Introduction
Gastric cancer (GC) is the fourth most commonly
diagnosed cancer and the second leading cause of cancer-associated
mortality worldwide (1). Despite its
early detection and current improvements in surgery with
perioperative chemotherapy, GC, with a median overall survival (OS)
of 12 months, remains difficult to treat and form a prognosis due
to the lack of specific symptoms during the early stages of disease
and metastasis (1,2). Recurrence is likely to occur during the
follow-up period as effective therapies are unavailable. At
present, only the pathological and clinical stages of disease are
accepted as conventional criteria to aid the treatment selection
process. To improve the OS of patients with this disease, the
identification of novel prognostic markers and potential
therapeutic targets is urgently required.
Apoptosis is a process of programmed cell death;
defects in apoptosis can lead to the development of cancer or
autoimmune disease. It has been reported that carcinogenesis could
be suppressed via apoptotic pathways through inducible proteins,
including inhibitors of apoptosis or FLICE-like inhibitory protein
(3). The cysteinyl aspartate
specific proteinase (caspase/CASP) family of proteins can also
induce apoptosis. CASPs are a group of proteases with similar
structures in the cytoplasm, comprising small and large catalytic
subunits plus a prodomain. Their active sites contain cysteine
residues, which specifically cleave the peptide bonds of aspartic
acid residues of target proteins (4). CASPs are involved in cell growth and
differentiation, as well as the regulation of eukaryotic apoptosis
(4,5). According to the homology of its
protease sequence, CASPs can be divided into three subfamilies:
CASP1 subfamily, including CASPs 1, 4, 5, 11, 12 and 14, which
serve as inflammatory proteases; the CASP2 subfamily, including
CASPs 2 and 9, which are upstream CASPs that process
proinflammatory cytokines or induce apoptosis, and the CASP3
subfamily, which includes the effector proteases, CASPs 3, 6, 7, 8
and 10 (6,7). The CASP family-mediated signaling
pathway involves a cascade of enzymatic reactions; each component
forms a complex network structure. It has been suggested that CASPs
may be key for the development of more effective anticancer
therapies (4). The vital role of
these proteins in medicine has been reported and their substrates
may be considered as anticancer drug targets as chemotherapeutic
agents induce the death of malignant cells by acting upstream of
CASPs (5); however, the prognostic
value of the CASP family of proteins in GC has not yet been
determined.
Based on highly specialized complexes in response to
various proinflammatory and proapoptotic signals, the CASP protein
family may be stimulated in the intrinsic and extrinsic pathway.
For the former pathway, with chemical stimuli and a lack of growth
factors or radiation, increases in the membrane permeability of
mitochondria and the transport of cytochrome c from its
interspace into the cytosol lead to the formation of the
apoptosome, which recruits apoptotic peptidase activating factor 1
(Apaf 1), proCASP-9 and ATP (4,8). In
addition, cellular damage signaling via p53 or other sensors to
antagonize the Bcl-2 family of proteins results in increased
permeabilization of the mitochondria (5). On the contrary, in the extrinsic
pathway, tumor necrosis factor (TNF)-α or Fas ligand engages their
own cell membrane receptor, the TNF receptor (TNFR) or Fas, causing
the oligomerization of the TNFR superfamily member 1A-associated
death domain (TRADD) or Fas-associated death domain (FADD),
respectively. Subsequently, a death-inducing signaling complex
forms, which associates with CASPs to induce certain effects
(4,9).
The present study performed a comprehensive analysis
using the Kaplan-Meier plotter (KM plotter) tool to determine
whether the expression of CASPs genes is associated with the
prognosis of GC and the various clinicopathological features of
patients with this disease. The results suggested that CASP mRNA
expression levels may be crucial prognostic biomarkers and that
anticancer drug targeting and the regulation of gene expression may
improve the therapeutic effects of treatment.
Materials and methods
Survival analysis
The association between the mRNA expression profile
of members of the CASP protein family and the OS of patients with
GC was analyzed with KM plotter (http://kmplot.com/analysis/); expression profiles
(dataset numbers: GSE14210, GSE15459, GSE22377, GSE29272 and
GSE62254) were downloaded from the Gene Expression Omnibus and The
Cancer Genome Atlas (10,11), with 876 GC patient samples divided
into high- and low-expression groups based on the median expression
levels of each gene. The Affymetrix GeneChip platform provided
probes for each gene (12). Only
samples measured on Human Genome U133 plus 2.0 arrays were included
in the analysis. Information of the databases were validated by
literature searches for various prognostic markers in GC in PubMed
for the prediction of OS using univariate and multivariate Cox
proportional hazards regression analyses (10).
Determination of CASP gene prognostic
values
The 12 selected CASP genes comprising the array
were: CASP1, CASP2, CASP3, CASP4, CASP5, CASP6, CASP7, CASP8,
CASP9, CASP10, CASP12 and CASP14, which serve vital roles in the
caspase pathway. The prognostic value of each caspase ligand was
evaluated via K-M survival plots, using GC tumor data, or by using
several clinical GC criteria and classifications, including the
Lauren classification (intestinal type, diffuse type and mixed
type) and clinicopathological features [mortality, human epidermal
growth factor receptor-2 (HER2) expression status, pathological
stages, sex, treatment strategy and differentiation degree]. The
treatment strategies were sorted into the surgery alone group,
fluorouracil (5-FU)-based adjuvant group and other adjuvant group;
for pathological stages, patients were classified into four stages.
The number-at-risk was also determined from analysis. Hazard ratios
(HRs) with 95% confidence interval (95% CI) and log-rank P-values
were calculated. P<0.05 was considered to indicate a
statistically significant difference.
Results
Association between OS of all patients
based on mRNA expression and Lauren classification
The data of the 12 CASP genes present in the KM
plotter database were employed for the analysis of OS. The OS for
876 patients with GC was investigated in association with 9 genes
(CASP1, CASP3, CASP4, CASP5, CASP6, CASP7, CASP8, CASP9 and
CASP10). Separate curves were plotted to determine the association
between OS and the Lauren classification criteria, including
intestinal type (n=320), diffuse type (n=241) and mixed type
(n=32). Investigations into CASP2, CASP12 and CASP14 were conducted
with 631 patient samples analyzed on Human Genome U133 plus 2.0
arrays which could only be included when using the selected probes,
containing 269, 240 and 29 samples of intestinal, diffuse and mixed
types respectively.
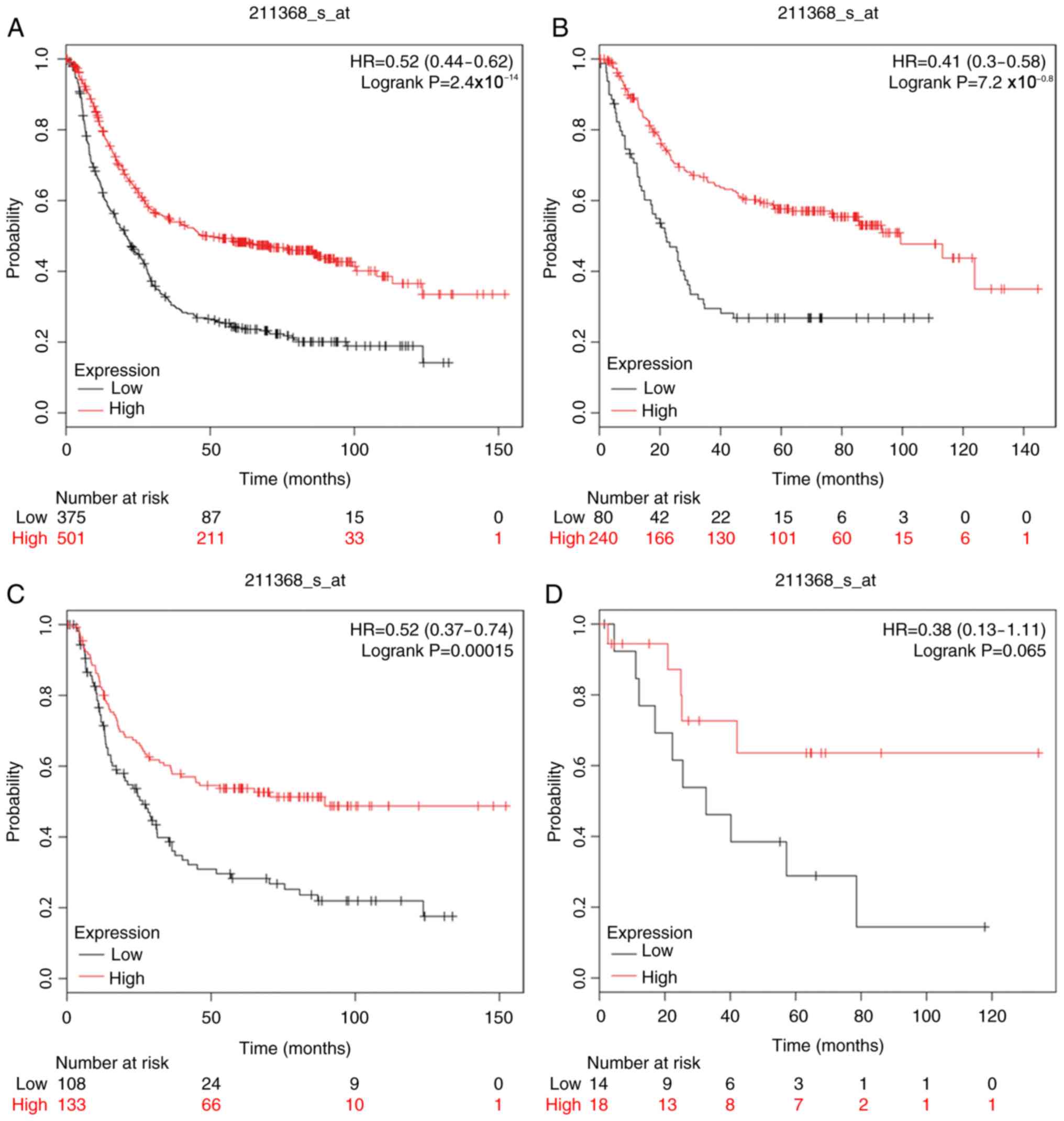
The prognostic value of CASP1 was initially assessed
in the database (Affymetrix ID: 211368_s_at). As presented in
Fig. 1, increased mRNA expression of
CASP1 was associated with better OS in all patients with GC
[HR=0.52 (0.44–0.62), P=2.4×10−14]; improved OS was
related to intestinal type [HR=0.41 (0.3–0.58),
P=7.2×10−8] and diffuse type [HR=0.52 (0.37–0.74),
P=1.5×10−4].
