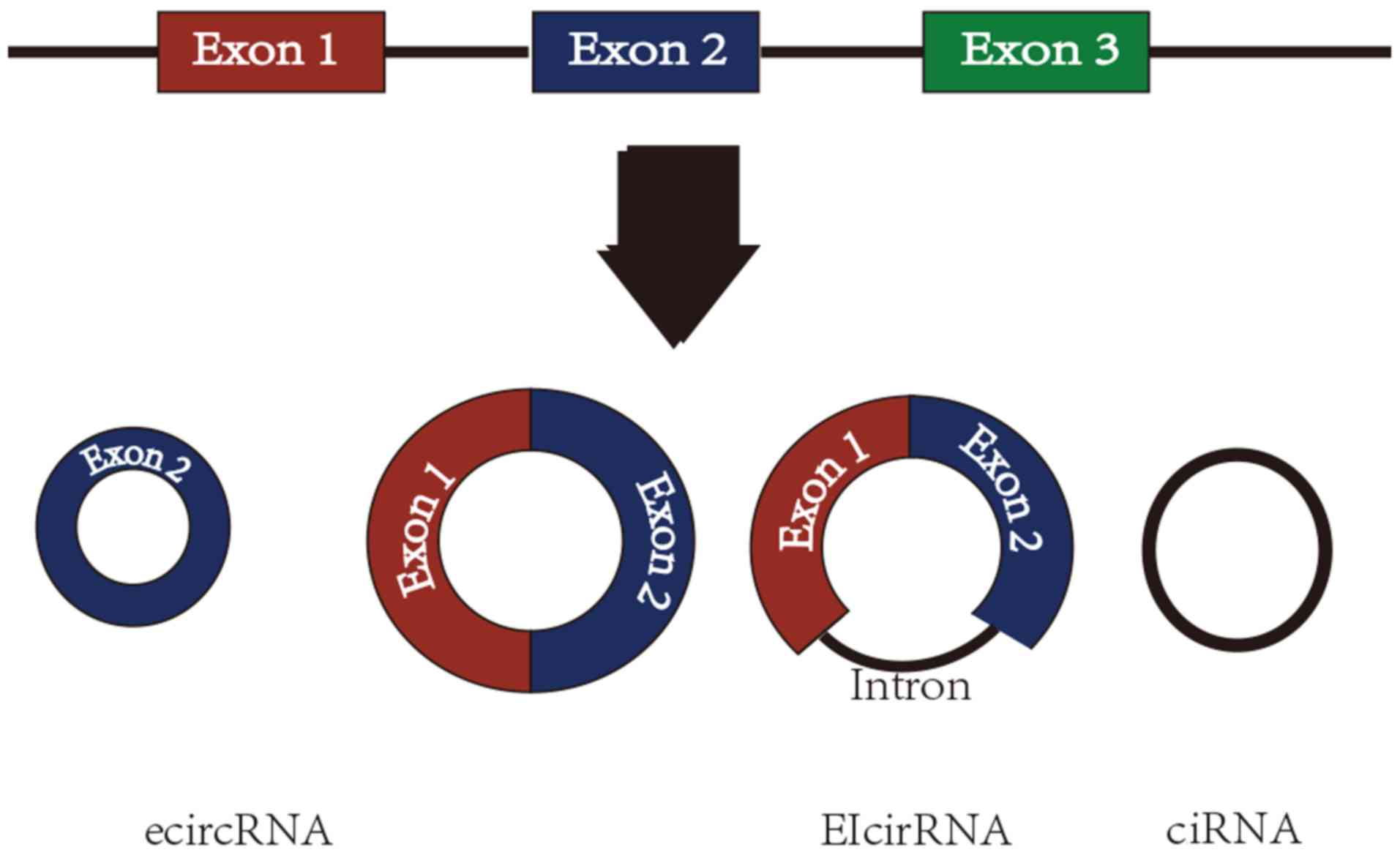
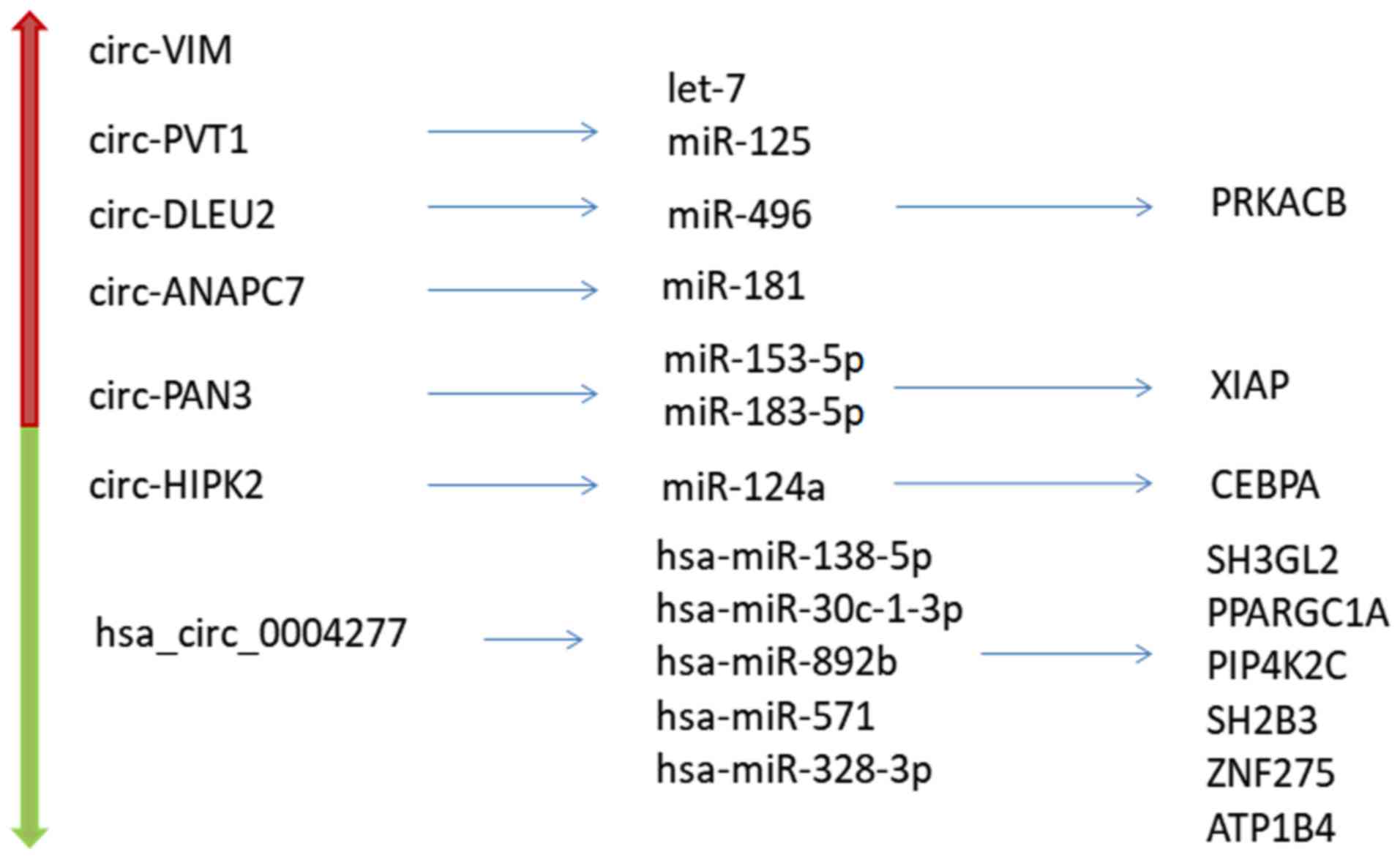
|
1
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu Z, Yan Y, Zeng S, Dai S, Chen X, Wei J
and Gong Z: Circular RNAs: Clinical relevance in cancer.
Oncotarget. 9:1444–1460. 2017.PubMed/NCBI
|
|
6
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kelly S, Greenman C, Cook PR and
Papantonis A: Exon skipping is correlated with exon
circularization. J Mol Biol. 427:2414–2417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kolakofsky D: Isolation and
characterization of sendai virus DI-RNAs. Cell. 8:547–555. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Z, Xie Q, He D, Ling Y, Li Y, Li J
and Zhang H: Circular RNA: New star, new hope in cancer. BMC
Cancer. 18:8342018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
16
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The output of protein-coding genes
shifts to circular RNAs when the Pre-mRNA processing machinery is
limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abe N, Hiroshima M, Maruyama H, Nakashima
Y, Nakano Y, Matsuda A, Sako Y, Ito Y and Abe H: Rolling circle
amplification in a prokaryotic translation system using small
circular RNA. Angew Chem Int Ed Engl. 52:7004–7008. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rybak-Wolf A, Stottmeister C, Glazar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Enuka Y, Lauriola M, Feldman ME, Sas-Chen
A, Ulitsky I and Yarden Y: Circular RNAs are long-lived and display
only minimal early alterations in response to a growth factor.
