Introduction
During cell differentiation and development, RNA
levels are inevitably regulated in response to environmental cues.
Among the different mechanisms of RNA regulation, RNA degradation
is an important method of gene expression control (1). In eukaryotic cells, the processes of
mRNA degradation are generally divided into the 5′ to 3′ and the 3′
to 5′pathways (1). In yeast, mRNA
degradation also proceeds via two pathways: Rapid 5′ to
3′degradation by removing the 5′-cap structure, and slower 3′ to
5′degradation mediated by the exosome complex (2). In human cells, exosome-mediated 3′ to
5′degradation is the predominant pathway (3–5). The
exosome complex (EXOSC), also known as the polymyositis/scleroderma
complex, is a multi-protein intracellular complex capable of
degrading various types of RNA. The core of the exosome is a ring
to which other proteins are attached (6). Exosome component 2 (EXOSC2), as a
peripheral section of exosome component 9 (EXOSC9), combines with
exosome component 4 (EXOSC4) and exosome component 7 (EXOSC7) to
stabilize the ring of RNase PH-domain subunits (7). Furthermore, EXOSC4, exosome component 8
(EXOSC8) and EXOSC9 bind to adenylate-uridylate-rich
element-containing RNAs (8). EXOSC10
is involved in the maturation of 5.8S ribosomal (r)RNA (9). In 2017, two study groups demonstrated
that RNA stability regulation has an important role in
hematopoietic stem and progenitor cell differentiation and T-cell
homeostasis (10,11).
Mantle cell lymphoma (MCL) comprises 3–10% of all
non-Hodgkin B-cell lymphoma cases, as a distinct entity with a t
(q13;q32) translocation event (12–18).
Among patients with MCL, 60–65-year-old males represent the
predominant patient type, and the majority of cases are diagnosed
at the advanced stage with extranodal involvement (15). At present, various treatments are
used for MCL: Chemotherapy; bone marrow transplantation;
radiotherapy; immunotherapy; and targeted therapy. However, to
date, no consensus regarding the optimal treatment has been reached
among physicians, and rapid relapses hamper their curative effect
(19). As a result, MCL has a
relatively short median survival time of 5–7 years (20–22). To
evaluate and select appropriate treatments for patients with MCL,
several previous studies have investigated various approaches to
stratify patients, including gene expression analysis of 20
‘proliferation signature’ genes (23), a PCR-based 5-gene model (24), the antigen Ki-67 (Ki-67)
proliferation index (25) and the
Mantle Cell International Prognostic Index (MIPI) (26). However, while all of these prognostic
factors are associated with survival to a certain extent, none of
them has been proven to be an effective tool for the selection of
therapy. Therefore, it is necessary to explore the molecular
mechanisms of MCL progression further, and to identify novel
treatment approaches for MCL. It has been suggested that mutations
in exosome component 3 (EXOSC3) may cause spinal motor neuron
disease and cerebellar atrophy (27). However, little is known about the
prognostic significance of EXOSC family genes in MCL. In the
present study, the prognostic significance and biological
implication of EXOSC genes in MCL were investigated by using a
Bioinformatics analysis of gene expression data.
Materials and methods
Data sources
Affymetrix Human Genome U133 Plus 2.0 Array datasets
were obtained from the NCBI Gene Expression Omnibus (GEO) database
(28). The datasets GSE93291 and
GSE36000 were used, containing the gene expression data of 123 and
38 MCL patients, respectively.
Gene expression analysis
The gene expression data in each probe set of all
the arrays were computed using the robust multiarray averaging
algorithm. Log2-transformed values were used to represent the
relative RNA expression value of each probe set or gene. An
unpaired Student's t-test was used to identify the differentially
expressed genes. P<0.05 was considered to indicate a
statistically significant difference. Upregulated or downregulated
differentially expressed genes were defined as genes with a log2
(fold change) of >1 or <-1, respectively.
Pearson's correlation coefficient was used to
calculate the correlation coefficient of the expression level of 10
EXOSC genes in MCL. The 123 patients with MCL were divided into two
groups using a fuzzy clustering method in the ggplot2 package
(version 3.1.1; ggplot2.tidyverse.org), based on the gene expression
of the 10 EXOSC genes. To determine the association of EXOSC gene
expression with survival of patients with MCL, Kaplan-Meier curves
were calculated and log-rank tests was performed. Comparison
between the expression levels of FA complementation group A (FANCA)
and INO80 complex subunit D between EXO.index-high and -low groups
was performed using an unpaired Student's t-test.