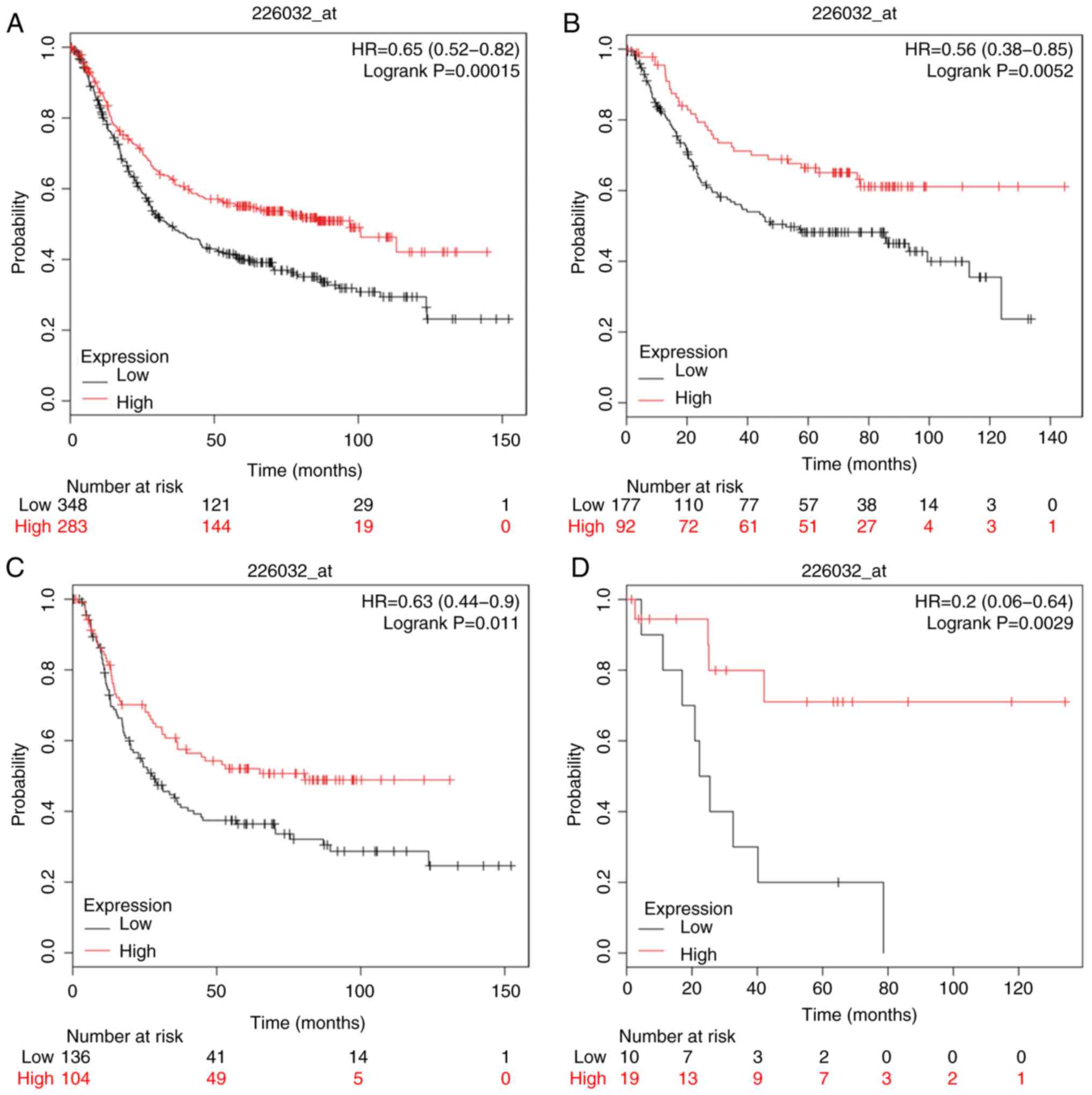
For CASP2 (Affymetrix ID: 226032_at), increased mRNA
expression was positively associated with OS in all patients with
GC [HR=0.65 (0.52–0.82), P=1.5×10−4; Fig. 2], corresponding to the outcome of the
intestinal, diffuse and mixed types in GC [HR=0.56 (0.38–0.85),
P=5.3×10−3; HR=0.63 (0.44–0.9), P=0.011; HR=0.2
(0.06–0.64), P=2.9×10−3; respectively].
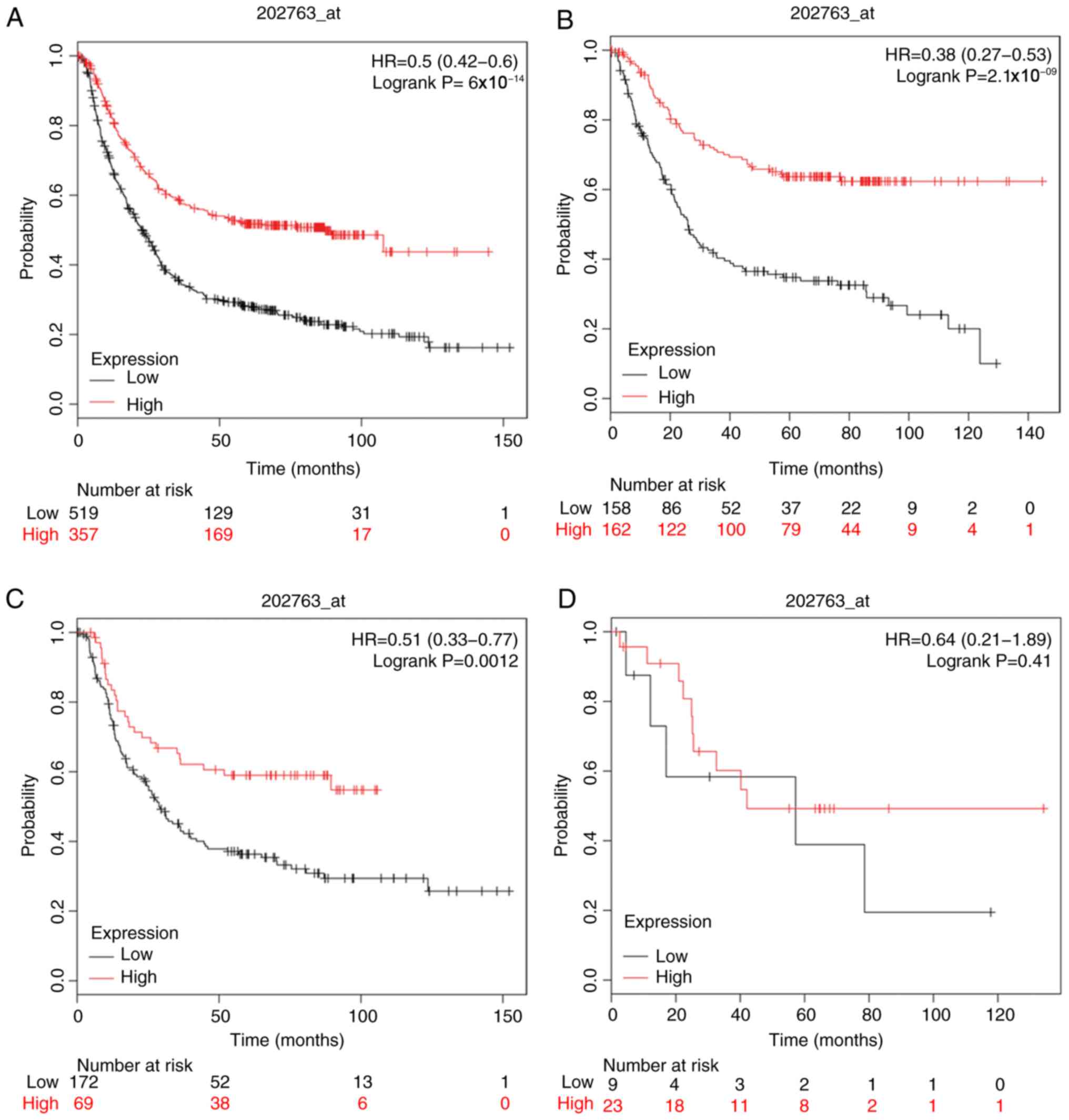
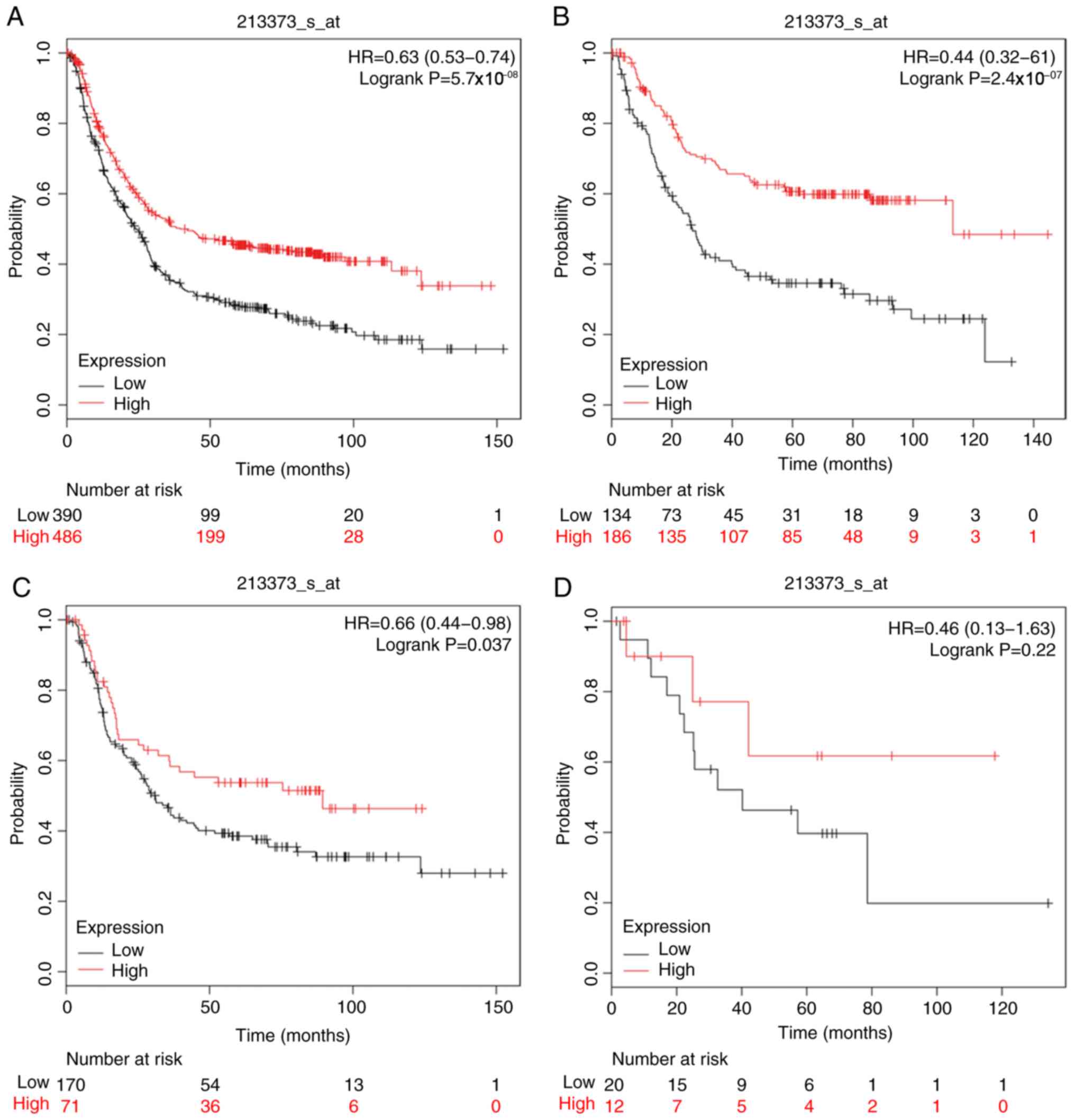
The prognostic significance of CASP3, CASP4, CASP5,
CASP6, CASP7 and CASP8 expression was investigated using the
corresponding Affymetrix databases: 202763_at, 209310_s_at,
207500_at, 209790_s_at, 207181_s_at and 213373_s_at. The mRNA
expression levels of the aforementioned CASPs were associated with
favorable OS in patients with GC [HR=0.5 (0.42–0.6),
P=6×10−14; HR=0.65 (0.53–0.8), P=5.9×10−5;
HR=0.66 (0.56–0.78), P=1.7×10−6; HR=0.68 (0.57–0.81),
P=1.2×10−5; HR=0.44 (0.36–0.54), P=2.2×10−16
and HR=0.63 (0.53–0.74), P=5.7×10−8; respectively], as
well as patients of intestinal and diffuse GC types, but not mixed
type GC (Figs. 3–8).
