Introduction
Hepatitis B virus (HBV) infection is a global health
concern that causes >1,000,000 deaths worldwide per year
(1). HBV-infected patients face the
risk of developing cirrhosis, hepatic decompensation and
hepatocellular carcinoma (HCC) (1,2). HCC is
the fifth most common type of cancer and the third highest cause of
cancer-associated mortality worldwide, next to lung and stomach
cancer (3). The high mortality
associated with HCC is due to its unresponsiveness to treatment and
a delay in recognizing symptoms (2,4). Chronic
hepatitis B infection is a leading precursor of HCC; however,
little is currently known about the pathogenesis of HCC.
Covalently closed circular DNA (cccDNA) is an
important intermediate in the life cycle of HBV. It does not
directly participate in HBV replication, but maintains a stable
pool within the hepatocyte nucleus (5). A previous study demonstrated that HCC
often develops as HBV replication intensifies during the late stage
of hepatitis B (6), a period in
which cccDNA becomes predominant in quantity (7). Therefore, it may be possible to predict
the involvement of cccDNA in HBV-associated HCC by measuring the
levels of intrahepatic cccDNA in paired tumor and peri-tumor
tissues.
HBV-encoded X protein (HBx) is a key viral
oncoprotein produced during the development of HBV-associated HCC
(8,9). It regulates signaling pathways by
interacting with a variety of proteins (10). For example, HBx can be specifically
modified by E3 ubiquitin ligases to upregulate its expression and
promote transcription, cell proliferation and tumor growth
(11).
The human male specific lethal 2 (MSL2) ortholog is
an E3 ubiquitin ligase that ubiquitylates the tumor suppressor p53,
as well as histone H2B to mediate transcriptional control (12). Gao et al (13) reported that HBx-mediated upregulation
of MSL2 activates HBV cccDNA in hepatoma cells to promote
hepatocarcinogenesis, forming a HBx/MSL2/cccDNA/HBV positive
feedback loop. The present study aimed to compare the cccDNA, MSL2
mRNA and HBx mRNA levels in tumor and peri-tumor tissues, and to
compare differences in cccDNA and MSL2 mRNA levels between
HBx-positive and HBx-negative patients. Furthermore, the present
study investigated the effect of antiviral therapy on these
indicators.
Materials and methods
Patients
The present study included a total of 53 patients,
including three patients without hepatitis B as a control. The 50
HBV-associated patients with HCC were divided into three groups
according to hepatitis B envelope antigen (HBeAg) status, tumor
size and HCC recurrence for comparison of cccDNA contents. The
contents of cccDNA, MSL2 mRNA or HBx mRNA in the control patients
were determined separately and one patient was included as a
control in each analysis. The extraction procedures for cccDNA,
MSL2 mRNA and HBx mRNA were based on the sample to be assayed.
cccDNA, MSL2 mRNA, HBx mRNA were stored at −20°C from a single
HBV-negative sample, based on a fixed control patient, and were
added to each PCR analysis; fold-changes were determined based on
the relative levels of cccDNA, MSL2 mRNA and HBx mRNA. A total of
50 patients with HBV-associated HCC and 3 patients with HCC without
hepatitis B who had undergone curative liver resection or liver
transplantation at the Department of Surgery, Seoul National
University Hospital (Seoul, South Korea) between October 2016 and
March 2018 were included in the present study. Samples were
prepared with tumor and peri-tumor liver tissues collected from
these patients. The history of antiviral therapy before surgery and
tumor recurrence following surgery were obtained via each patient's
medical records, follow-up until September 2018. The diagnosis of
HCC was evaluated by two experienced pathologists at Seoul National
University Hospital and analysis was conducted in a blinded manner.
Patients with HCC with evidence of HBV-infected history were
enrolled in this study, and patients who had consumed alcohol or
had been infected with hepatitis C or hepatitis D viruses were
excluded from this study. In addition, patients with
cholangiocarcinoma were excluded from this study. Tumor and
peri-tumor tissues were resected, and peri-tumor tissue were
obtained >1 cm from the edge of the tumor, and rapidly frozen
and stored at −80°C. Written consent, approving the use of tissues
for research purpose following surgery, was obtained from each
patient included in the present study. The experimental protocol
was approved by the Institutional Review Board of Seoul National
University Hospital (IRB no. H-1809-001-967).
Detection of HBsAg, HBeAg, and
HBV-DNA
The hepatitis B surface antigen (HBsAg) and HBeAg
levels were assessed by the chemiluminescence enzyme immunoassay
(CLEIA) method using a commercially available enzyme immunoassay
kit (Lumipulse, Fuji Rebio, Inc.), according to the manufacturer's
protocols. Serum concentrations of HBV-DNA were determined using a
PCR HBV monitoring kit (Roche Diagnostics K.K.), which had a
quantitative range between 2.6 and 7.6 log copies/ml.
Isolation of intrahepatic total DNA
and cccDNA
Total genomic DNA was extracted from ~20-30 mg of
liver tissue using the QIAamp DNA Mini kit (Qiagen GmbH) according
to the manufacturer's protocol. The concentration of total DNA was
determined at 260 nm with a spectrophotometer (Eppendorf).
