEGF is linked with a poor
prognosis in a variety of human malignancies (30). Ustuner et al (31) indicated that low serum VEGF is a
significant and independent prognostic factor in patients with
SCLC. E-cadherin is distributed in various types of epithelial cell
and regulates cell adhesion. The most important epithelial to
mesenchymal transition (EMT) tumor marker is the downregulation or
silencing of E-cadherin, which is considered the prerequisite for
the ability of epithelial cells to invade and metastasize (32). A low expression or deficiency of
E-cadherin may induce EMT, leading to the invasion and metastasis
of non-SCLC cells (33). The NM23
gene is a complementary DNA that was first isolated from a mouse
melanoma cell line and has a negative regulatory role in tumor
metastasis (34). The downregulation
of NM23 has been reported to be closely associated with metastasis
and poor prognosis in patients with various tumor types, including
lung cancer, melanoma and breast cancer (35–37).
To the best of our knowledge, there have been no
previous reports of the interactions between NSE and the
pro-metastatic gene VEGF, the metastasis suppressor gene NM23 and
E-cadherin. The western blotting results obtained in the present
study indicated that the silencing of NSE repressed the migration
of SCLC cells, accompanied by the upregulation of E-cadherin and
NM23 and downregulation of VEGF. These results suggest that NSE may
contribute to the formation of metastasis from SCLC, however, the
potential specific regulatory mechanisms require further
investigation.
The present study also attempted to analyze the
correlation between NSE levels and prognostic value in patients
with SCLC. However, due to the limited number of samples and the
fact that the majority of the patients with SCLC were at an
advanced stage, and thus not suitable for surgery, it was not
possible to perform the corresponding investigation. This is a
limitation of the present study and warrants investigation in the
future.
In conclusion, the results of the present study
revealed for the first time, to the best of our knowledge, the
functional contribution of NSE in the progression of SCLC. These
results indicate that NSE may serve as a therapeutic target in
addition to its use as a biomarker in SCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Guangdong Province (grant no.
2017A030310337), the Guangdong Special Fund Project of Fundamental
and Applied Research (grant no. 2018A030313160) and the Guangzhou
Planned Project of Science and Technology (grant no.
201804010221).
Availability of data and materials
All data generated and analyzed during this study
are included in this manuscript.
Authors' contributions
XL, SL, JF, JH, CW, LY and GL designed and performed
the study. XL, SL, XF, YW and MG performed the experiments and
collected and analyzed the data. JF, JH and CW analyzed the results
and wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wistuba II, Gazdar AF and Minna JD:
Molecular genetics of small cell lung carcinoma. Semin Oncol 28 (2
Suppl 4). S3–S13. 2001.
|
|
3
|
van Meerbeeck JP, Fennell DA and De
Ruysscher DK: Small- cell lung cancer. Lancet. 378:1741–1755. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pelayo Alvarez M, Westeel V, Cortés-Jofré
M and Bonfill Cosp X: Chemotherapy versus best supportive care for
extensive small cell lung cancer. Cochrane Database Syst Rev.
CD0019902013.PubMed/NCBI
|
|
5
|
Shirasawa M, Fukui T, Kusuhara S, Hiyoshi
Y, Ishihara M, Kasajima M, Nakahara Y, Otani S, Igawa S, Yokoba M,
et al: Prognostic significance of the 8th edition of the TNM
classification for patients with extensive disease small cell lung
cancer. Cancer Manag Res. 10:6039–6047. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu G, DU X, Zhou X, Bao W, Chen L, Chen
J, Ji Y and Wang S: Prophylactic cranial irradiation in 399
patients with limited-stage small cell lung cancer. Oncol Lett.