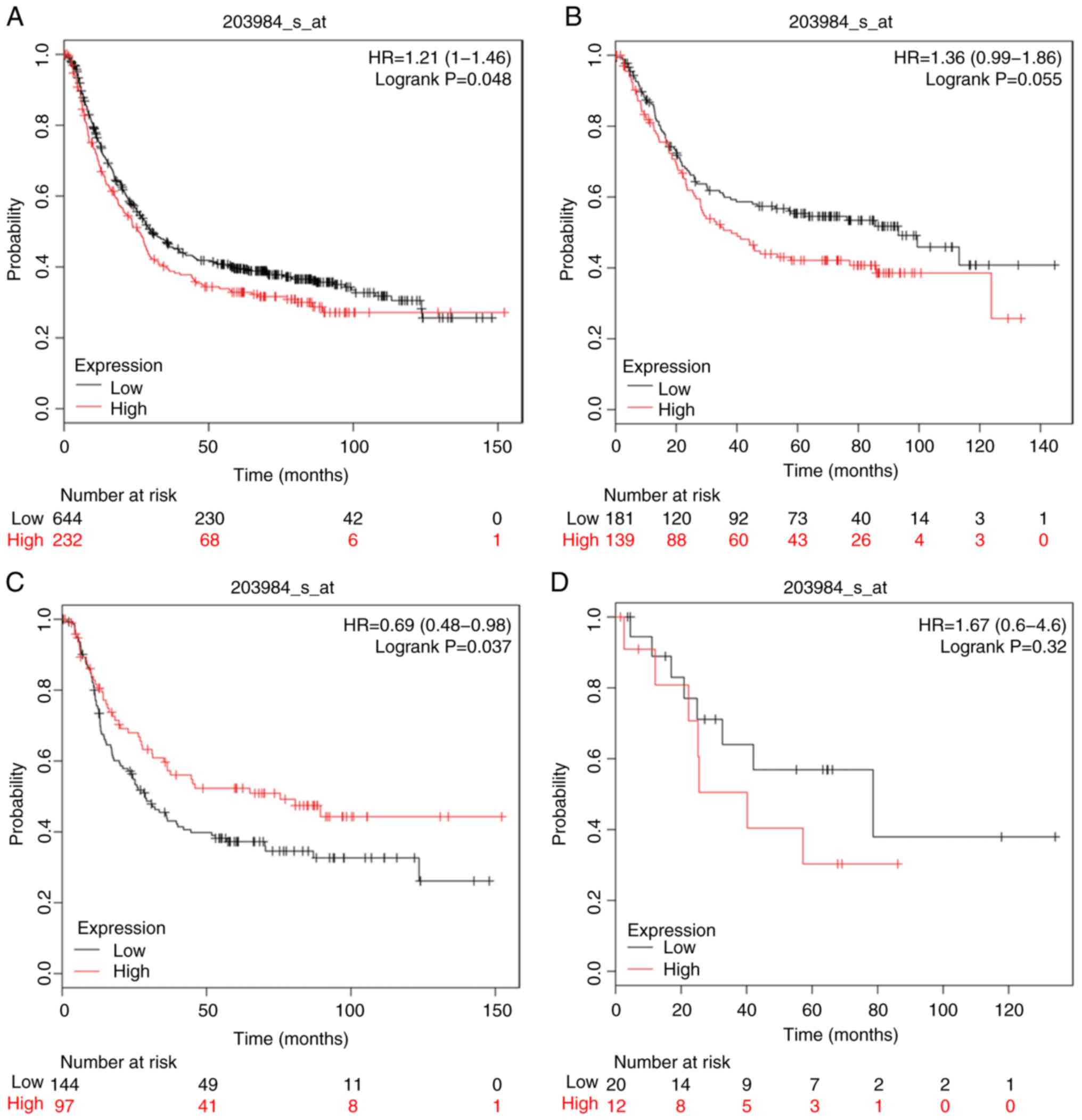
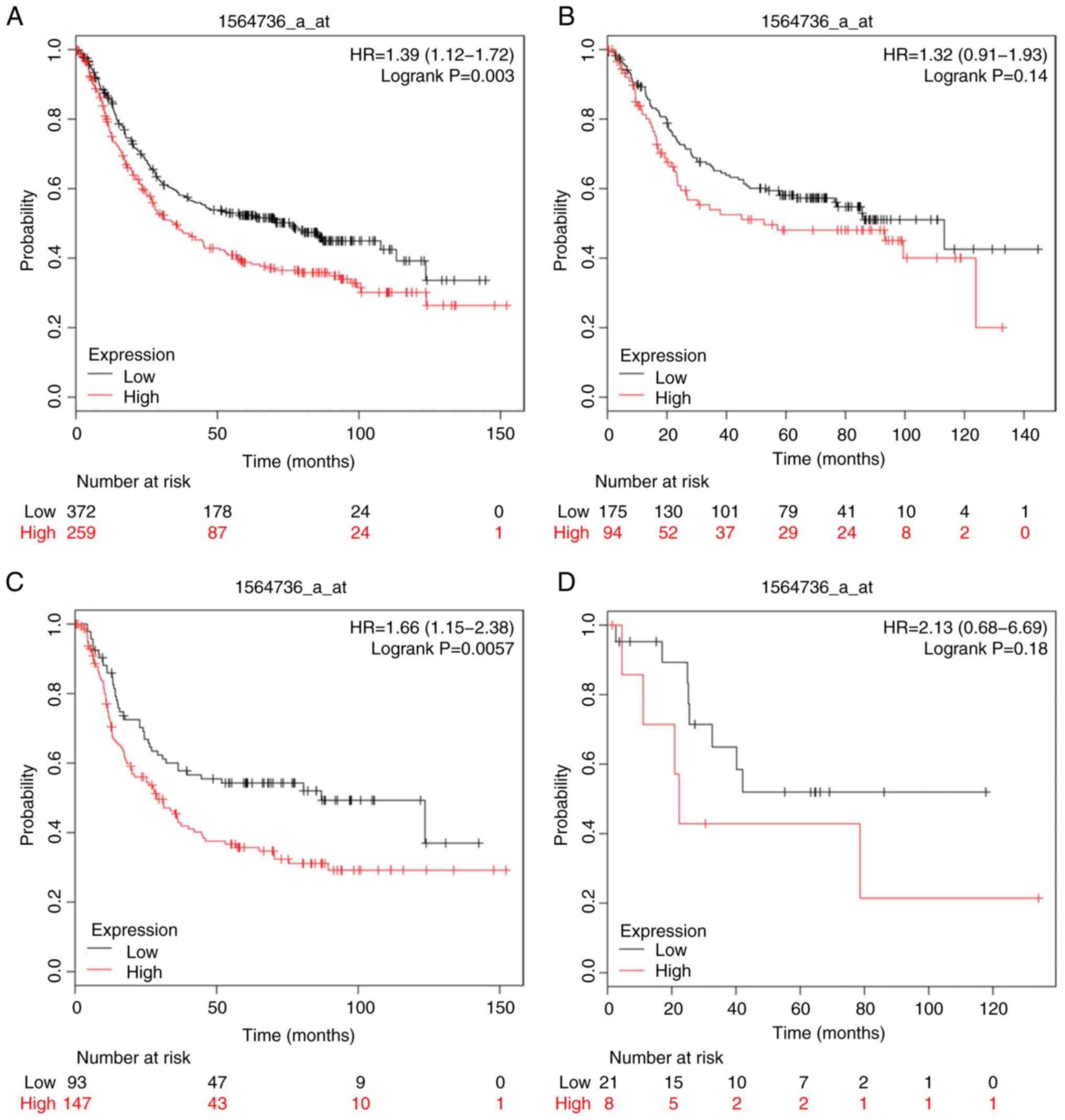
As presented in Figs.
9–11, increased expression of
CASP9 (Affymetrix ID: 203984_s_at) and CASP12 (Affymetrix ID:
1564736_a_at) was associated with poor OS in patients with GC
[HR=1.21 (1–1.46), P=4.8×10−2; HR=1.39 (1.12–1.72),
P=3×10−3]; CASP12 was linked to diffuse type GC [HR=1.66
(1.15–2.38), P=5.7×10−3]. Unexpectedly, diffuse type
patients with high CASP9 mRNA expression levels exhibited improved
OS [HR=0.69 (0.48–0.98), P=3.7×10−3]. Of note,
intestinal and mixed GC types were not associated with OS for
either gene.