Plasmid-safe™ ATP-dependent DNase (Epicentre; Illumina, Inc.)
incubation with Ambion™ RNase A (Thermo Fisher Scientific, Inc.)
for 30 min at 70°C was used to hydrolyze linear double-stranded
DNA, linear and closed circular single-stranded DNA, for the
isolation of double-stranded closed circular DNA, including HBV
cccDNA (14).
cccDNA detection using quantitative
PCR (qPCR)
Intrahepatic levels of cccDNA in tumor and
peri-tumor tissues were compared. cccDNA was isolated by digestion
from 300 mg of total DNA with Plasmid-safe™ ATP-dependent DNase and
diluted with 20 ml of diethyl pyrocarbonate water. A total of 1 ml
of cccDNA was then used for qPCR amplification (7500 Real-time PCR
Instrument system; Applied Biosystems; Thermo Fisher Scientific,
Inc.), which was performed using TOPreal™ qPCR PreMIX SYBR Green
(Enzynomics). β-actin amplicons were used as the internal reference
for subsequent PCR analysis. Each sample was assayed three times to
determine the mean cycle threshold (Ct) values for HBV cccDNA and
β-actin. Relative transcriptional fold-changes were calculated as
2−∆∆Ct (15). The
detected cccDNA levels in patients with HBV-associated HCC were
presented as a fold-change relative to that in the one HBV-negative
control patient. The thermocycling was performed as follows: 95°C
for 10 min, followed by 45 cycles of 95°C for 15 sec, 63°C for 30
sec, 72°C for 25 sec and 95°C for 15 sec (16). Optimal sensitivity was obtained by
combining two forward primers with a reverse primer. The sequences
of the PCR primers are listed in Table
I (17).
 | Table I.Primers used for qPCR and reverse
transcription-qPCR for the detection of hepatitis B virus cccDNA,
MSL2, HBx and β-actin. |
Table I.
Primers used for qPCR and reverse
transcription-qPCR for the detection of hepatitis B virus cccDNA,
MSL2, HBx and β-actin.
| Gene | Primer
sequence |
|---|
| cccDNA | Forward,
5′-GCGGWCTCCCCGTCTGTGCC-3′; 5′-GTCTGTGCCTTCTCATCTGC-3′ |
|
| Reverse,
5′-GTCCATGCCCCAAAGCCAACC-3′ |
| MSL2 | Forward,
5′-ACAGTGAGAAAGTTCAGCCA-3′ |
|
| Reverse,
5′-AGCACGCCCACATTTACT-3′ |
| HBx | Forward,
5′-ATGGCTGCTAGGCTGTGC-3′ |
|
| Reverse,
5′-TTAGGCAGAGGGGAAAAAGTTG-3′ |
| β-actin | Forward,
5′-GTGCACCTGACTCCTGAGGAGA-3′ |
|
| Reverse,
5′-CCTTGATACCAACCTGCCCAG-3′ |
MSL2 and HBx mRNA detection using
reverse-transcription qPCR (RT-qPCR)
Total RNA was extracted from tumor and peri-tumor
tissues using TRIzol® reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. A First-Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.) was used to reverse
transcribe total RNA into cDNA. Synthesis conditions were as
follows: 25°C for 10 min, 37°C for 120 min and 85°C for 5 min. cDNA
was used as the template for qPCR using a TOPreal™ qPCR PreMIX SYBR
Green. β-actin was used as an internal control for normalization.
Relative transcriptional fold-changes were calculated using the
2−∆∆Ct method (15).
Thermocycling was performed as follows: 95°C for 10 min, followed
by 45 cycles of 95°C for 15 sec, 63°C for 30 sec, 72°C for 25 sec
and 95°C for 15 sec. All primers used are listed in Table I. MSL2 and HBx mRNA levels in
patients with HBV-associated HCC were presented as fold-changes
relative to that in the fixed one HBV negative control patient.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 23; IBM Corp.). Variables with normal and skewed
distribution of paired samples were analyzed using paired Student's
t-tests and Wilcoxon signed ranks tests, respectively. The
Mann-Whitney U test was used to analyze continuous variables.
Correlation analysis was performed using Spearman's correlation
test. P<0.05 was considered to indicate a statistically
significant difference. Data are expressed as the mean ± standard
deviation, with bars in the graphs representing standard
deviation.
Results
Clinical characteristics of
patients
A total of 50 patients with HBV-associated HCC were
included in the present study (40 males and 10 females; mean age,
58±10 years; age range, 34–80). Of these patients, 49 were
HBsAg-positive, and one exhibited seroconversion following
antiviral treatment; 31 patients had received different degrees of
antiviral therapy and 19 had not; 14 patients were HBeAg-positive
(28%) and 36 were HBeAg-negative (72%). Postoperative HCC
recurrence occurred in 17 patients (34%), and 33 (66%) did not
present with recurrence; 30 patients (60%) had a tumor size ≥3 cm,
whereas 20 patients (40%) had a tumor size <3 cm (Table II).
 | Table II.Clinical characteristics of patients
with HCC (n=50). |
Table II.
Clinical characteristics of patients
with HCC (n=50).