Nucleic Acids Res. 44:1370–1383. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Yang L and Chen LL: The biogenesis,
functions and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S and Han L: Circular RNAs as promising
biomarkers in cancer: Detection, function, and beyond. Genome Med.
11:152019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao Y and Zhao F: Computational strategies
for exploring circular RNAs. Trends Genet. 34:389–400. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong R, Ma XK, Li GW and Yang L: CIRCpedia
v2: An updated database for comprehensive circular RNA annotation
and expression comparison. Genomics Proteomics Bioinformatics.
16:226–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44:D209–D215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu SM, Liu H, Huang PJ, Chang IY, Lee CC,
Yang CY, Tsai WS and Tan BC: circlncRNAnet: An integrated web-based
resource for mapping functional networks of long or circular forms
of noncoding RNAs. Gigascience. 7:1–10. 2018.
|
|
35
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan C, Lei X, Fang Z, Jiang Q and Wu FX:
CircR2Disease: A manually curated database for experimentally
supported circular RNAs associated with various diseases. Database
(Oxford). Jan 1–2018.(Epub ahead of print). doi:
10.1093/database/bay044. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J,
Xiang Y, Liu L, Zhong S, Han L and He C: Comprehensive
characterization of tissue-specific circular RNAs in the human and
mouse genomes. Brief Bioinform. 18:984–992. 2017.PubMed/NCBI
|
|
38
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46:D925–D929.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen B, Wang Z, Li Z, Song H and Ding X:
Circular RNAs: An emerging landscape in tumor metastasis. Am J
Cancer Res. 9:630–643. 2019.PubMed/NCBI
|
|
40
|
Maia J, Caja S, Strano Moraes MC, Couto N
and Costa-Silva B: Exosome-based cell-cell communication in the
tumor microenvironment. Front Cell Dev Biol. 6:182018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fanale D, Taverna S, Russo A and Bazan V:
Circular RNA in exosomes. Adv Exp Med Biol. 1087:109–117. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu H, Gong Z, Shen Y, Fang Y and Zhong S:
Circular RNA expression in extracellular vesicles isolated from
serum of patients with endometrial cancer. Epigenomics. 10:187–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang ZK, Yao FY, Xu JQ, Deng Z, Su RG,
Peng YP, Luo Q and Li JM: Microarray expression profile of circular
RNAs in peripheral blood mononuclear cells from active tuberculosis
patients. Cell Physiol Biochem. 45:1230–1240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Yong K, Wong DT and Xiao X: The landscape of MicroRNA,
Piwi-interacting RNA and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu J, Han Q, Gu Y, Ma J, Mcgrath M, Qiao
F, Chen B, Song C and Ge Z: Circular RNA PVT1 expression and its
roles in acute lymphoblastic leukemia. Epigenomics. 10:723–732.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Galasso M, Costantino G, Pasquali L,
Minotti L, Baldassari F, Corra F, Agnoletto C and Volinia S:
Profiling of the predicted circular RNAs in ductal in situ and
invasive breast cancer: A pilot study. Int J Genomics.
2016:45038402016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Szymczyk A, Macheta A and Podhorecka M:
Abnormal microRNA expression in the course of hematological
malignancies. Cancer Manag Res. 10:4267–4277. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hassan S and Smith M: Acute myeloid
leukaemia. Hematology. 19:493–494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Estey E and Dohner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Short NJ, Rytting ME and Cortes JE: Acute
myeloid leukaemia. Lancet. 392:593–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dolnik A, Engelmann JC,
Scharfenberger-Schmeer M, Mauch J, Kelkenberg-Schade S, Haldemann
B, Fries T, Krönke J, Kühn MW, Paschka P, et al: Commonly altered
genomic regions in acute myeloid leukemia are enriched for somatic
mutations involved in chromatin remodeling and splicing. Blood.
120:e83–e92. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wu S, Du Y, Beckford J and Alachkar H:
Upregulation of the EMT marker vimentin is associated with poor
clinical outcome in acute myeloid leukemia. J Transl Med.