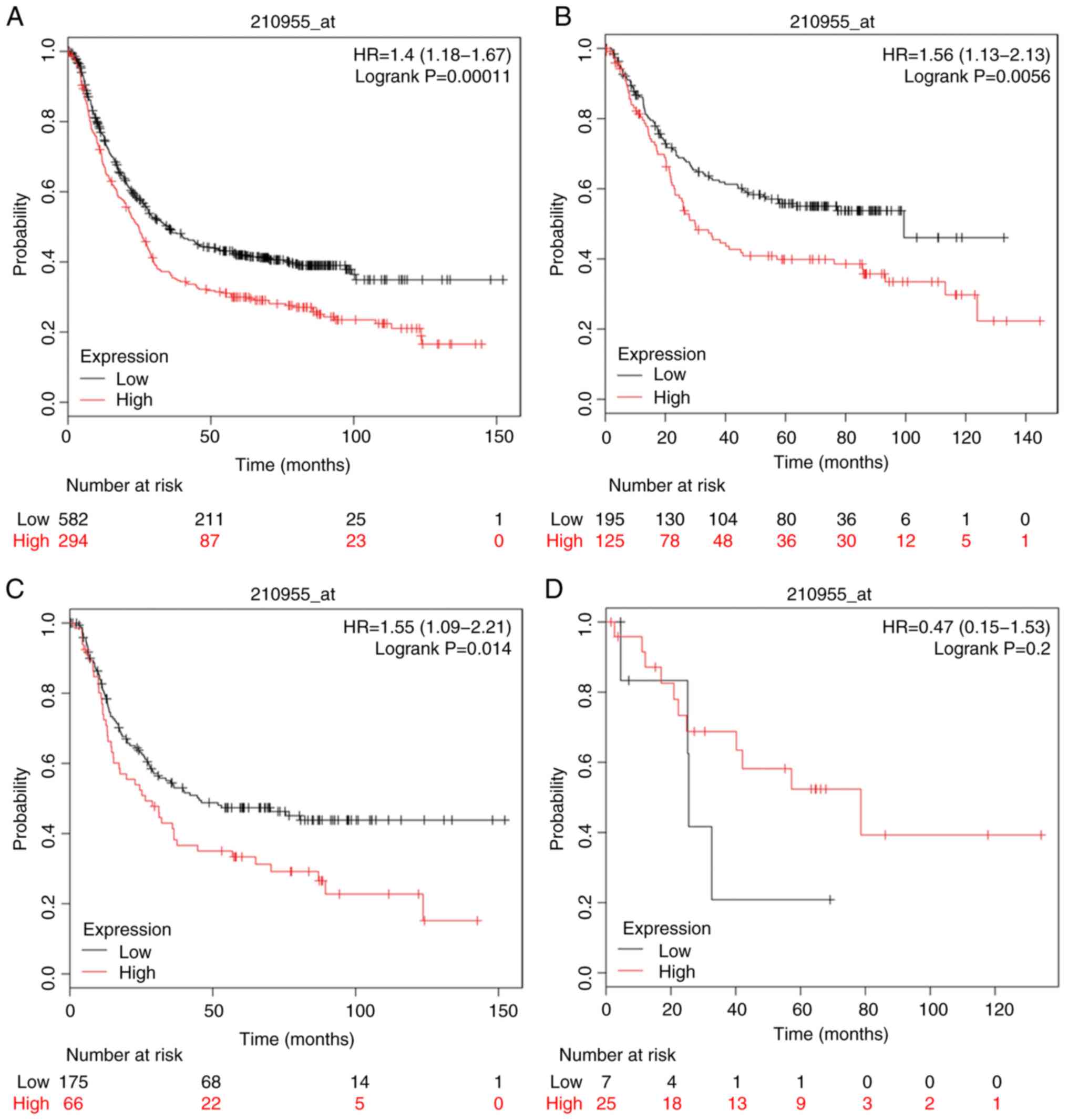
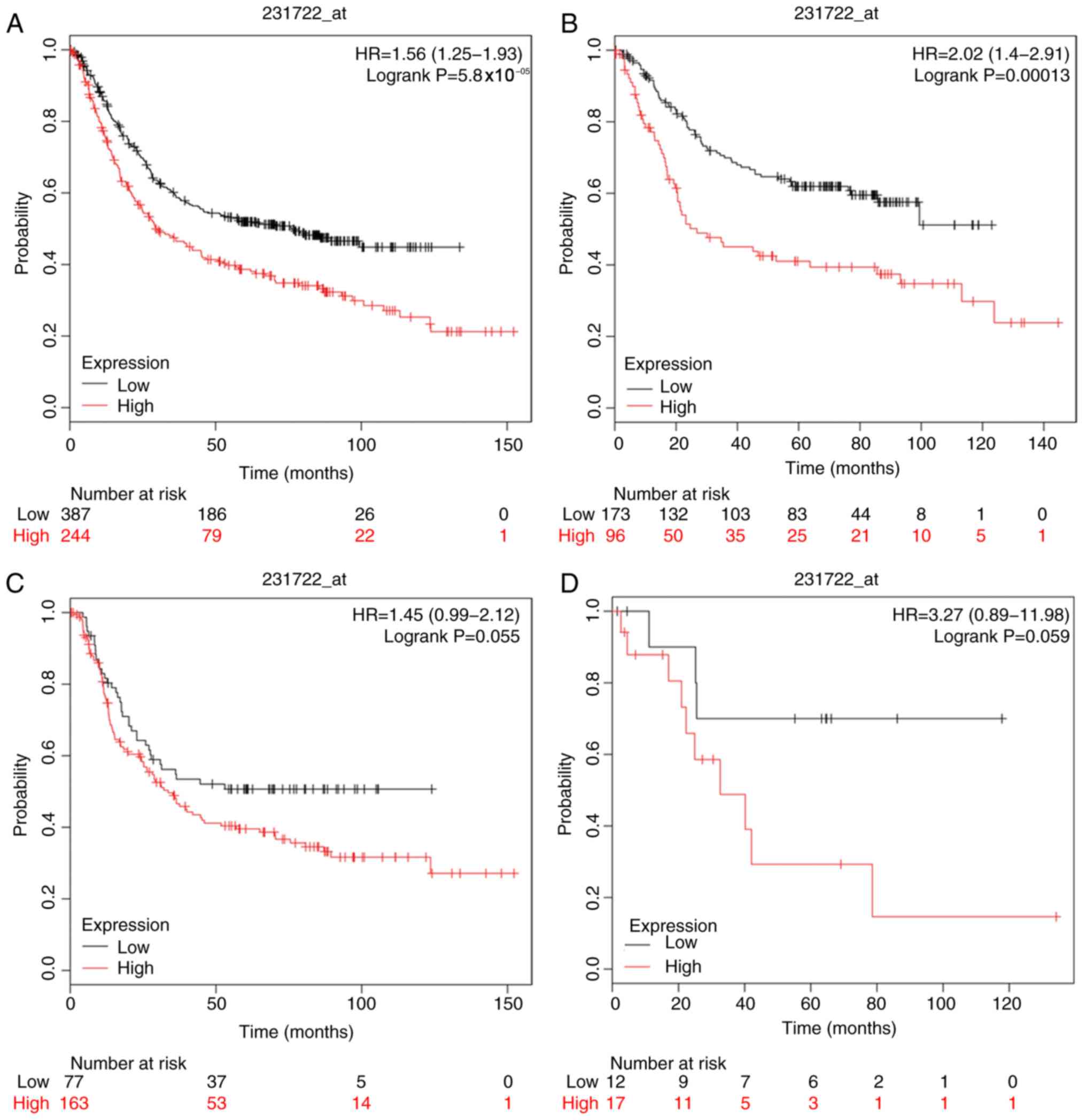
Furthermore, the prognostic value of CASP10 and
CASP14 was investigated using Affymetrix databases (210955_at and
231722_at, respectively). Upregulated CASP10 and CASP14 mRNA
expression was significantly associated with poor OS in patients
with GC [HR=1.4 (1.18–1.67), P=1.1×10−4; HR=1.56
(1.25–1.93), P=5.8×10−5]; intestinal type GC was also
related to poor OS [HR=1.56 (1.13–2.13), P=5.9×10−3;
HR=2.02 (1.4–2.91), P=1.3×10−4] (Figs. 10 and 12). In addition, analysis of mixed type GC
indicated that the expression of CASP10 and CASP14 above or below
the median level could not be applied to distinguish patients of
distinct prognostic groups.
Association between OS based on mRNA
expression and clinicopathological features
To further examine the association between the
expression of individual CASP genes and other clinicopathological
parameters, the links between OS and mortality (Table I), pathological grade (Table II), HER2 expression status (Table III), treatment strategy (Table IV), sex (Table V) and differentiation degree
(Table VI), were determined.
 | Table I.Association between CASP gene
expression levels and mortality at 50 months in patients with
gastric cancer. |
Table I.
Association between CASP gene
expression levels and mortality at 50 months in patients with
gastric cancer.
| CASP | Expression | N | Surviving
patients | Mortality rate | P-value |
|---|
| CASP1 | Low | 375 | 87 | 0.768000 |
<0.001a |
|
| High | 501 | 211 | 0.578842 |
|
| CASP2 | Low | 348 | 121 | 0.652299 |
<0.001a |
|
| High | 283 | 144 | 0.491166 |
|
| CASP3 | Low | 519 | 129 | 0.751445 |
<0.001a |
|
| High | 357 | 169 | 0.526611 |
|
| CASP4 | Low | 655 | 198 | 0.697710 |
<0.001a |
|
| High | 221 | 100 | 0.547511 |
|
| CASP5 | Low | 447 | 121 | 0.729306 |
<0.001a |
|
| High | 429 | 177 | 0.587413 |
|
| CASP6 | Low | 483 | 139 | 0.712215 |
<0.001a |
|
| High | 393 | 159 | 0.595420 |
|
| CASP7 | Low | 591 | 150 | 0.746193 |
<0.001a |
|
| High | 285 | 148 | 0.480702 |
|
| CASP8 | Low | 390 | 99 | 0.746154 |
<0.001a |
|
| High | 486 | 199 | 0.590535 |
|
| CASP9 | Low | 644 | 230 | 0.642857 |
<0.001a |
|
| High | 232 | 68 | 0.706897 |
|
| CASP10 | Low | 582 | 211 | 0.637457 |
<0.001a |
|
| High | 294 | 87 | 0.704082 |
|
| CASP12 | Low | 372 | 178 | 0.521505 |
<0.001a |
|
| High | 259 | 87 | 0.664093 |
|
| CASP14 | Low | 387 | 186 | 0.519380 |
<0.001a |
|
| High | 244 | 79 | 0.676230 |
|
 | Table II.Association between CASP gene
expression levels and OS in patients with gastric cancer based on
pathological stages. |
Table II.
Association between CASP gene
expression levels and OS in patients with gastric cancer based on
pathological stages.
| CASPs | Pathological
grade | N | HR (95% CI) | P-value |
|---|
| CASP1 | I | 67 | 0.28
(0.1–0.75) | 0.007a |
|
| II | 140 | 0.48
(0.24–0.96) | 0.034a |
|
| III | 305 | 0.43
(0.32–0.57) |
<0.001a |
|
| IV | 148 | 0.6
(0.41–0.88) | 0.009a |
| CASP2 | I | 62 | 0.62
(0.21–1.86) | 0.390 |
|
| II | 135 | 0.44
(0.23–0.84) | 0.010a |
|
| III | 197 | 0.63
(0.43–0.93) | 0.018a |
|
| IV | 140 | 0.51
(0.32–0.81) | 0.004a |
| CASP3 | I | 67 | 0.32
(0.11–0.88) | 0.020a |
|
| II | 140 | 0.32
(0.14–0.71) | 0.003a |
|
| III | 305 | 0.42
(0.29–0.62) |
<0.001a |
|
| IV | 148 | 0.55
(0.34–0.87) | 0.010a |
| CASP4 | I | 67 | 0.34
(0.12–0.95) | 0.032a |
|
| II | 140 | 0.58
(0.32–1.06) | 0.071 |
|
| III | 305 | 0.49
(0.35–0.68) |
<0.001a |
|
| IV | 148 | 0.83
(0.57–1.23) | 0.360 |
| CASP5 | I | 67 | 2.33
(0.84–6.45) | 0.096 |
|
| II | 140 | 0.44
(0.24–0.81) | 0.007a |
|
| III | 305 | 0.63
(0.45–0.88) | 0.007a |
|
| IV | 148 | 0.51
(0.34–0.77) | 0.001a |
| CASP6 | I | 67 | 0.55
(0.19–1.6) | 0.270 |
|
| II | 140 | 0.68
(0.36–1.31) | 0.250 |
|
| III | 305 | 0.59
(0.42–0.83) | 0.002a |
|
| IV | 148 | 0.79
(0.5–1.23) | 0.290 |
| CASP7 | I | 67 | 0.27
(0.09–0.84) | 0.016a |
|
| II | 140 | 0.42
(0.23–0.78) | 0.005a |
|
| III | 305 | 0.36
(0.24–0.54) |
<0.001a |
|
| IV | 148 | 0.47
(0.31–0.72) |
<0.001a |
| CASP8 | I | 67 | 0.27
(0.08–0.94) | 0.027a |
|
| II | 140 | 1.57
(0.85–2.89) | 0.150 |
|
| III | 305 | 0.47
(0.35–0.63) |
<0.001a |
|
| IV | 148 | 0.67
(0.43–1.04) | 0.072 |
| CASP9 | I | 67 | 3.73
(1.31–10.62) | 0.009a |
|
| II | 140 | 1.54
(0.85–2.79) | 0.160 |
|
| III | 305 | 1.48
(1.09–2.01) | 0.011a |
|
| IV | 148 | 0.78
(0.49–1.23) | 0.280 |
| CASP10 | I | 67 | 2.35
(0.8–6.85) | 0.110 |
|
| II | 140 | 0.54 (0.29–1) | 0.046a |
|
| III | 305 | 1.59
(1.19–2.12) | 0.002a |
|
| IV | 148 | 1.51
(1.01–2.25) | 0.042a |
| CASP12 | I | 62 | 0.51
(0.17–1.59) | 0.240 |
|
| II | 135 | 0.48
(0.25–0.91) | 0.022a |
|
| III | 197 | 1.38
(0.94–2.02) | 0.094 |
|
| IV | 140 | 0.81
(0.54–1.21) | 0.300 |
| CASP14 | I | 62 | 2.36
(0.78–7.15) | 0.120 |
|
| II | 135 | 0.56
(0.27–1.16) | 0.110 |
|
| III | 197 | 2.21
(1.45–3.37) |
<0.001a |
|
| IV | 140 | 1.85
(1.22–2.8) | 0.003a |
 | Table III.Association between CASP gene
expression and OS in patients with gastric cancer based on HER2
expression status. |
Table III.