| Characteristic | n (%) |
|---|
| Sex |
|
|
Male | 40 (80) |
|
Female | 10 (20) |
| Pre-op serum HBV
DNA |
|
|
Positive | 42 (84) |
|
Negative | 8 (16) |
| Pre-op HBeAg
status |
|
|
Positive | 14 (28) |
|
Negative | 36 (72) |
| Pre-op antiviral
treatment |
|
| No
antiviral treatment | 19 (38) |
|
Antiviral treatment | 31 (62) |
| Operation
method |
|
|
Curative resection | 45 (90) |
| Liver
transplantation | 5 (10) |
|
HBV
recurrence | 2 (40a) |
|
HCC
recurrence | 3 (60a) |
| Post-op HCC
recurrence | 17 (34) |
| Lung or lymphatic
metastasis | 5 (10) |
| Tumor size, cm |
|
| ≥3 | 30 (60) |
|
<3 | 20 (40) |
Comparison of cccDNA in tumor and
peri-tumor tissues
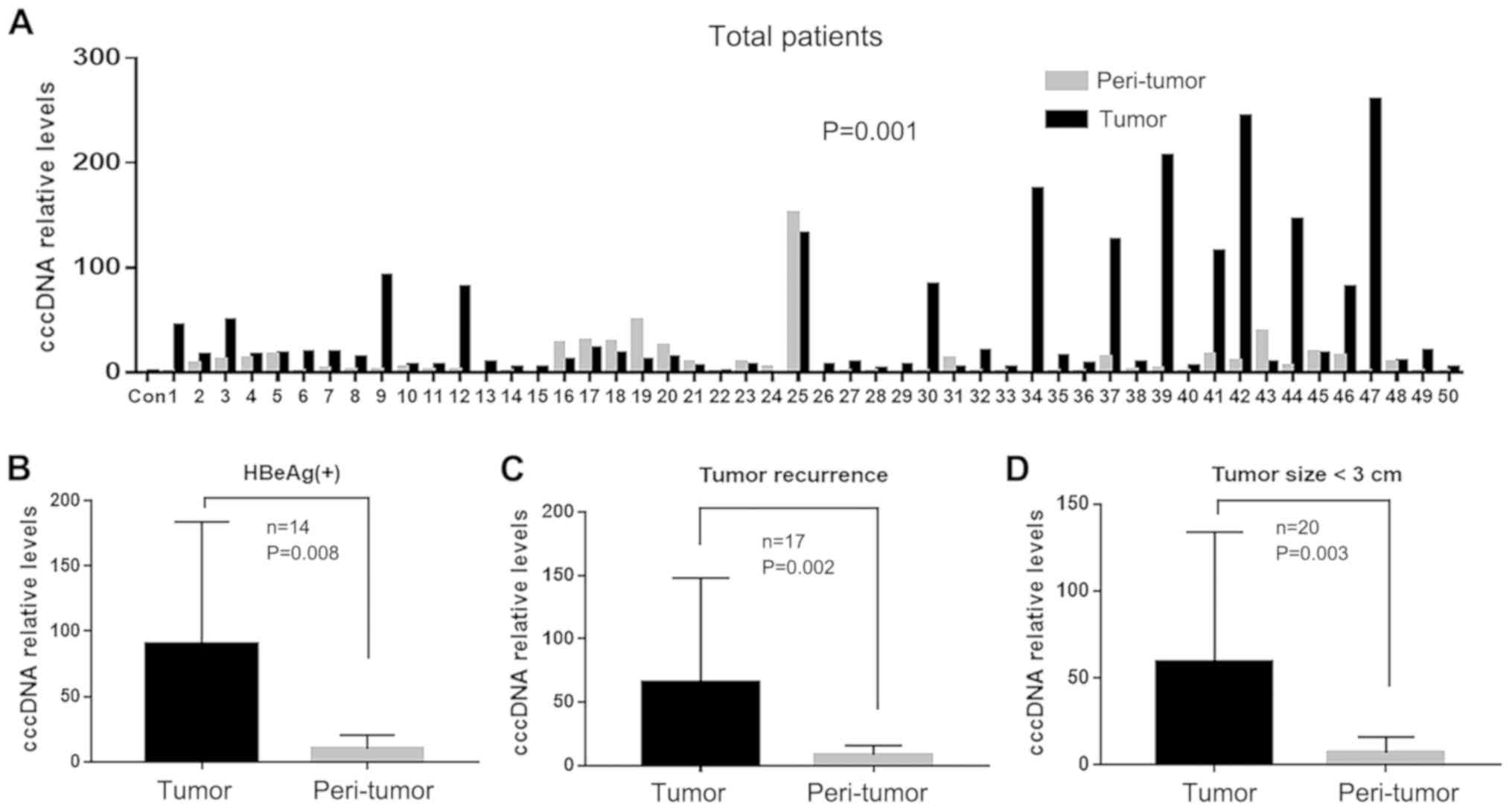
For 50 patients with HBV, intrahepatic cccDNA levels
were significantly higher in tumor tissues compared with those in
peri-tumor tissues (44.68±65.14 vs. 11.47±23.03, P=0.001; Fig. 1A). Furthermore, the difference in
intrahepatic cccDNA levels was more apparent in the tumor tissues
compared with the peri-tumor tissues in the HBeAg-positive
(90.07±93.94 vs. 10.05±10.48, respectively; P=0.008; Fig. 1B), tumor recurrence (65.78±79.85 vs.
8.55±7.16; P=0.002; Fig. 1C) and
<3 cm tumor size (59.65±72.67 vs. 7.02±9.02; P=0.003; Fig. 1D) groups.
Comparison of cccDNA/MSL2/HBx levels
in tumor and peri-tumor tissues of patients with HCC exhibiting
liver cirrhosis
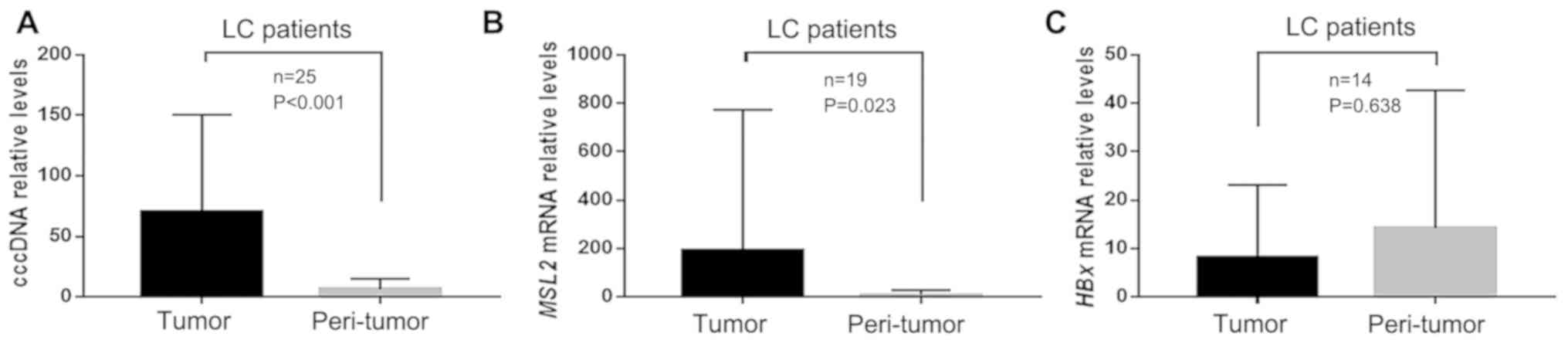
From the cohort of 50 patients with HBV-associated
HCC in the present study, 25 with preoperative cirrhosis were
included in this analysis. Of these 25 patients with cirrhosis, six
samples could not be paired due to lack of tumor tissue after
analysis of cccDNA when analyzing MSL2 mRNA and HBx mRNA.