Definition of EXO.index for survival
prediction
An EXO.index was defined to predict survival of
patients with MCL. The EXO.index was calculated by using the
following formula:
EXO.indexj=Hj/Fj, where
EXO.indexj represented the index of EXOSC genes of the
jth sample to predict survival. Hj represented the
product of the expression of harmful genes (hazard ratio of >1)
with P<0.05 in the jth sample. A total of 5 of the 10 EXOSC
genes (EXOSC1, EXOSC2, EXOSC4, EXOSC5 and EXOSC7) with a hazard
ratio of >1 were included. Fj represented the product
of the expression of favorable genes (hazard ratio of <1) of the
jth sample. Of the 10 EXOSC genes, 1 gene, EXOSC3, with a hazard
ratio of <1 was included.
Gene ontology (GO) analysis
The Database for Annotation and Visualization and
Integrated Discovery tool was used with default parameters for GO
analysis (29–31). Enriched GO terms (P<0.05)
presented in the figures were manually curated, and only
non-redundant GO terms in the biological process category were
provided (32).
R software (version 3.1.3; www.r-project.org) was used for data analysis. Values
are expressed as the mean ± standard error of the mean in scatter
plots. P<0.05 was considered to indicate a statistically
significant difference.
Results
Survival in MCL predicted by 6 of 10
EXOSC genes
To investigate the association between the EXOSC
genes (EXOSC1-EXOSC10) and the survival of patients with MCL, a