11:2654–2660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chan BA and Coward JI: Chemotherapy
advances in small-cell lung cancer. J Thorac Dis. 5 (Suppl
5):S565–S578. 2013.PubMed/NCBI
|
|
8
|
Oze I, Ito H, Nishino Y, Hattori M,
Nakayama T, Miyashiro I, Matsuo K and Ito Y: Trends in small-cell
lung cancer survival in 1993–2006 based on population-based cancer
registry data in Japan. J Epidemiol. Nov 17–2018.(Epub ahead of
print). doi: 10.2188/jea.JE20180112.
|
|
9
|
Harmsma M, Schutte B and Ramaekers FC:
Serum markers in small cell lung cancer: Opportunities for
improvement. Biochim Biophys Acta. 1836:255–272. 2013.PubMed/NCBI
|
|
10
|
Bremnes RM, Sundstrom S, Aasebø U, Kaasa
S, Hatlevoll R and Aamdal S; Norweigian Lung Cancer StudyGroup, :
The value of prognostic factors in small cell lung cancer: Results
from a randomised multicenter study with minimum 5 year follow-up.
Lung Cancer. 39:303–313. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ono A, Naito T, Ito I, Watanabe R, Shukuya
T, Kenmotsu H, Tsuya A, Nakamura Y, Murakami H, Kaira K, et al:
Correlations between serial pro-gastrin-releasing peptide and
neuron-specific enolase levels, and the radiological response to
treatment and survival of patients with small-cell lung cancer.
Lung Cancer. 76:439–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao WX and Luo JF: Serum neuron-specific
enolase levels were associated with the prognosis of small cell
lung cancer: A meta-analysis. Tumour Biol. 34:3245–3248. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sharma RA, Wotherspoon AC, Cook G, Morgan
GJ and Huddart RA: Neuron-specific enolase expression in multiple
myeloma. Lancet OncoL. 7:9602006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang L, Zhou JG, Yao WX, Tian X, Lv SP,
Zhang TY, Jin SH, Bai YJ and Ma H: Systematic review and
meta-analysis of the efficacy of serum neuron-specific enolase for
early small cell lung cancer screening. Oncotarget. 8:64358–64372.
2017.PubMed/NCBI
|
|
15
|
Molina R, Auge JM, Escudero JM, Marrades
R, Viñolas N, Carcereny E, Ramirez J and Filella X: Mucins CA 125,
CA 19.9, CA 15.3 and TAG-72.3 as tumor markers in patients with
lung cancer: Comparison with CYFRA 21-1, CEA, SCC and NSE. Tumour
Biol. 29:371–380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu S, Guo H, Kong L, Li H, Zhang Y, Zhu H
and Yu J: The prognostic factors in the elderly patients with small
cell lung cancer: A retrospective analysis from a single cancer
institute. Int J Clin Exp Pathol. 8:11033–11041. 2015.PubMed/NCBI
|
|
17
|
Buil-Bruna N, López-Picazo JM,
Moreno-Jiménez M, Martín-Algarra S, Ribba B and Trocóniz IF: A
population pharmacodynamic model for lactate dehydrogenase and
neuron specific enolase to predict tumor progression in small cell
lung cancer patients. AAPS J. 16:609–619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fizazi K, Cojean I, Pignon JP, Rixe O,
Gatineau M, Hadef S, Arriagada R, Baldeyrou P, Comoy E and Le
Chevalier T: Normal serum neuron specific enolase (NSE) value after
the first cycle of chemotherapy: An early predictor of complete
response and survival in patients with small cell lung carcinoma.
Cancer. 82:1049–1055. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Zhang W, Yin W, Xiao Y, Zhou C, Hu
Y and Geng S: The prognostic value of the serum neuron specific
enolase and lactate dehydrogenase in small cell lung cancer
patients receiving first-line platinum-based chemotherapy. Medicine
(Baltimore). 96:e82582017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu S, Cai X, Xia L, Jiang C, Chen P, Wang
X, Zhang B and Zhao HY: Chloroquine exerts antitumor effects on NB4
acute promyelocytic leukemia cells and functions synergistically
with arsenic trioxide. Oncol Lett. 15:2024–2030. 2018.PubMed/NCBI
|
|
21
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lv XB, Jiao Y, Qing Y, Hu H, Cui X, Lin T,
Song E and Yu F: miR-124 suppresses multiple steps of breast cancer
metastasis by targeting a cohort of pro-metastatic genes in vitro.