Therefore, HBx and MSL2 mRNA levels were measured in 19 paired
samples. Of these, 14 samples were HBx-positive. In the
HBx-positive patients, the cccDNA and MSL2 mRNA levels were
compared in tumor and peri-tumor tissues. The results revealed that
the level of cccDNA was significantly higher in tumor tissue
compared with that in peri-tumor tissue, in patients with HCC
exhibiting liver cirrhosis (70.42±78.56 vs. 6.36±8.47; P<0.001;
Fig. 2A). Similarly, the level of
MSL2 mRNA was also higher in tumor tissue compared with that in
peri-tumor tissue (191.78±566.32 vs. 9.77±18.56; P=0.023; Fig. 2B). However, the level of HBx mRNA was
not significantly different between the two tissue types
(8.24±14.38 vs. 14.26±27.40; P=0.638; Fig. 2C).
Comparison of cccDNA/MSL2/HBx levels
in tumor and peri-tumor tissues of patients receiving or not
receiving antiviral therapy
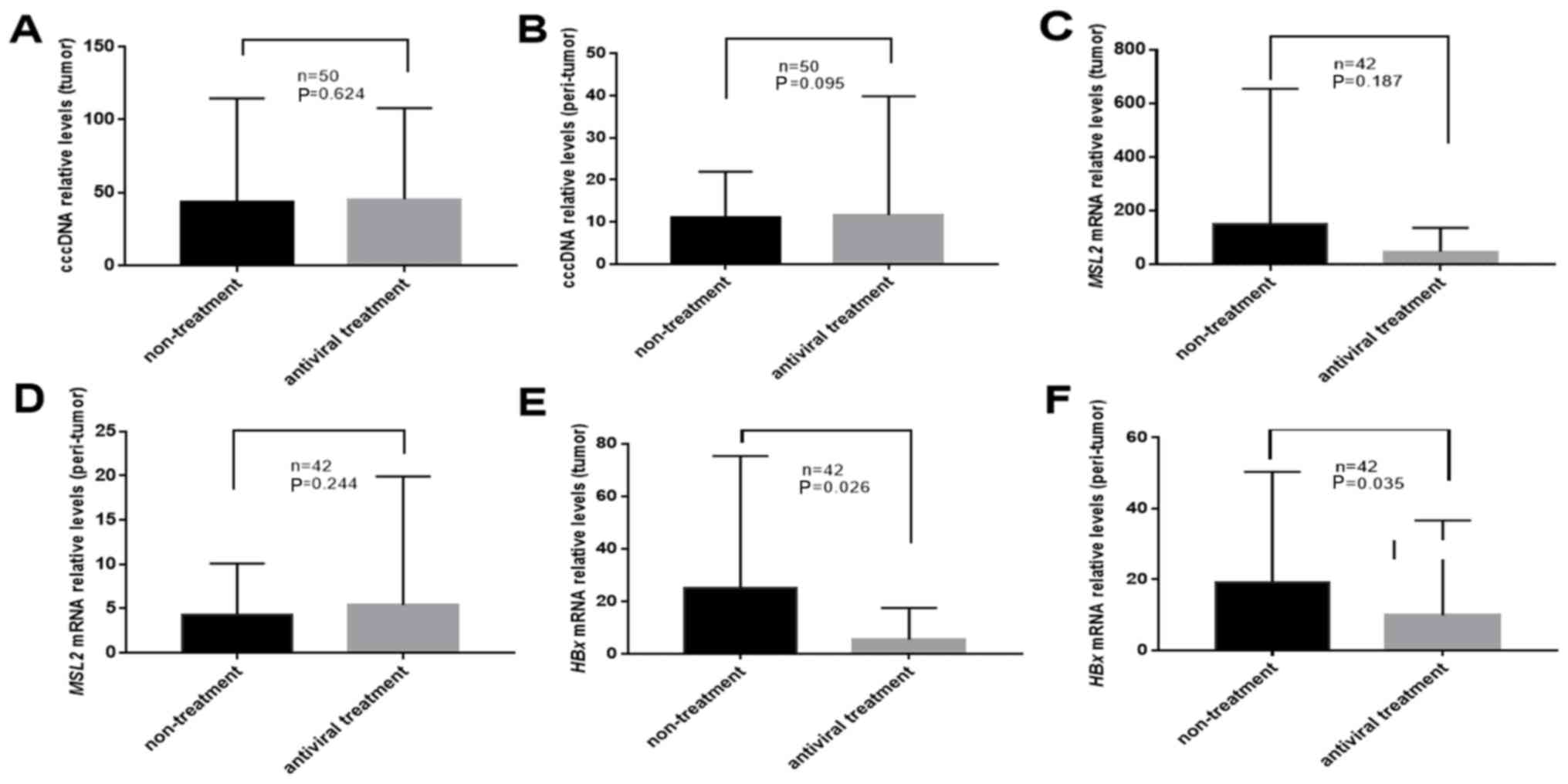
Of the 50 patients, 31 had undergone antiviral
treatment prior to surgery. The results of the present study
demonstrated that in tumor and peri-tumor tissues, antiviral
therapy did not significantly change the levels of cccDNA
(43.54±69.09 vs. 45.38±61.49; P=0.624 and 11.19±10.47 vs.