total of 123 MCL expression profiles from the GEO dataset GSE93291
were analyzed. It was identified that the expression levels of 6
out of 10 EXOSC genes were significantly associated with the
survival of patients with MCL (P<0.05; log-rank test). The 10
EXOSC genes were categorized based on the hazard ratio values. The
genes with a hazard ratio of <1 were defined as ‘favorable
genes’, as they had a positive influence on the survival of MCL.
Only EXOSC3, with a hazard ratio of 0.76 [95% confidence interval
(CI), 0.61–0.94], was a significant ‘favorable gene’. Conversely,
the genes with a hazard ratio of >1 were defined as ‘harmful
genes’ due to having a negative effect on the survival of MCL.
Based on this, 5 of the 10 EXOSC genes (EXOSC1, EXOSC2, EXOSC4,
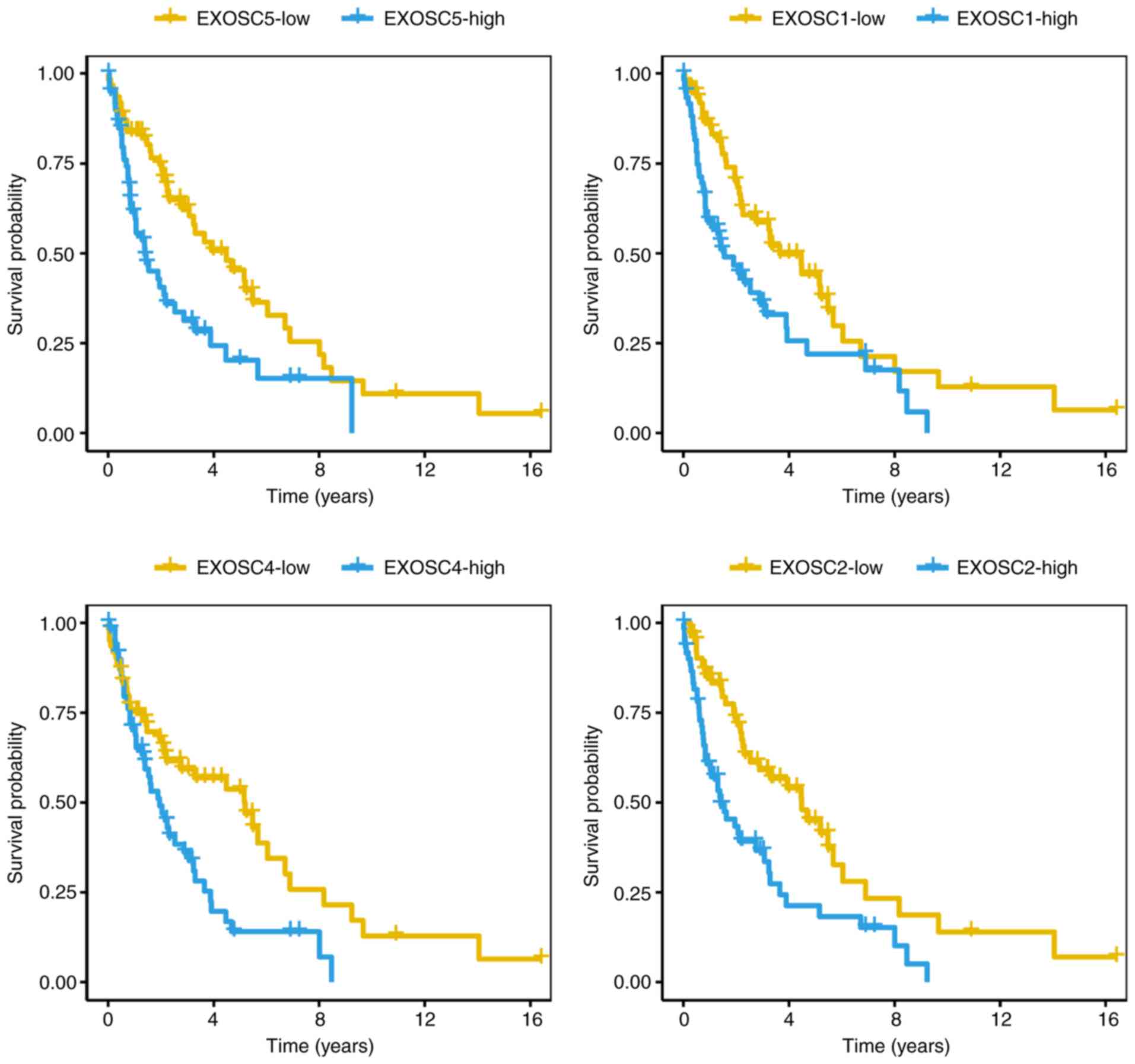
EXOSC5 and EXOSC7) were classified as ‘harmful genes’ (Fig. 1A and B). With a hazard ratio of 5.87
(95% CI, 2.72–12.69), EXOSC1 was the most significant gene among
the ‘harmful genes’. The Kaplan-Meier curves of overall survival
for the 123 patients with MCL were then generated to compare 4
EXOSC genes with the log-rank test values (Fig. 2; EXOSC5, P=7.0×10−7;
EXOSC1, P=5.7×10−6; EXOSC4, P=1.1×10−4;
EXOSC2, P=2.7×10−3; log-rank test), indicating that the
‘harmful’ EXOSC genes were able to predict the survival of patients
with MCL.
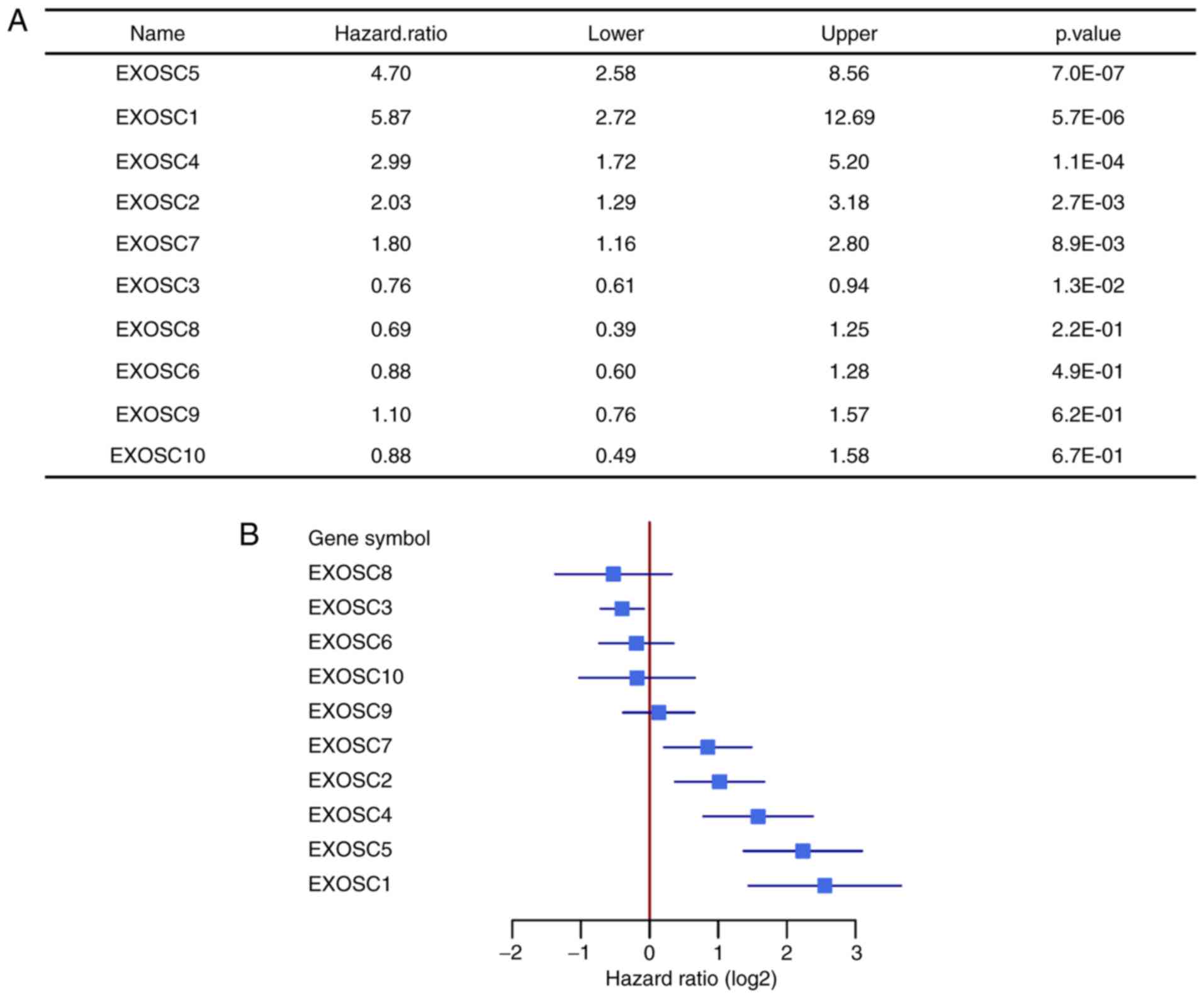 | Figure 1.Survival analysis and forest plot of