CHIN J CANCER. 30:821–830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan HJ, Tan Y and Gu W: Neuron specific
enolase and prognosis of non-small cell lung cancer: A systematic
review and meta-analysis. J BUON. 19:153–156. 2014.PubMed/NCBI
|
|
24
|
Proshchina AE, Krivova YS, Barabanov VM
and Saveliev SV: Ontogeny of neuro-insular complexes and islets
innervation in the human pancreas. Front Endocrinol (Lausanne).
5:572014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang D, Wang M, Sui A, Wang Y, Yang R,
Wang Z, Zhao Y, Jiao W and Shen Y: Prospective validation of
quantitative NSE mRNA in pleural fluid of non-small cell lung
cancer patients. Med Oncol. 30:6992013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jang SM, Kim JW, Kim CH, Kim D, Rhee S and
Choi KH: p19(ras) Represses proliferation of non-small cell lung
cancer possibly through interaction with Neuron-Specific Enolase
(NSE). Cancer Lett. 289:91–98. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gronowitz JS, Bergström R, Nôu E, Påhlman
S, Brodin O, Nilsson S and Källander CF: Clinical and serologic
markers of stage and prognosis in small cell lung cancer. A
multivariate analysis. Cancer. 66:722–732. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jett JR, Schild SE, Kesler KA and
Kalemkerian GP: Treatment of small cell lung cancer: Diagnosis and
management of lung cancer, 3rd ed: American College of Chest
Physicians evidence-based clinical practice guidelines. Chest. 143
(Suppl 5):e400S–e419S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lurje G, Zhang W, Schultheis AM, Yang D,
Groshen S, Hendifar AE, Husain H, Gordon MA, Nagashima F, Chang HM
and Lenz HJ: Polymorphisms in VEGF and IL-8 predict tumor
recurrence in stage III colon cancer. Ann Oncol. 19:1734–1741.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ustuner Z, Saip P, Yasasever V, Vural B,
Yazar A, Bal C, Ozturk B, Ozbek U and Topuz E: Prognostic and
predictive value of vascular endothelial growth factor and its
soluble receptors, VEGFR-1 and VEGFR-2 levels in the sera of small
cell lung cancer patients. Med Oncol. 25:394–399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Berx G and van Roy F: Involvement of
members of the cadherin superfamily in cancer. Cold Spring Harb
Perspect Biol. 1:a31292009. View Article : Google Scholar
|
|
33
|
Li LI, Lv Y, Zhang Y, He L and Zhang H:
Expression and clinical significance of Oct-4 and E-cad in
non-small-cell lung cancer. Oncol Lett. 11:234–236. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Carotenuto M, de Antonellis P, Chiarolla
CM, Attanasio C, Damiani V, Boffa I, Aiese N, Pedone E, Accordi B,
Basso G, et al: A therapeutic approach to treat prostate cancer by
targeting Nm23-H1/h-Prune interaction. Naunyn Schmiedebergs Arch
Pharmacol. 388:257–269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Esposito S, Russo MV, Airoldi I, Tupone
MG, Sorrentino C, Barbarito G, Di Meo S and Di Carlo E: SNAI2/Slug
gene is silenced in prostate cancer and regulates neuroendocrine
differentiation, metastasis-suppressor and pluripotency gene
expression. Oncotarget. 6:17121–17134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang CS, Shih MK, Chuang CH and Hu ML:
Lycopene inhibits cell migration and invasion and upregulates
Nm23-H1 in a highly invasive hepatocarcinoma, SK-Hep-1 cells. J
Nutr. 135:2119–2123. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim YI, Park S, Jeoung DI and Lee H: Point
mutations affecting the oligomeric structure of Nm23-H1 abrogates
its inhibitory activity on colonization and invasion of prostate
cancer cells. Biochem Biophys Res Commun. 307:281–289. 2003.
View Article : Google Scholar : PubMed/NCBI
|