11.64±27.77; P=0.095; Fig. 3A and B,
respectively) or MSL2 mRNA (159.79±502.34 vs. 46.28±85.56; P=0.187
and 4.36±5.81 vs. 5.34±13.96; P=0.244; Fig. 3C and D, respectively). The levels of
HBx mRNA in tumor and peri-tumor tissues from patients treated with
antivirals were significantly lower compared with those in
untreated patients (26.05±50.05 vs. 5.36±11.59; P=0.026 and
19.65±31.10 vs. 9.51±25.74; P=0.035; Fig. 3E and F, respectively).
Comparison between HBx, cccDNA and
MSL2 in tumor and peri-tumor tissues of HBx-positive patients
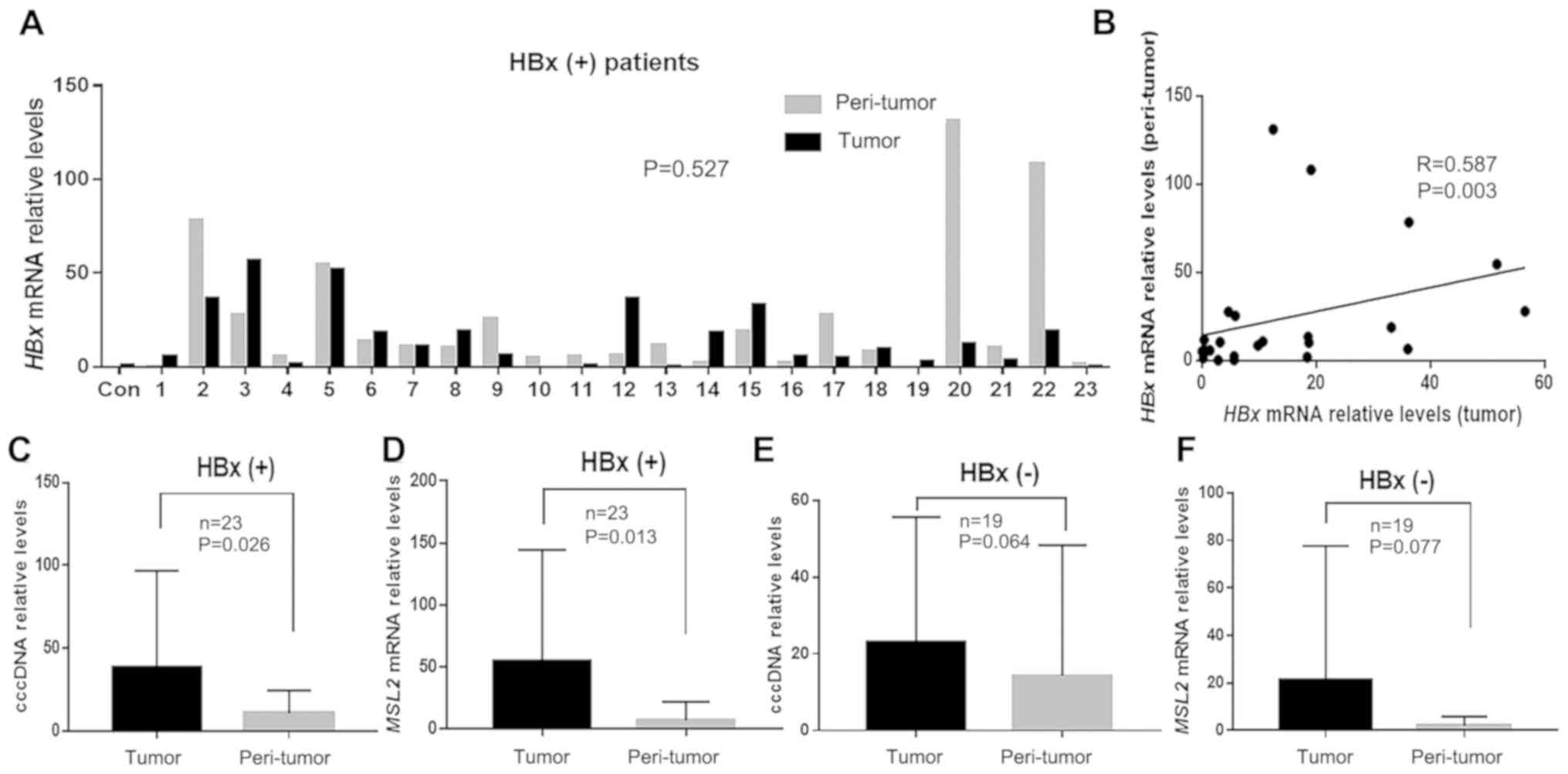
When analyzing MSL2 mRNA and HBx mRNA, 8 of the
total 50 patients could not be paired due to the lack of tumor
tissues after analysis of cccDNA. Therefore, HBx mRNA and MSL2 mRNA
levels were measured in 42 paired samples. Of these, HBx mRNA was
only detected in 23 pairs. In these 23 HBx-positive patients, there
was no significant difference in HBx mRNA levels observed between
the tumor and peri-tumor tissues (23.98±42.70 vs. 24.24±34.70;
P=0.527; Fig. 4A). However, HBx mRNA
levels in tumor and peri-tumor tissues were positively correlated
(r=0.587; P=0.003; Fig. 4B).
Furthermore, the levels of cccDNA and MSL2 mRNA in the HBx-positive
group were significantly higher in tumor tissues compared with
those in peri-tumor tissues (38.76±56.85 vs. 10.49±12.60; P=0.026
and 55.17±87.50 vs. 8.09±14.79; P=0.013; Fig. 4C and D, respectively). However, in
the HBx-negative group, these two parameters were not significantly
different between tumor and peri-tumor tissues (31.90±46.27 vs.