16:1702018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yi YY, Yi J, Zhu X, Zhang J, Zhou J, Tang
X, Lin J, Wang P and Deng ZQ: Circular RNA of vimentin expression
as a valuable predictor for acute myeloid leukemia development and
prognosis. J Cell Physiol. 234:3711–3719. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Storlazzi CT, Fioretos T, Surace C, Lonoce
A, Mastrorilli A, Strombeck B, D'Addabbo P, Iacovelli F, Minervini
C, Aventin A, et al: MYC-containing double min in hematologic
malignancies: Evidence in favor of the episome model and exclusion
of MYC as the target gene. Hum Mol Genet. 15:933–942. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chinen Y, Sakamoto N, Nagoshi H, Taki T,
Maegawa S, Tatekawa S, Tsukamoto T, Mizutani S, Shimura Y,
Yamamoto-Sugitani M, et al: 8q24 amplified segments involve novel
fusion genes between NSMCE2 and long noncoding RNAs in acute
myelogenous leukemia. J Hematol Oncol. 7:682014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qin S, Zhao Y, Lim G, Lin H and Zhang X
and Zhang X: Circular RNA PVT1 acts as a competing endogenous RNA
for miR-497 in promoting non-small cell lung cancer progression.
Biomed Pharmacother. 111:244–250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Verduci L, Ferraiuolo M, Sacconi A, Ganci
F, Vitale J, Colombo T, Paci P, Strano S, Macino G, Rajewsky N and
Blandino G: The oncogenic role of circPVT1 in head and neck
squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD
transcription-competent complex. Genome Biol. 18:2372017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Panda AC, Grammatikakis I, Kim KM, De S,
Martindale JL, Munk R, Yang X, Abdelmohsen K and Gorospe M:
Identification of senescence-associated circular RNAs (SAC-RNAs)
reveals senescence suppressor CircPVT1. Nucleic Acids Res.
45:4021–4035. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C
and Ma D: Characterization of hsa_circ_0004277 as a new biomarker
for acute myeloid leukemia via circular RNA Profile and
bioinformatics analysis. Int J Mol Sci. 18(pii): E5972017.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hirsch S, Blätte TJ, Grasedieck S,
Cocciardi S, Rouhi A, Jongen-Lavrencic M, Paschka P, Krönke J,
Gaidzik VI, Döhner H, et al: Circular RNAs of the nucleophosmin
(NPM1) gene in acute myeloid leukemia. Haematologica.
102:2039–2047. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Okamoto M, Hirai H, Taniguchi K, Shimura
K, Inaba T, Shimazaki C, Taniwaki M and Imanishi J: Toll-like
receptors (TLRs) are expressed by myeloid leukaemia cell lines, but
fail to trigger differentiation in response to the respective TLR
ligands. Br J Haematol. 147:585–587. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Nagai Y, Garrett KP, Ohta S, Bahrun U,
Kouro T, Akira S, Takatsu K and Kincade PW: Toll-like receptors on
hematopoietic progenitor cells stimulate innate immune system
replenishment. Immunity. 24:801–812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wu DM, Wen X, Han XR, Wang S, Wang YJ,
Shen M, Fan SH, Zhang ZF, Shan Q, Li MQ, et al: Role of circular
RNA DLEU2 in human acute myeloid leukemia. Mol Cell Biol. 38(pii):
e00259–18. 2018.PubMed/NCBI
|
|
71
|
Chen H, Liu T, Liu J, Feng Y, Wang B, Wang
J, Bai J, Zhao W, Shen Y, Wang X, et al: Circ-ANAPC7 is upregulated
in acute myeloid leukemia and appears to target the MiR-181 family.
Cell Physiol Biochem. 47:1998–2007. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ
and Wu WB: CircPAN3 mediates drug resistance in acute myeloid
leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol.