Association between CASP gene
expression and OS in patients with gastric cancer based on HER2
expression status.
| CASP | HER2 status | N | Low | High | HR (95% CI) | P-value |
|---|
| CASP1 | Negative | 532 | 139 | 188 | 0.47
(0.38–0.6) |
1.0×10−10a |
|
| Positive | 344 | 147 | 197 | 0.62
(0.48–0.81) |
2.8×10−4a |
| CASP2 | Negative | 429 | 245 | 184 | 0.58
(0.44–0.76) |
8.0×10−5a |
|
| Positive | 202 | 150 | 52 | 0.67
(0.43–1.04) |
7.2×10−2 |
| CASP3 | Negative | 532 | 283 | 249 | 0.43
(0.34–0.55) |
1.8×10−12a |
|
| Positive | 344 | 246 | 98 | 0.67
(0.49–0.91) |
9.0×10−3a |
| CASP4 | Negative | 532 | 398 | 134 | 0.56
(0.42–0.74) |
5.3×10−5a |
|
| Positive | 344 | 150 | 194 | 0.87
(0.67–1.13) |
3.0×10−1 |
| CASP5 | Negative | 532 | 236 | 296 | 0.63
(0.51–0.79) |
5.9×10−5a |
|
| Positive | 344 | 238 | 106 | 0.6 (0.44–0.8) |
5.7×10−4a |
| CASP6 | Negative | 532 | 192 | 340 | 0.68
(0.55–0.86) |
1.1×10−3a |
|
| Positive | 344 | 179 | 165 | 0.61
(0.47–0.79) |
2.2×10−4a |
| CASP7 | Negative | 532 | 351 | 181 | 0.39
(0.3–0.51) |
8.2×10−13a |
|
| Positive | 344 | 256 | 88 | 0.55
(0.4–0.77) |
3.2×10−4a |
| CASP8 | Negative | 532 | 239 | 293 | 0.54
(0.43–0.67) |
3.5×10−8a |
|
| Positive | 344 | 256 | 88 | 0.69
(0.51–0.95) |
2.3×10−2a |
| CASP9 | Negative | 532 | 325 | 207 | 1.22
(0.97–1.52) |
9.2×10−2 |
|
| Positive | 344 | 92 | 252 | 0.72
(0.55–0.96) |
2.5×10−2a |
| CASP10 | Negative | 532 | 375 | 157 | 1.53
(1.22–1.93) |
2.7×10−4a |
|
| Positive | 344 | 183 | 161 | 1.21
(0.93–1.57) |
1.5×10−1 |
| CASP12 | Negative | 429 | 192 | 237 | 1.56
(1.18–2.04) |
1.3×10−3a |
|
| Positive | 202 | 136 | 66 | 1.42
(0.96–2.1) |
7.7×10−2 |
| CASP14 | Negative | 429 | 147 | 282 | 1.59
(1.18–2.14) |
2.3×10−3a |
|
| Positive | 202 | 96 | 106 | 2.09
(1.43–3.07) |
1.1×10−4a |
 | Table IV.Association between CASP gene
expression and OS in patients with gastric cancer based on
treatment strategy. |
Table IV.
Association between CASP gene
expression and OS in patients with gastric cancer based on
treatment strategy.
| CASP | Treatment | Cases | HR (95% CI) | P-value |
|---|
| CASP1 | Surgery alone | 380 | 0.56
(0.4–0.78) |
6.5×10−4a |
|
| 5-FU-based
adjuvant | 153 | 0.51
(0.36–0.72) |
1.4×10−4a |
|
| Other adjuvant | 76 | 0.41 (0.17–1) |
4.2×10−2a |
| CASP2 | Surgery alone | 380 | 0.68
(0.51–0.91) |
9.3×10−3a |
|
| 5-FU-based
adjuvant | 34 | 3.42
(1.2–9.77) |
1.5×10−2a |
|
| Other adjuvant | 76 | 2.59
(1.07–6.25) |
2.8×10−2a |
| CASP3 | Surgery alone | 380 | 0.66
(0.48–0.91) |
1.1×10−2a |
|
| 5-FU-based
adjuvant | 153 | 1.25
(0.88–1.77) |
2.0×10−1 |
|
| Other adjuvant | 76 | 0.35
(0.12–1.05) |
5.0×10−2 |
| CASP4 | Surgery alone | 380 | 0.65
(0.46–0.92) |
1.4×10−2a |
|
| 5-FU-based
adjuvant | 153 | 1.51
(1.04–2.2) |
3.1×10−2a |
|
| Other adjuvant | 76 | 0.52
(0.2–1.36) |
1.7×10−1 |
| CASP5 | Surgery alone | 380 | 0.6 (0.45–0.8) |
4.7×10−4a |
|
| 5-FU-based
adjuvant | 153 | 1.67
(1.16–2.4) |
5.2×10−3a |
|
| Other adjuvant | 76 | 0.39
(0.16–0.95) |
3.1×10−2a |
| CASP6 | Surgery alone | 380 | 0.75
(0.55–1.02) |
6.5×10−2 |
|
| 5-FU-based
adjuvant | 153 | 1.25
(0.87–1.78) |
2.3×10−1 |
|
| Other adjuvant | 76 | 0.34
(0.14–0.84) |
1.4×10−2a |
| CASP7 | Surgery alone | 380 | 0.42
(0.3–0.58) |
5.2×10−8a |
|
| 5-FU-based
adjuvant | 153 | 1.28
(0.86–1.92) |
2.3×10−1 |
|
| Other adjuvant | 76 | 0.26
(0.1–0.67) |
2.9×10−3a |
| CASP8 | Surgery alone | 380 | 0.81
(0.6–1.09) |
1.6×10−1 |
|
| 5-FU-based
adjuvant | 153 | 1.27 (0.9–1.8) |
1.8×10−1 |
|
| Other adjuvant | 76 | 0.31
(0.13–0.75) |
5.8×10−3a |
| CASP9 | Surgery alone | 380 | 0.81
(0.58–1.14) |
2.2×10−1 |
|
| 5-FU-based
adjuvant | 153 | 1.52
(1.07–2.16) |
1.9×10−2a |
|
| Other adjuvant | 76 | 4.03
(1.66–9.78) |
8.6×10−4a |
| CASP10 | Surgery alone | 380 | 1.27
(0.94–1.71) |
1.3×10−1 |
|
| 5-FU-based
adjuvant | 153 | 1.34
(0.94–1.91) |
1.1×10−1 |
|
| Other adjuvant | 76 | 0.58
(0.24–1.41) |
2.2×10−1 |
| CASP12 | Surgery alone | 380 | 1.55
(1.15–2.08) |
3.7×10−3a |
|
| 5-FU-based
adjuvant | 34 | 1.71
(0.56–5.2) |
3.4×10−1 |
|
| Other adjuvant | 76 | 3.52
(1.17–10.55) |
1.7×10−2a |
| CASP14 | Surgery alone | 380 | 1.34
(0.97–1.84) |
7.5×10−2 |
|
| 5-FU-based
adjuvant | 34 | 2.25
(0.65–7.78) |
1.9×10−1 |
|
| Other adjuvant | 76 | 0.46
(0.19–1.12) |
7.8×10−2 |
 | Table V.Association between CASP gene
expression and OS in patients with gastric cancer patients based on
sex. |
Table V.
Association between CASP gene
expression and OS in patients with gastric cancer patients based on
sex.
| CASPs | Sex | N | HR (95% CI) | P-value |
|---|
| CASP1 | Male | 545 | 0.54
(0.44–0.67) |
1.6×10−8a |
|
| Female | 236 | 0.39
(0.27–0.56) |
1.3×10−7a |
| CASP2 | Male | 349 | 0.62
(0.46–0.83) |
1.5×10−3a |
|
| Female | 187 | 0.58
(0.38–0.89) |
1.1×10−2a |
| CASP3 | Male | 545 | 0.47
(0.37–0.6) |
3.4×10−10a |
|
| Female | 236 | 0.45
(0.3–0.66) |
3.1×10−5a |
| CASP4 | Male | 545 | 0.6
(0.46–0.78) |
1.6×10−4a |
|
| Female | 236 | 0.66
(0.46–0.95) |
2.5×10−3a |
| CASP5 | Male | 545 | 0.69
(0.56–0.85) |
5.8×10−4a |
|
| Female | 236 | 0.53
(0.37–0.77) |
7.7×10−4a |
| CASP6 | Male | 545 | 0.67
(0.54–0.84) |
2.9×10−4a |
|
| Female | 236 | 0.55
(0.35–0.84) |
5.2×10−3a |
| CASP7 | Male | 545 | 0.4 (0.3–0.53) |
1.6×10−11a |
|
| Female | 236 | 0.32
(0.21–0.49) |
2.5×10−8a |
| CASP8 | Male | 545 | 0.57
(0.46–0.71) |
1.7×10−7a |
|
| Female | 236 | 0.59
(0.38–0.91) |
1.6×10−2a |
| CASP9 | Male | 545 | 1.25
(0.98–1.58) |
7.0×10−2 |
|
| Female | 236 | 1.29
(0.88–1.88) |
1.9×10−1 |
| CASP10 | Male | 545 | 1.5
(1.21–1.86) |
1.9×10−4a |
|
| Female | 236 | 1.46
(1.03–2.07) |
3.3×10−2a |
| CASP12 | Male | 349 | 1.72
(1.28–2.33) |
3.3×10−4a |
|
| Female | 187 | 1.43
(0.93–2.19) |
1.0×10−1 |
| CASP14 | Male | 349 | 1.71
(1.24–2.36) |
9.4×10−4a |
|
| Female | 187 | 1.73
(1.06–2.81) |
2.6×10−2a |
 | Table VI.Association of CASP gene expression
with OS in patients with gastric cancer based on differentiation
degree. |
Table VI.