13.82±33.23; P=0.064 and 20.95±55.00 vs. 1.19±2.19; P=0.077;
Fig. 4E and F, respectively).
Discussion
HBV is a small enveloped DNA virus that replicates
via an RNA intermediate (18).
Following infection, which is hepatocyte-specific, the capsid is
transported to the nucleus and the relaxed circular (rc) DNA is
released and converted into the persistent form, cccDNA. This
cccDNA serves as a template for the transcription of different
viral RNAs (18,19). The 3.5-kb pregenomic RNA is
encapsulated and reverse-transcribed into new rcDNA. Then,
capsid-containing rcDNA is enveloped and released as newly formed
virions or redirected toward the nucleus to establish a cccDNA
pool. The long half-life of the cccDNA ensures the persistence of
HBV in infected cells (18,20).
As the majority of patients receive different
degrees of antiviral treatment following the diagnosis of hepatitis
B, viral replication activity is inhibited in certain patients
(21). Furthermore, new antiviral
technology can detect low levels of HBV DNA and HBsAg at follow-up,
even in those patients with negative HBsAg and HBV DNA loads
(20,22). A previous study has reported that
HBsAg, HBeAg and HBV DNA are negative in patients with
seroclearance, but that cccDNA is positive in the liver tissues of
all patients (23). As cccDNA
maintains a stable pool in the hepatocyte nucleus, antiviral
therapy cannot completely remove HBV (18). In addition, studies on liver
transplantation for HBV-associated HCC have reported that high
levels of cccDNA in tissues can lead to post-operative HBV
recurrence despite the use of high-dose hepatitis B immunoglobulin
prophylaxis during liver transplantation (24). Furthermore, univariate analyses have
previously revealed significant associations between HCC recurrence
and HBV reinfection (25).
Therefore, the present study hypothesized that cccDNA may be
involved in the development of HBV-associated HCC.
To investigate this hypothesis, the present study
compared cccDNA content in tumor and peri-tumor tissues by dividing
patients with HCC into three groups based on HBeAg status, tumor
size and post-operative HCC recurrence. The results revealed that
HBV cccDNA was present in both tumor and peri-tumor tissues, and
that levels were higher in the former. It was also observed that
cccDNA levels were significantly higher in tumor tissues compared
with levels in peri-tumor tissues of the HBeAg-positive, <3 cm
tumor size and HCC recurrence groups. HCC recurred in 8 of the 14
(57%) HBeAg-positive patients within 18 months of surgery.
Therefore, it was speculated that HBV activity is associated with
the formation of early tumors (tumor size, <3 cm) and tumor
recurrence, and that this association may be due to the presence of
cccDNA levels in liver tissues. Levels of HBeAg as a serological
marker can indicate the state of viral replication activity in
patients with chronic hepatitis B (26). A number of studies have reported that
viral replication is more robust in HBeAg-positive patients, which
leads to inflammatory liver injury (27–29) and
a higher risk of tumorigenesis compared with HBeAg-negative
patients (29). It has also been
suggested that HBeAg-positive patients are at risk of early
post-operative HCC recurrence (30–32).
Therefore, the present study hypothesized that HBV cccDNA may exert
a tumorigenic effect on HBV-associated HCC, particularly in
HBeAg-positive patients with strong viral activity. Furthermore,
cccDNA, MSL2 mRNA and HBx mRNA levels were compared in tumor and
peri-tumor tissues in patients with HBV-associated HCC who
exhibited preoperative cirrhosis. It was revealed that, in patients
with cirrhosis, the levels of cccDNA and MSL2 mRNA were also
significantly higher in tumor tissues compared with levels in
peri-tumor tissues. However, HBx mRNA was not significantly
different between the two tissue types. These results further
indicate that cccDNA and MSL2 are oncogenic during the development
of HBV-associated HCC.
The present study assessed whether the cccDNA, MSL2
and HBx levels were changed by antiviral therapy. The results
revealed no significant difference in cccDNA and MSL2 mRNA levels
between the antiviral treatment and untreated groups. However, HBx
mRNA in the tumor and peri-tumor tissues of the antiviral treatment
group was significantly lower compared with that in the untreated
group. These results suggest that antiviral therapy can affect the
tumorigenic effect of cccDNA and MSL2 by decreasing the amount of
HBx. Therefore, the present study further verified the association
between levels of cccDNA and MSL2 mRNA in the HBx-positive and
HBx-negative groups.
The present study detected the levels of HBx and
MSL2 mRNA in 42 pairs of tumor and peri-tumor tissue. The results
revealed that HBx mRNA was detectable in 23 of these tissue pairs,
but not in the other 19. The HBx mRNA levels in 23 pairs of tumor
and peri-tumor tissues were not significantly different; however,
in terms of expression, there was a positive correlation between
tumor and peri-tumor tissues. Furthermore, in the HBx-positive
group, the levels of cccDNA and MSL2 mRNA were significantly higher
in tumor tissues compared with those in peri-tumor tissues.
However, in the HBx-negative group, the levels of these markers
were not significantly different between these two types of tissue.
These results suggest that in the presence of HBx, cccDNA and MSL2
may regulate the formation of HBV-associated HCC. Gao et al
(13) reported that HBx-elevated
MSL2 mRNA led to the activation of HBV cccDNA in hepatoma cells,
which promoted hepatocarcinogenesis and formed a positive feedback
loop of HBx/MSL2/cccDNA/HBV. However, the molecular mechanism
underlying the involvement of cccDNA in patients with
HBV-associated HCC receiving antiviral therapy remains unclear.