70:42–54.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
de Thé H and Chen Z: Acute promyelocytic
leukaemia: Novel insights into the mechanisms of cure. Nat Rev
Cancer. 10:775–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li S, Ma Y, Tan Y, Ma X, Zhao M, Chen B,
Zhang R, Chen Z and Wang K: Profiling and functional analysis of
circular RNAs in acute promyelocytic leukemia and their dynamic
regulation during all-trans retinoic acid treatment. Cell Death
Dis. 9:6512018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Huang R, Zhang Y, Han B, Bai Y, Zhou R,
Gan G, Chao J, Hu G and Yao H: Circular RNA HIPK2 regulates
astrocyte activation via cooperation of autophagy and ER stress by
targeting MIR124-2HG. Autophagy. 13:1722–1741. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hackanson B, Bennett KL, Brena RM, Jiang
J, Claus R, Chen SS, Blagitko-Dorfs N, Maharry K, Whitman SP,
Schmittgen TD, et al: Epigenetic modification of CCAAT/enhancer
binding protein alpha expression in acute myeloid leukemia. Cancer
Res. 68:3142–3151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kaleem B, Shahab S, Ahmed N and Shamsi TS:
Chronic myeloid leukemia-prognostic value of mutations. Asian Pac J
Cancer Prev. 16:7415–7423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhou T, Medeiros LJ and Hu S: Chronic
myeloid leukemia: Beyond BCR-ABL1. Curr Hematol Malig Rep.
13:435–445. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Okcanoğlu TB and Gündüz C: Circular RNAs
in leukemia. Biomed Rep. 1–5. 2019.
|
|
80
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Deininger MW, Goldman JM and Melo JV: The
molecular biology of chronic myeloid leukemia. Blood. 96:3343–3356.
2000.PubMed/NCBI
|
|
82
|
McGahon A, Bissonnette R, Schmitt M,
Cotter KM, Green DR and Cotter TG: BCR-ABL maintains resistance of
chronic myelogenous leukemia cells to apoptotic cell death. Blood.
83:1179–1187. 1994.PubMed/NCBI
|
|
83
|
Mauro MJ: Defining and managing imatinib
resistance. Hematol Am Soc Hematol Educ Program. 219–225. 2006.
View Article : Google Scholar
|
|
84
|
Pan Y, Lou J, Wang H, An N, Chen H, Zhang
Q and Du X: CircBA9.3 supports the survival of leukaemic cells by
up-regulating c-ABL1 or BCR-ABL1 protein levels. Blood cells Mol
Dis. 73:38–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu J, Kong F, Lou S, Yang D and Gu L:
Global identification of circular RNAs in chronic myeloid leukemia
reveals hsa_circ_0080145 regulates cell proliferation by sponging
miR-29b. Biochem Biophy Res Commun. 504:660–665. 2018. View Article : Google Scholar
|
|
86
|
Musolino C, Oteri G, Allegra A, Mania M,
D'Ascola A, Avenoso A, Innao V, Allegra AG and Campo S: Altered
microRNA expression profile in the peripheral lymphoid compartment
of multiple myeloma patients with bisphosphonate-induced
osteonecrosis of the jaw. Ann Hematol. 97:1259–1269. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Du XJ, Lu JM and Sha Y: MiR-181a inhibits
vascular inflammation induced by ox-LDL via targeting TLR4 in human
macrophages. J Cell Physiol. 233:6996–7003. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Litwińska Z and Machaliński B: miRNAs in
chronic myeloid leukemia: Small molecules, essential function. Leuk
Lymphoma. 58:1297–1305. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zenz T, Mertens D, Kuppers R, Döhner H and
Stilgenbauer S: From pathogenesis to treatment of chronic
lymphocytic leukaemia. Nat Rev Cancer. 10:37–50. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Rai KR and Jain P: Chronic lymphocytic
leukemia (CLL)-then and now. Am J Hematol. 91:330–340. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Xia L, Wu L, Bao J, Li Q, Chen X, Xia H
and Xia R: Circular RNA circ-CBFB promotes proliferation and
inhibits apoptosis in chronic lymphocytic leukemia through
regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res
Commun. 503:385–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Franiak-Pietryga I, Maciejewski H, Ziemba
B, Appelhans D, Voit B, Robak T, Jander M, Treliński J, Bryszewska
M and Borowiec M: Blockage of Wnt/β-catenin signaling by
nanoparticles reduces survival and proliferation of CLL cells in
vitro-preliminary study. Macromol Biosci. 172017.doi:
10.1002/mabi.201700130.
|
|
93
|
Dahl M, Daugaard I, Andersen MS, Hansen
TB, Grønbæk K, Kjems J and Kristensen LS: Enzyme-free digital
counting of endogenous circular RNA molecules in B-cell
malignancies. Lab Invest. 98:1657–669. 2018. View Article : Google Scholar : PubMed/NCBI
|