Association of CASP gene expression
with OS in patients with gastric cancer based on differentiation
degree.
| CASP | Differentiation
degree | N | HR (95% CI) | P-value |
|---|
| CASP1 | Poor | 165 | 0.48
(0.31–0.74) |
<0.001a |
|
| Moderate | 67 | 0.64
(0.33–1.23) | 0.180 |
|
| Good | 32 | 0.57
(0.19–1.7) | 0.310 |
| CASP2 | Poor | 121 | 0.72
(0.44–1.18) | 0.190 |
|
| Moderate | 67 | 1.91
(0.9–4.07) | 0.086 |
|
| Good | – | – | – |
| CASP3 | Poor | 165 | 0.85
(0.57–1.27) | 0.430 |
|
| Moderate | 67 | 0.58
(0.28–1.19) | 0.130 |
|
| Good | 32 | 0.21
(0.08–0.54) |
<0.001a |
| CASP4 | Poor | 165 | 0.79
(0.5–1.22) | 0.290 |
|
| Moderate | 67 | 2.08
(1.05–4.1) | 0.031a |
|
| Good | 32 | 0.55
(0.22–1.33) | 0.180 |
| CASP5 | Poor | 165 | 0.65
(0.41–1.04) | 0.069 |
|
| Moderate | 67 | 2.01
(1.02–3.95) | 0.040a |
|
| Good | 32 | 0.47
(0.17–1.3) | 0.140 |
| CASP6 | Poor | 165 | 1.4 (0.93–2.1) | 0.110 |
|
| Moderate | 67 | 0.54
(0.28–1.05) | 0.064 |
|
| Good | 32 | 0.48
(0.16–1.42) | 0.170 |
| CASP7 | Poor | 165 | 0.81
(0.5–1.33) | 0.410 |
|
| Moderate | 67 | 0.46
(0.19–1.11) | 0.079 |
|
| Good | 32 | 0.49
(0.21–1.16) | 0.097 |
| CASP8 | Poor | 165 | 0.57
(0.37–0.87) | 0.008a |
|
| Moderate | 67 | 2.19
(1.05–4.57) | 0.032a |
|
| Good | 32 | 2.86
(0.95–8.59) | 0.051 |
| CASP9 | Poor | 165 | 0.83
(0.55–1.23) | 0.350 |
|
| Moderate | 67 | 1.65
(0.82–3.31) | 0.160 |
|
| Good | 32 | 1.67
(0.65–4.32) | 0.280 |
| CASP10 | Poor | 165 | 1.5 (1–2.24) | 0.046a |
|
| Moderate | 67 | 1.7
(0.71–4.08) | 0.230 |
|
| Good | 32 | 0.38 (0.15–1) | 0.042a |
| CASP12 | Poor | 121 | 0.69
(0.41–1.16) | 0.160 |
|
| Moderate | 67 | 0.58
(0.27–1.23) | 0.150 |
|
| Good | – | – | – |
| CASP14 | Poor | 121 | 0.65
(0.36–1.17) | 0.150 |
|
| Moderate | 67 | 1.61
(0.84–3.08) | 0.150 |
|
| Good | – | – | – |
The association between the expression levels of
CASPs and the 50-month mortality rate was investigated in patients
with GC. In Table I, high expression
levels of CASP7 revealed the lowest mortality rate of 0.4807.
Conversely, low expression levels of CASP1 were associated with the
highest mortality rate of 0.7680.
As presented in Table
II, increased expression of CASP1, CASP3 and CASP7 was linked
to OS in patients with GC of pathological grade I. CASP2 and CASP5
were associated with stage II–IV GC, while CASP4 and CASP8 were
associated with stage I and III GC, and CASP6 was linked to stage
III GC. In addition, CASP10 and CASP14 were revealed to be
associated with unfavorable OS in stages III and IV of GC; CASP9
was linked to poor OS in stage I and III GC. On the contrary,
CASP10 overexpression corresponded to improved OS in patients with
stage II GC.
In the present study, the association between CASP
expression and HER2 status was investigated in GC patients
(Table III). Positive and negative
HER2 expression were linked to favorable OS and CASP1, CASP3,
CASP5, CASP5, CASP6 and CASP8 mRNA expression. Conversely, CASP2
and CASP4 was associated with negative HER2 status in patients with
GC. Analysis of CASP14 with positive or negative HER2 expression
was associated with unfavorable OS; CASP10 and CASP12 indicated
poorer OS with negative HER2 expression.
As for the treatment strategy, CASP1 was
significantly associated with better OS, in all three groups
(surgery alone, 5-FU-based adjuvant and other adjuvant; P<0.001;
Table IV). The 5-FU-based adjuvant
group exhibited an increased HR with CASP2, CASP4, CASP5 and CASP9
mRNA expression. Overexpression of other CASPs was associated with
improved OS with each treatment (Table
IV).
As presented in Table
V, sex, expression status and the expression of CASP1-8, were
associated with better OS in male and female patients with GC.
Nevertheless, CASP10 and CASP14 were associated with poor OS
regardless of sex.
Based on the differentiation degree of GC (Table VI), increased CASP2, CASP4, CASP5
and CASP8 expression under moderate differentiation was associated
with poor OS. GC of a poor differentiation degree with upregulated
CASP1 and CASP8 expression was positively associated with OS.
Discussion
In the present study, the KM plotter was employed to
determine the association between CASP gene expression and OS in
GC. The results revealed that CASP1-8 were associated with improved
OS for patients with GC, whereas high expression of CASPs 9, 10, 12
and 14 were linked to poor OS. In the CASP3 subfamily, all
components except CASP10 were positively associated with improved
OS in GC.
Caspase 8, as an initiator, is activated by
associating with death-inducing signaling complex of the
extracellular pathway and cleaves procaspases 3, 4, 7, 9 and 10
and/or activates Bid. This leads to mitochondrial permeabilization
executed by BCL2-associated X protein and BCL2 antagonist/killer
protein (5). As for the association
between CASPs and cancer, Du et al (13) explored the effects of a
six-nucleotide deletion polymorphism of the CASP8 gene in the
digestive tract on the risk of cancer development; all genetic
models exhibited protection against the development of GC. The
antitumor effects of CASPs were also investigated by Kanehara et
al (14), in which the degree of
CASP8 activation could indicate the anticancer potential of this
CASP. Apart from GC, CASP8 has been reported to sense
proliferation-associated DNA damage and may serve as an indicator
of liver cancer (15). Of note,
apoptosis induced by aspirin and vitamin E succinate in GC has been
reported to involve CASP8 (16,17).
CASPs 3, 6 and 7 act as executioners of apoptosis,
causing programmed cell death by hydrolysis of the caspase target
protein.
CASP3 acts on poly(ADP-ribose)polymerase (PARP),
DNA-PK, Rho GDP-dissociation inhibitor (RhoGDI), sterol regulatory
element-binding protein (SREBP) and CKq and can be activated by
CASPs 8, 9 and 10. PARP is a multifunctional protein involved in
post-translational modifications in the majority of eukaryotic
cells. PARP has been considered to be a receptor for DNA damage as
it is activated by recognizing DNA fragments that are structurally
damaged. It performs poly ADP-ribosylation on a variety of nuclear
proteins; histones detach from DNA via the ADP-ribosylation of
histones for the binding of repair proteins, promoting the repair
of damaged DNA. In vivo, PARP is the main cleavage target of
CASP3. Human PARP is cleaved between Asp124 and Gly215, separating
its catalytic domain at the carboxyl end and the domain at the
amino terminus; thus, PARP loses its enzymatic activity. The
cleavage of PARP has been considered to be an important indicator
of apoptosis and an indicator of CASP3 activation. Additionally,
PARP can be activated by CASP7. A variety of therapeutic agents,
including resveratrol, berberine, homoharringtonine and silibinin
can inhibit interactions between CASP3 and PARP and are applied for
the treatment of breast cancer, human epidermoid carcinoma,
leukemia and human bladder transitional cell carcinoma (18–21).
DNA-PK is a key protein kinase involved the process of genomic DNA
damage repair via non-homologous end joining and maintains telomere
stability. DNA damage reparation can affect the sensitivity of
genotoxic drugs to cancer cells. Thus, in clinical settings,
inhibiting the activity of DNA-PK may be an effective anticancer
strategy. DNA-PK and PARP inhibitors have been applied as
chemo-/radio-sensitizers in Ewing sarcoma (22). Similarly, Alikarami et al
(23) indicated that inhibition of
DNA-PK increased the chemosensitivity of B-cell precursor acute
lymphoblastic leukemia to doxorubicin. The negative association
between the content of DNA-PK and CASP3 supports the results of the
present study in which CASP3 expression may improve the OS of
patients with GC. RhoGDI2, is a member of RhoGDI family of
proteins. In GC, RhoGDI2 is correlated with cancer growth,
metastasis and chemoresistance (24). Regarding the underlying mechanism,
RhoGDI2 upregulates vascular endothelial growth factor (VEGF)-C
expression, which promotes cancer cell invasion and induces
cisplatin resistance. Of note, RhoGDI2 positively regulates Rac1
activity, which can suppress VEGF-C expression (24). Furthermore, RhoGDI2 is associated
with 5-FU resistance (25). As a
potential therapeutic target, during drug-induced apoptosis,
RhoGDI2 is cleaved by CASP3 (26).
RhoGDI-signaling and mevalonate, as well as mitochondrial
metabolism, could be targeted by bergamot natural products to
eradicate cancer stem cells (27).
CASP6 can be activated by CASPs 7 and 3 and act on
lamin A, the main component of the basal lamina. It is a dynamic
network located beneath the nuclear membrane and serves an
important mechanical role, and can directly or indirectly interact
with chromatin. Lamin A serves a major role in maintaining
chromatin structure, transcription, DNA replication and apoptosis.
Wu et al (28) indicated that
downregulation of lamin A was an independent risk factor for the
poor prognosis of GC; however, the effects of CASP6 and lamin A on
GC require further investigation. As for CASP7, SREBP-1 and −2 may
bind to the proximal promoter region of the CASP7 gene, inducing
its expression (29).
CASP10 may affect the apoptosis of GC cells by
acting as an initiator, which has been associated with poor OS. It
can be activated by procaspase 8 and TRADD in the extrinsic pathway
and acts on CASP3. Additionally, CASP10 has been associated with
cancer. For instance, in myeloma, the survival of cancer cells is
dependent in CASP10 as myeloma cells require a basal level of
autophagy to survive; however, CASP10 regulates this response to
prevent cell death, promoting disease progression (30).
The CASP1 subfamily, including CASPs 1, 4, 5, 11, 12
and 14, exhibited a positive association with improved OS. CASP1
can activate CASPs 3 and 4, as well as interleukin (IL)-1 and
IL-18; conversely, CASP4 can also activate CASP1. CASP3 has been
reported to act as an executioner. Therefore, CASP1 can indirectly
promote apoptosis, indicating a positive association with improved
OS in GC. Regarding ILs, active CASP1 processes pro-IL-1β and
pro-IL-18 to initiate immune responses; IL-18, counteracts the
effects of IL-1β, which has proinflammatory activities similar to
another CASP1 substrate, in order to control disease by preventing
hyperactivation of the immune response in the gastric tissues
(31).
As for CASP4, this protein can be activated by
declines in adenosine deaminase activity and could induce gastric
epithelial cell apoptosis, promoting the formation of gastric
ulcers; however, a small population of cells may undergo malignant
transformation (32). The diagnostic
value of adenosine deaminase is of less importance as its activity
in GC did not significantly differ to that in normal mucosa
(33). CASP4 could act as a novel
diagnostic and prognostic biomarker of non-small cell lung cancer
as reported by Terlizzi et al (34); however, further investigation is
required.