Furthermore, regional differences are also an influencing factor
for liver cancer. Therefore, the molecular mechanism underlying the
effects of antiviral therapy on tumorigenesis were analyzed from
the perspective of cccDNA, MSL2 and HBx in patients from the Seoul
National University College of Medicine. The results from the
present study are also consistent with those in previous studies,
as cccDNA was significantly higher in the tumor tissues compared
with that in peri-tumor tissues. Furthermore, it was demonstrated
that antiviral therapy can inhibit HBx mRNA expression, but does
not directly affect tumor cccDNA and MSL2 levels. These findings
provide a guide for future research on antiviral therapy for
HBV-associated HCC.
One limitation of the present study was that the
specimens collected were taken only once from explanted liver
samples during surgery. Since biopsy specimens were not obtained
during pre- and postoperative follow-up, changes in cccDNA levels
in the liver tissue during tumorigenesis could not be determined,
and levels of cccDNA, MSL2 mRNA and HBx mRNA during tumorigenesis
could only be inferred from intraoperative specimens. In addition,
due to insufficient samples in the present study, we were unable to
observe expression of MSL2 and HBx at the protein level. Thus,
further research is required in order to address this issue.
Another limitation of the present study is that the association
between cccDNA and MSL2 mRNA was not analyzed at the sequence
level. Certain key mutations in HBV cccDNA have been reported to
have prognostic value for patients with HBV-associated HCC
(33–35). Therefore, further research should
include a genetic analysis of HBV cccDNA mutations to fully
elucidate the association between HBV cccDNA and HBV-associated
HCC.
In summary, cccDNA may promote the development of
HBV-associated HCC, particularly in HBeAg-positive patients with
high levels of viral replication. This phenomenon may occur as a
result of crosstalk between HBx, MSL2 and cccDNA. Decreasing HBx
levels via antiviral therapy may inhibit this process. However,
this should be investigated by performing additional experiments,
such as studying the interaction between cccDNA and MSL2, and
performing genetic analyses on HBV cccDNA mutations.
Acknowledgements
The authors would like to thank researchers, Ms
Kyung-ae Kim, Ms Soo-in Seo and Ms Min-Young Park, Seoul National
University Hospital, for providing technical assistance in the
present study.
Funding
The present study was supported by the Seoul
National University Hospital research fund (grant no.
3020140190).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XJ, NY, KL and KS designed the study. XJ and SL
performed the experiments. XJ, SH, HK, NY, KL and KS analyzed the
data. XJ, SH and KS wrote the article. All authors approved the
final published version of this article.
Ethics approval and consent to
participate
The experimental protocol was approved by the
Institutional Review Board of Seoul National University Hospital
(IRB no. H-1809-001-967). Written consent, approving the use of
tissues for research following surgery, was obtained from each
patient included in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
cccDNA
|
covalently closed circular DNA
|
|
HBV
|
hepatitis B virus
|
|
HBsAg
|
hepatitis B surface antigen
|
|
HBeAg
|
hepatitis B e antigen
|
|
HBV DNA
|
hepatitis B virus total DNA
|
|
HBx
|
HBV-encoded X protein
|
|
HCC
|
hepatocellular carcinoma
|
|
MSL2
|
male specific lethal 2
|
References
|
1
|
Aljarbou AN: The emergent concern of
Hepatitis B globally with special attention to Kingdom of Saudi
Arabia. Int J Health Sci (Qassim). 7:333–340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allweiss L and Dandri M: The role of
cccDNA in HBV maintenance. Viruses. 9(pii): E1562017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lupberger J and Hildt E: Hepatitis B
virus-induced oncogenesis. World J Gastroenterol. 13:74–81. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blum HE: Hepatocellular carcinoma: Therapy
and prevention. World J Gastroenterol. 11:7391–7400.
2005.PubMed/NCBI
|
|
5
|
Wu M, Li J, Yue L, Bai L, Li Y, Chen J,
Zhang X and Yuan Z: Establishment of Cre-mediated HBV recombinant
cccDNA (rcccDNA) cell line for cccDNA biology and antiviral
screening assays. Antiviral Res. 152:45–52. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mazet-Wagner AA, Baclet MC, Loustaud-Ratti
V, Denis F and Alain S: Real-time PCR quantitation of hepatitis B
virus total DNA and covalently closed circular DNA in peripheral
blood mononuclear cells from hepatitis B virus-infected patients. J
Virol Methods. 138:70–79. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wong DK, Yuen MF, Yuan H, Sum SS, Hui CK,
Hall J and Lai CL: Quantitation of covalently closed circular
hepatitis B virus DNA in chronic hepatitis B patients. Hepatology.
40:727–737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ng SA and Lee C: Hepatitis B virus X gene
and hepatocarcinogenesis. J Gastroenterol. 46:974–990. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang X, Zhang H and Ye L: Effects of
hepatitis B virus X protein on the development of liver cancer. J
Lab Clin Med. 147:58–66. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding R, Han S, Lu Y, Guo C, Xie H, Zhang
N, Song Z, Cai L, Liu J and Dou K: Overexpressed Id-1 is associated
with patient prognosis and HBx expression in hepatitis B
virus-related hepatocellular carcinoma. Cancer Biol Ther.
10:299–307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rabut G and Peter M: Function and
regulation of protein neddylation. ‘Protein modifications: Beyond
the usual suspects’ review series. EMBO Rep. 9:969–976. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villa R, Forne I, Muller M, Imhof A,
Straub T and Becker PB: MSL2 combines sensor and effector functions
in homeostatic control of the Drosophila dosage compensation
machinery. Mol Cell. 48:647–654. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao Y, Feng J, Yang G, Zhang S, Liu Y, Bu
Y, Sun M, Zhao M, Chen F, Zhang W, et al: Hepatitis B virus X
protein-elevated MSL2 modulates hepatitis B virus covalently closed
circular DNA by inducing degradation of APOBEC3B to enhance
hepatocarcinogenesis. Hepatology. 66:1413–1429. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bowden S, Jackson K, Littlejohn M and
Locarnini S: Quantification Of HBV covalently closed circular DNA
from liver tissue by real-time PCR. Methods Mol Med. 95:41–50.
2004.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duan BW, Lu SC, Lai W, Liu XE and Liu Y:
The detection of (total and ccc) HBV DNA in liver transplant
recipients with hepatitis B vaccine against HBV reinfection. Hum
Vaccin Immunother. 11:2490–2494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim JW, Lee SH, Park YS, Hwang JH, Jeong
SH, Kim N and Lee DH: Replicative activity of hepatitis B virus is
negatively associated with methylation of covalently closed
circular DNA in advanced hepatitis B virus infection.
Intervirology. 54:316–325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schreiner S and Nassal M: A role for the
Host DNA damage response in hepatitis B virus cccDNA formation-and
beyond? Viruses. 9(pii): E1252017. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu S, Li N, Zhou PC, Huang Y, Zhou RR and
Fan XG: Detection of HBV DNA and antigens in HBsAg-positive
patients with primary hepatocellular carcinoma. Clin Res Hepatol
Gastroenterol. 41:415–423. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chuaypen N, Sriprapun M, Praianantathavorn
K, Payungporn S, Wisedopas N, Poovorawan Y and Tangkijvanich P:
Kinetics of serum HBsAg and intrahepatic cccDNA during pegylated
interferon therapy in patients with HBeAg-positive and
HBeAg-negative chronic hepatitis B. J Med Virol. 89:130–138. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pichoud C, Berby F, Stuyver L, Petit MA,
Trepo C and Zoulim F: Persistence of viral replication after
anti-HBe seroconversion during antiviral therapy for chronic
hepatitis B. J Hepatol. 32:307–316. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roche B, Feray C, Gigou M, Roque-Afonso
AM, Arulnaden JL, Delvart V, Dussaix E, Guettier C, Bismuth H and
Samuel D: HBV DNA persistence 10 years after liver transplantation
despite successful anti-HBS passive immunoprophylaxis. Hepatology.
38:86–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suzuki F, Miyakoshi H, Kobayashi M and
Kumada H: Correlation between serum hepatitis B virus core-related
antigen and intrahepatic covalently closed circular DNA in chronic
hepatitis B patients. J Med Virol. 81:27–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chauhan R, Lingala S, Gadiparthi C, Lahiri
N, Mohanty SR, Wu J, Michalak TI and Satapathy SK: Reactivation of
hepatitis B after liver transplantation: Current knowledge,
molecular mechanisms and implications in management. World J
Hepatol. 10:352–370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song GW, Ahn CS, Lee SG, Hwang S, Kim KH,
Moon DB, Ha TY, Jung DH, Park GC, Kang SH, et al: Correlation
between risk of hepatitis B virus recurrence and tissue expression
of covalently closed circular DNA in living donor liver transplant
recipients treated with high-dose hepatitis B immunoglobulin.
Transplant Proc. 46:3548–3553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nguyen MH and Keeffe EB: Are hepatitis B e
antigen (HBeAg)-positive chronic hepatitis B and HBeAg-negative
chronic hepatitis B distinct diseases? Clin Infect Dis.
47:1312–1314. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ganem D and Prince AM: Hepatitis B virus
infection-natural history and clinical consequences. N Engl J Med.
350:1118–29. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Q, Lin L, Yoo S, Wang W, Blank S,
Fiel MI, Kadri H, Luan W, Warren L, Zhu J and Hiotis SP: Impact of
non-neoplastic vs intratumoural hepatitis B viral DNA and
replication on hepatocellular carcinoma recurrence. Br J Cancer.
115:841–847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang HI, Lu SN, Liaw YF, You SL, Sun CA,
Wang LY, Hsiao CK, Chen PJ, Chen DS and Chen CJ; Taiwan
Community-Based Cancer Screening Project Group, : Hepatitis B e
antigen and the risk of hepatocellular carcinoma. N Engl J Med.
347:168–174. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun HC, Zhang W, Qin LX, Zhang BH, Ye QH,
Wang L, Ren N, Zhuang PY, Zhu XD, Fan J and Tang ZY: Positive serum
hepatitis B e antigen is associated with higher risk of early
recurrence and poorer survival in patients after curative resection
of hepatitis B-related hepatocellular carcinoma. J Hepatol.
47:684–690. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kubo S, Hirohashi K, Yamazaki O, Matsuyama
M, Tanaka H, Horii K, Shuto T, Yamamoto T, Kawai S, Wakasa K, et
al: Effect of the presence of hepatitis B e antigen on prognosis
after liver resection for hepatocellular carcinoma in patients with
chronic hepatitis B. World J Surg. 26:555–560. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shirabe K, Kanematsu T, Matsumata T,
Adachi E, Akazawa K and Sugimachi K: Factors linked to early
recurrence of small hepatocellular carcinima after hepatectomy:
Univariate and multivarate analysis. Hepatology. 14:802–805. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiao F, Long L, Ding S, Xie X, Jia L and
Lu F: Complete genome sequencing and clinical analysis of
intrahepatic hepatitis B virus cccDNA from HCC. Microb Pathog.
109:49–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang G, Han M, Chen F, Xu Y, Chen E, Wang
X, Liu Y, Sun J, Hou J, Ning Q and Wang Z: Hepatitis B virus
genotype B and mutations in basal core promoter and pre-core/core
genes associated with acute-on-chronic liver failure: A multicenter
cross-sectional study in China. Hepatol Int. 8:508–516. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu H, Zhao M, Lou G, Zheng M, Cao Q and
Chen Z: New point mutations in surface and core genes of hepatitis
B virus associated with acute on chronic liver failure identified
by complete genomic sequencing. PLoS One. 10:e01231392015.
View Article : Google Scholar : PubMed/NCBI
|