CASP5 is an inflammatory caspase; together with
CASP11, these proteins serve a role in the immune system. The
association between inflammation and cancer has been reported, and
it is widely accepted that the tumor-promoting inflammatory
environment is one of the major hallmarks of cancer (35). In combination with the results of the
present study that CASPs 5 and 11 were linked to improved OS, it
was proposed that these proteins may serve roles in inflammation.
In addition, several studies have reported supporting findings.
Viganò et al (36) revealed
that CASP5 was a key determinant of one-step inflammasome
activation mediated by IL-1α and IL-1β release in human monocytes,
following treatment with lipopolysaccharide. Regarding CASP11,
knockout experiments revealed that in CASP11−/− mice,
the levels of IL-1β and IL-18 in the colon were significantly
reduced compared with wild-type mice. This suggested a possible
mechanism in which CASP11 may attenuate acute experimental colitis.
CASP11 has been reported to serve an important role in suppressing
the development of acute colitis, but may also be involved in
chronic relapsing-remitting colitis and inflammation-driven colon
tumorigenesis (37).
CASP12 has unique characteristics that are not
mutual to other family members, as is activated via endoplasmic
reticulum (ER) stress-induced cell death, rather than the intrinsic
and extrinsic pathways. ER stress is the consequence of abnormal
calcium absorption and is released from the ER. The activation of
CASP12 via its cleavage may proceed by proteases present on the
surface of the ER membrane, such as calpain. Intracellular calcium
elevation accounts for the activation and transport of calpain and
other ER-associated proteins to the ER surface. As an alternative
method of activation, CASP7 on the ER surface is capable of
cleaving procaspase12 (38,39). With respect to downstream substrates,
procaspase 9 is a substrate that is cleaved at the processing site,
transmitting proteolytic signals to CASP3 (38). The direct activation of CASP9 may be
associated with poor OS. Additionally, CASP12 as a truncated or
full-length proenzyme may arise from the single nucleotide
polymorphism of CASP12, impairing the innate immune and
inflammatory responses to infection in carcinoma, increasing the
risk of sepsis (40).
Van de Craen et al (41) revealed that procaspase 14 can be
weakly cleaved by CASP8 into active fragments, which did not result
in the subsequent cleavage of the classical CASP family substrates,
including CASPs 3, 6 or 7. CASP14, another divergent member of its
family, has a short prodomain and possesses a variety of unique
properties principally involved in epithelial cell differentiation,
rather than apoptosis and inflammation. Overexpression of CASP14
has been detected in epithelial malignant tumors, indicating that
CASP14 may be vital in carcinogenesis and cancer progression
(42,43). Furthermore, CASP14 has been reported
to be an anti-apoptotic protein, bound with cytochrome c to
reduce cisplatin sensitivity in vitro. As CASPs and
cytochrome c have recently emerged as innovative targets for
antitumor drugs, including camptothecin, adriamycin and cisplatin
with aim to destabilize the mitochondrial membrane and activate
procaspases, CASP14 may inhibit drug-induced DNA fragmentation and
cell death associated with increased chemoresistance (44).
Within the CASP2 subfamily, CASP2 was associated
with favorable OS for GC, while CASP9 was linked to poor OS.
Functioning similarly to CASP10, CASP2 activates CASP3 and PARP to
directly or indirectly initiate apoptosis, which is mainly
regulated by the CRADD-caspase 2 cascade pathway. Possessing a
similar dual-domain structure to FADD, CRADD contains an N-terminal
caspase homology domain that affects CASP2, along with a
carboxy-terminal death domain which interacts with
receptor-interacting protein; interactions between CASP2 and CRADD
are mediated by its caspase recruitment domain (45). It has been reported that trichostatin
A can induce CRADD to activate CASP2-dependent apoptosis not only
in GC cells, but also in prostate cancer cells by inhibiting
histone deacetylase, which suggests a novel therapeutic approach
for the treatment of GC (46,47). In
addition, the results of the present study were consistent with the
report of Yoo et al (48),
which revealed the deficient expression of CASPs 2, 6 and 7 in GC
cells compared with normal mucosal cells of the stomach.
Furthermore, CASP2 is involved in other processes, including p53
regulation, cell cycle regulation and the DNA damage response, as
well as cholesterol and triacylglycerol homeostasis, in which this
protein is regulated by SREBP-2 (49–51).
Those processes relate to the exhibited increasing HR in the
5-FU-based adjuvant group in Table
IV and it could provide a novel research direction.
With regards to the CASP9 pathway, the apoptosome
(cytochrome c, Apaf 1 and procaspase 9) facilitates the
oligomerization and subsequent auto-proteolysis of procaspase 9 to
CASP9, leading to the activation of CASP3 and CASP7 (52). In addition, modified Bid proteins
could transmit a signal via the intrinsic pathway of CASP
activation to interact with procaspase 9 when low concentrations of
CASP8 are insufficient to activate CASP3 (53). Overexpression of phosphorylated CASP9
(p-CASP9), an anti-apoptotic protein, in malignant GC cells could
be an inhibitory mechanism of apoptosis mediated by CASP9,
suggesting the protumorigenic role of CASP9 in the development of
GC (54). This report of apoptosis
resistance supports the findings of the present study. Numerous
studies have proposed that the clearance of GC cells could be
conducted by activating CASP9-induced apoptosis (55,56),
which may be linked to the favorable OS of patients with diffuse
type GC observed in the present study. In light of these previous
studies, the mechanism of CASP9 in apoptosis may involve the
antagonism between p-CASP9 and CASP9, in which CASP9 is
phosphorylated at Thr125, mediated by the mitogen-activated protein
kinase pathway.
The Lauren classification is one of the most widely
applicable classification systems in GC. It was adopted in the
present study for its high repeatability and convenience in clinic,
especially for its value for prognostic analysis (57,58),
whereas previous studies merely observed the specific relation
between CASP expression and the Lauren classification, which
provides a new study direction to investigate its mechanism.
Several limitations of the present study should be
considered. Further research is required to account for the effects
of treatment strategies, pathological stages, HER2 expression
status, sex and differentiation degree on the prognostic value of
CASPs. While certain hypotheses have been proposed based on the
literature and clinical statistics; further investigation is
required using animal models and performing clinical trials. For
instance, the number of mixed type patients is small due to the
limited data in KM plotter. Additionally, although the association
between mRNA expression and prognosis was explored, analysis should
be conducted at the protein level. To this end, the authors aim to
validate the findings of the present study in the future, using an
independent cohort and RNA sequencing.
In conclusion, in the present study the prognostic
value of the mRNA expression profile of CASPs in patients with GC
was assessed using the KM plotter database. Among them,
overexpression of CASP1-8 at the mRNA level was linked to improved
OS, whereas upregulated expression of CASPs 9, 10, 12 and 14 was
associated with poor OS. The findings of the present study may
provide insight into the relationship between each molecule and
their role in cancer. The present study may serve as a basis for
the development of novel therapeutic strategies using CASPs as
powerful and precise prognostic predictors in GC, in which
potential targeting agents may be applied for the treatment of this
disease; however, further study is required.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data of the mRNA expression levels and overall
survival can be found in the online KM plotter database (http://kmplot.com/analysis/).
Authors' contributions
ZW, FN and FY designed the study. ZC and XZ obtained
the data. FY, ZC, XZ and JC performed data processing and
statistical analysis. ZW and FN wrote the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
5-FU
|
5-Fluorouracil
|
|
Apaf1
|
apoptotic peptidase activating factor
1
|
|
Bcl-2
|
B-cell lymphoma 2
|
|
Bid
|
BH3 interacting domain death
agonist
|
|
caspase
|
cysteinyl aspartate specific
proteinase
|
|
CASP1
|
caspase 1
|
|
CASP2
|
caspase 2
|
|
CASP3
|
caspase 3
|
|
CASP4
|
caspase 4
|
|
CASP5
|
caspase 5
|
|
CASP6
|
caspase 6
|
|
CASP7
|
caspase 7
|
|
CASP8
|
caspase 8
|
|
CASP9
|
caspase 9
|
|
CASP10
|
caspase 10
|
|
CASP12
|
caspase 12
|
|
CASP14
|
caspase 14
|
|
CI
|
Confidence intervals
|
|
CRADD
|
CASP2 and RIPK1 domain containing
adaptor with death domain
|
|
ER
|
endoplasmic reticulum
|
|
FADD
|
Fas associated via death domain
|
|
GC
|
Gastric cancer
|
|
HER2
|
Human epidermal growth factor
receptor-2
|
|
HGU133
|
Human Genome U133
|
|
HR
|
Hazard ratio
|
|
KM plotter
|
Kaplan-Meier plotter
|
|
MAPK
|
mitogen-activated protein kinase
|
|
OS
|
Overall survival
|
|
P
|
Probability
|
|
P-caspase 9
|
phosphorylated caspase 9
|
|
TRADD
|
TNFRSF1A associated via death
domain
|
References
|
1
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lordick F and Hoffmeister A: Treatment of
gastric cancer. Internist (Berl). 55:15–16, 18-22. 2014.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. Biomed Res Int. 2014:1508452014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frejlich E, Rudno-Rudzińska J, Janiszewski
K, Salomon L, Kotulski K, Pelzer O, Grzebieniak Z, Tarnawa R and
Kielan W: Caspases and their role in gastric cancer. Adv Clin Exp
Med. 22:593–602. 2013.PubMed/NCBI
|
|
5
|
Crawford ED and Wells JA: Caspase
substrates and cellular remodeling. Annu Rev Biochem. 80:1055–1087.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chowdhury I, Tharakan B and Bhat GK:
Caspases-an update. Comp Biochem Physiol B Biochem Mol Biol.
151:10–27. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fan TJ, Han LH, Cong RS and Liang J:
Caspase family proteases and apoptosis. Acta Biochim Biophys Sin
(Shanghai). 37:719–727. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mace PD, Riedl SJ and Salvesen GS: Caspase
enzymology and activation mechanisms. Methods Enzymol. 544:161–178.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boatright KM and Salvesen GS: Mechanisms
of caspase activation. Curr Opin Cell Biol. 15:725–731. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dalma-Weiszhausz DD, Warrington J,
Tanimoto EY and Miyada CG: The affymetrix GeneChip platform: An
overview. Methods Enzymol. 410:3–28. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du H, Song GX, Fang MZ, Shu YQ, Zhao X and
Zhu LJ: A meta-analysis of caspase-8 −652 6N del polymorphism and
digestive tract cancer risk. J Biomed Res. 33:173–180.
2019.PubMed/NCBI
|
|
14
|
Kanehara I, Nakata B and Hirakawa K:
Caspase-8 is scarcely silenced and its activity is well correlated
with the anticancer effect of tumor necrosis factor-related
apoptosis-inducing ligand in gastric cancer cells. Oncol Rep.
14:1249–1253. 2005.PubMed/NCBI
|
|
15
|
Boege Y, Malehmir M, Healy ME, Bettermann
K, Lorentzen A, Vucur M, Ahuja AK, Böhm F, Mertens JC, Shimizu Y,
et al: A dual role of caspase-8 in triggering and sensing
proliferation-associated DNA damage, a key determinant of liver
cancer development. Cancer cell. 32:342.e10–359.e10. 2017.
View Article : Google Scholar
|
|
16
|
Gu Q, Wang JD, Xia HH, Lin MC, He H, Zou
B, Tu SP, Yang Y, Liu XG, Lam SK, et al: Activation of the
caspase-8/Bid and Bax pathways in aspirin-induced apoptosis in
gastric cancer. Carcinogenesis. 26:541–546. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Wu K and Yu WP: The expression and
activity of caspase-8 in the process of vitamin E succinate-induced
apoptosis in human gastric carcinoma SGC-7901 cells. Zhonghua Yu
Fang Yi Xue Za Zhi. 37:112–114. 2003.(In Chinese). PubMed/NCBI
|
|
18
|
Mondal A and Bennett LL: Resveratrol
enhances the efficacy of sorafenib mediated apoptosis in human
breast cancer MCF7 cells through ROS, cell cycle inhibition,
caspase 3 and PARP cleavage. Biomed Pharmacother. 84:1906–1914.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mantena SK, Sharma SD and Katiyar SK:
Berberine inhibits growth, induces G1 arrest and apoptosis in human
epidermoid carcinoma A431 cells by regulating Cdki-Cdk-cyclin
cascade, disruption of mitochondrial membrane potential and
cleavage of caspase 3 and PARP. Carcinogenesis. 27:2018–2027. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yinjun L, Jie J, Weilai X and Xiangming T:
Homoharringtonine mediates myeloid cell apoptosis via upregulation
of pro-apoptotic bax and inducing caspase-3-mediated cleavage of
poly(ADP-ribose) polymerase (PARP). Am J Hematol. 76:199–204. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tyagi A, Agarwal C, Harrison G, Glode LM
and Agarwal R: Silibinin causes cell cycle arrest and apoptosis in
human bladder transitional cell carcinoma cells by regulating
CDKI-CDK-cyclin cascade, and caspase 3 and PARP cleavages.
Carcinogenesis. 25:1711–1720. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vormoor B, Schlosser YT, Blair H, Sharma
A, Wilkinson S, Newell DR and Curtin N: Sensitizing Ewing sarcoma
to chemo- and radiotherapy by inhibition of the DNA-repair enzymes
DNA protein kinase (DNA-PK) and poly-ADP-ribose polymerase (PARP)
1/2. Oncotarget. 8:113418–113430. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alikarami F, Safa M, Faranoush M, Hayat P
and Kazemi A: Inhibition of DNA-PK enhances chemosensitivity of
B-cell precursor acute lymphoblastic leukemia cells to doxorubicin.
Biomed Pharmacother. 94:1077–1093. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cho HJ, Kim IK, Park SM, Baek KE, Nam IK,
Park SH, Ryu KJ, Choi J, Ryu J, Hong SC, et al: VEGF-C mediates
RhoGDI2-induced gastric cancer cell metastasis and cisplatin
resistance. Int J Cancer. 135:1553–1563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Z, He XY, Li JF, Yu BQ, Chen XH, Ji
J, Zhang JN, Gu QL, Zhu ZG and Liu BY: RhoGDI2 confers resistance
to 5-fluorouracil in human gastric cancer cells. Oncol Lett.
5:255–260. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Essmann F, Wieder T, Otto A, Müller EC,
Dörken B and Daniel PT: GDP dissociation inhibitor D4-GDI (Rho-GDI
2), but not the homologous rho-GDI 1, is cleaved by caspase-3
during drug-induced apoptosis. Biochem J 346 Pt. 3:777–783. 2000.
View Article : Google Scholar
|
|
27
|
Fiorillo M, Peiris-Pagès M,
Sanchez-Alvarez R, Bartella L, Di Donna L, Dolce V, Sindona G,
Sotgia F, Cappello AR and Lisanti MP: Bergamot natural products
eradicate cancer stem cells (CSCs) by targeting mevalonate,
Rho-GDI-signalling and mitochondrial metabolism. Biochim Biophys
Acta Bioenerg. 1859:984–996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu Z, Wu L, Weng D, Xu D, Geng J and Zhao
F: Reduced expression of lamin A/C correlates with poor
histological differentiation and prognosis in primary gastric
carcinoma. J Exp Clin Cancer Res. 28:82009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gibot L, Follet J, Metges JP, Auvray P,
Simon B, Corcos L and Le Jossic-Corcos C: Human caspase 7 is
positively controlled by SREBP-1 and SREBP-2. Biochem J.
420:473–483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lamy L, Ngo VN, Emre NC, Shaffer AL III,
Yang Y, Tian E, Nair V, Kruhlak MJ, Zingone A, Landgren O and
Staudt LM: Control of autophagic cell death by caspase-10 in
multiple myeloma. Cancer Cell. 23:435–449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hitzler I, Sayi A, Kohler E, Engler DB,
Koch KN, Hardt WD and Müller A: Caspase-1 has both proinflammatory
and regulatory properties in Helicobacter infections, which are
differentially mediated by its substrates IL-1β and IL-18. J
Immunol. 188:3594–3602. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yaguchi T, Saito M, Yasuda Y and Nishizaki
T: Caspase-4 activation in association with decreased adenosine
deaminase activity may be a factor for gastric ulcer. Digestion.
81:62–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Namiot Z, Kemona A, Stasiewicz J,
Marcinkiewicz M, Namiot A and Gorski J: Adenosine deaminase
activity in gastric cancer. Cancer Lett. 82:95–98. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Terlizzi M, Colarusso C, De Rosa I, De
Rosa N, Somma P, Curcio C, Sanduzzi A, Micheli P, Molino A,
Saccomanno A, et al: Circulating and tumor-associated caspase-4: A
novel diagnostic and prognostic biomarker for non-small cell lung
cancer. Oncotarget. 9:19356–19367. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Raposo TP, Beirão BC, Pang LY, Queiroga FL
and Argyle DJ: Inflammation and cancer: Till death tears them
apart. Vet J. 205:161–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Viganò E, Diamond CE, Spreafico R,
Balachander A, Sobota RM and Mortellaro A: Human caspase-4 and
caspase-5 regulate the one-step non-canonical inflammasome
activation in monocytes. Nat Commun. 6:87612015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Williams TM, Leeth RA, Rothschild DE,
McDaniel DK, Coutermarsh-Ott SL, Simmons AE, Kable KH, Heid B and
Allen IC: Caspase-11 attenuates gastrointestinal inflammation and
experimental colitis pathogenesis. Am J Physiol Gastrointest Liver
Physiol. 308:G139–G150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rao RV, Ellerby HM and Bredesen DE:
Coupling endoplasmic reticulum stress to the cell death program.
Cell Death Differ. 11:372–380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nakagawa T and Yuan J: Cross-talk between
two cysteine protease families. Activation of caspase-12 by calpain
in apoptosis. J Cell Biol. 150:887–894. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Saleh M, Vaillancourt JP, Graham RK, Huyck
M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT,
Hotchkiss RS, et al: Differential modulation of endotoxin
responsiveness by human caspase-12 polymorphisms. Nature.
429:75–79. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Van de Craen M, Van Loo G, Pype S, Van
Criekinge W, Van den brande I, Molemans F, Fiers W, Declercq W and
Vandenabeele P: Identification of a new caspase homologue:
Caspase-14. Cell Death Differ. 5:838–846. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koenig U, Sommergruber W and Lippens S:
Aberrant expression of caspase-14 in epithelial tumors. Biochem
Biophys Res Commun. 335:309–313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krajewska M, Kim H, Shin E, Kennedy S,
Duffy MJ, Wong YF, Marr D, Mikolajczyk J, Shabaik A,
Meinhold-Heerlein I, et al: Tumor-associated alterations in
caspase-14 expression in epithelial malignancies. Clin Cancer Res.
11:5462–5471. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fang HY, Chen CY, Hung MF, Hsiao YT,
Chiang TC, Lin TY, Chang HW, Chow KC and Ko WJ: Caspase-14 is an
anti-apoptotic protein targeting apoptosis-inducing factor in lung
adenocarcinomas. Oncol Rep. 26:359–369. 2011.PubMed/NCBI
|
|
45
|
Ahmad M, Srinivasula SM, Wang L, Talanian
RV, Litwack G, Fernandes-Alnemri T and Alnemri ES: CRADD, a novel
human apoptotic adaptor molecule for caspase-2, and FasL/tumor
necrosis factor receptor-interacting protein RIP. Cancer Res.
57:615–619. 1997.PubMed/NCBI
|
|
46
|
Shen Q, Tang W, Sun J, Feng L, Jin H and
Wang X: Regulation of CRADD-caspase 2 cascade by histone
deacetylase 1 in gastric cancer. Am J Transl Res. 6:538–547.
2014.PubMed/NCBI
|
|
47
|
Taghiyev AF, Guseva NV, Glover RA, Rokhlin
OW and Cohen MB: TSA-induced cell death in prostate cancer cell
lines is caspase-2 dependent and involves the PIDDosome. Cancer
Biol Ther. 5:1199–1205. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yoo NJ, Lee JW, Kim YJ, Soung YH, Kim SY,
Nam SW, Park WS, Lee JY and Lee SH: Loss of caspase-2, −6 and −7
expression in gastric cancers. APMIS. 112:330–335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mandruzzato S, Brasseur F, Andry G, Boon T
and van der Bruggen P: A CASP-8 mutation recognized by cytolytic T
lymphocytes on a human head and neck carcinoma. J Exp Med.
186:785–793. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nicholson DW: Caspase structure,
proteolytic substrates, and function during apoptotic cell death.
Cell Death Differ. 6:1028–1042. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Logette E, Le Jossic-Corcos C, Masson D,
Solier S, Sequeira-Legrand A, Dugail I, Lemaire-Ewing S, Desoche L,
Solary E and Corcos L: Caspase-2, a novel lipid sensor under the
control of sterol regulatory element binding protein 2. Mol Cell
Biol. 25:9621–9631. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Acehan D, Jiang X, Morgan DG, Heuser JE,
Wang X and Akey CW: Three-dimensional structure of the apoptosome:
Implications for assembly, procaspase-9 binding, and activation.
Mol Cell. 9:423–432. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kluck RM, Bossy-Wetzel E, Green DR and
Newmeyer DD: The release of cytochrome c from mitochondria: A
primary site for Bcl-2 regulation of apoptosis. Science.
275:1132–1136. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yoo NJ and Lee SH, Jeong EG and Lee SH:
Expression of phosphorylated caspase-9 in gastric carcinomas.
APMIS. 115:354–359. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang H, Li H, Chen F, Luo J, Gu J, Wang H,
Wu H and Xu Y: Baicalin extracted from Huangqin (Radix Scutellariae
Baicalensis) induces apoptosis in gastric cancer cells by
regulating B cell lymphoma (Bcl-2)/Bcl-2-associated X protein and
activating caspase-3 and caspase-9. J Tradit Chin Med. 37:229–5.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chang-Qing F, Yi L, De-Guang W, Qing-Bin
S, Xiang-Min H, Na T and Jian-Hua L: Immune clearance gastric
carcinoma cells in ascites by activating caspase-9-induced
apoptosis. APMIS. 119:173–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen YC, Fang WL, Wang RF, Liu CA, Yang
MH, Lo SS, Wu CW, Li AF, Shyr YM and Huang KH: Clinicopathological
variation of lauren classification in gastric cancer. Pathol Oncol
Res. 22:197–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ma J, Shen H, Kapesa L and Zeng S: Lauren
classification and individualized chemotherapy in gastric cancer.
Oncol Lett. 11:2959–2964. 2016. View Article : Google Scholar : PubMed/NCBI
|