10 EXOSC genes of 123 patients with mantle cell lymphoma. (A)
Survival analysis of 10 EXOSC genes, classified by hazard ratio.
(B) Forest plot of 10 EXOSC genes, classified by hazard ratio. The
forest plot includes the lower and upper 95% confidence intervals.
EXOSC, exosome complex. EXOSC5, exosome component 5; EXOSC1,
exosome component 1; EXOSC4, exosome component 4; EXOSC2, exosome
component 2; EXOSC7, exosome component 7; EXOSC3, exosome component
3; EXOSC8, exosome component 8; EXOSC6, exosome component 6;
EXOSC9, exosome component 9; EXOSC10, exosome component 10. |
Correlation of the 10 EXOSC genes in
MCL
Correlation plots of the expression of the 10 EXOSC
genes in MCL are presented in Fig.
3. Of note, certain pairs of EXOSC genes exhibited a positive
correlation, including EXOSC4 and EXOSC5 (R=0.47), EXOSC1 and
EXOSC5 (R=0.44), EXOSC1 and EXOSC2 (R=0.42), and EXOSC4 and EXOSC7
(R=0.35). Other pairs of EXOSC genes had a negative correlation,
including EXOSC7 and EXOSC9 (Rr=−0.49), and EXOSC7 and EXOSC8
(R=−0.36). In addition, certain pairs of EXOSC genes exhibited no
correlation, including EXOSC1 and EXOSC8 (R=−0.02), and EXOSC6 and
EXOSC10 (R=−0.03). The 5 ‘harmful genes’, EXOSC1, EXOSC2, EXOSC4,
EXOSC5 and EXOSC7, exhibited a positive mutual correlation. EXOSC3,
as a ‘favorable gene’, had a weak positive correlation with EXOSC8
(R=0.24), a weak negative correlation with EXOSC1 (R=−0.26) and no
correlation with EXOSC5 (R=−0.08).
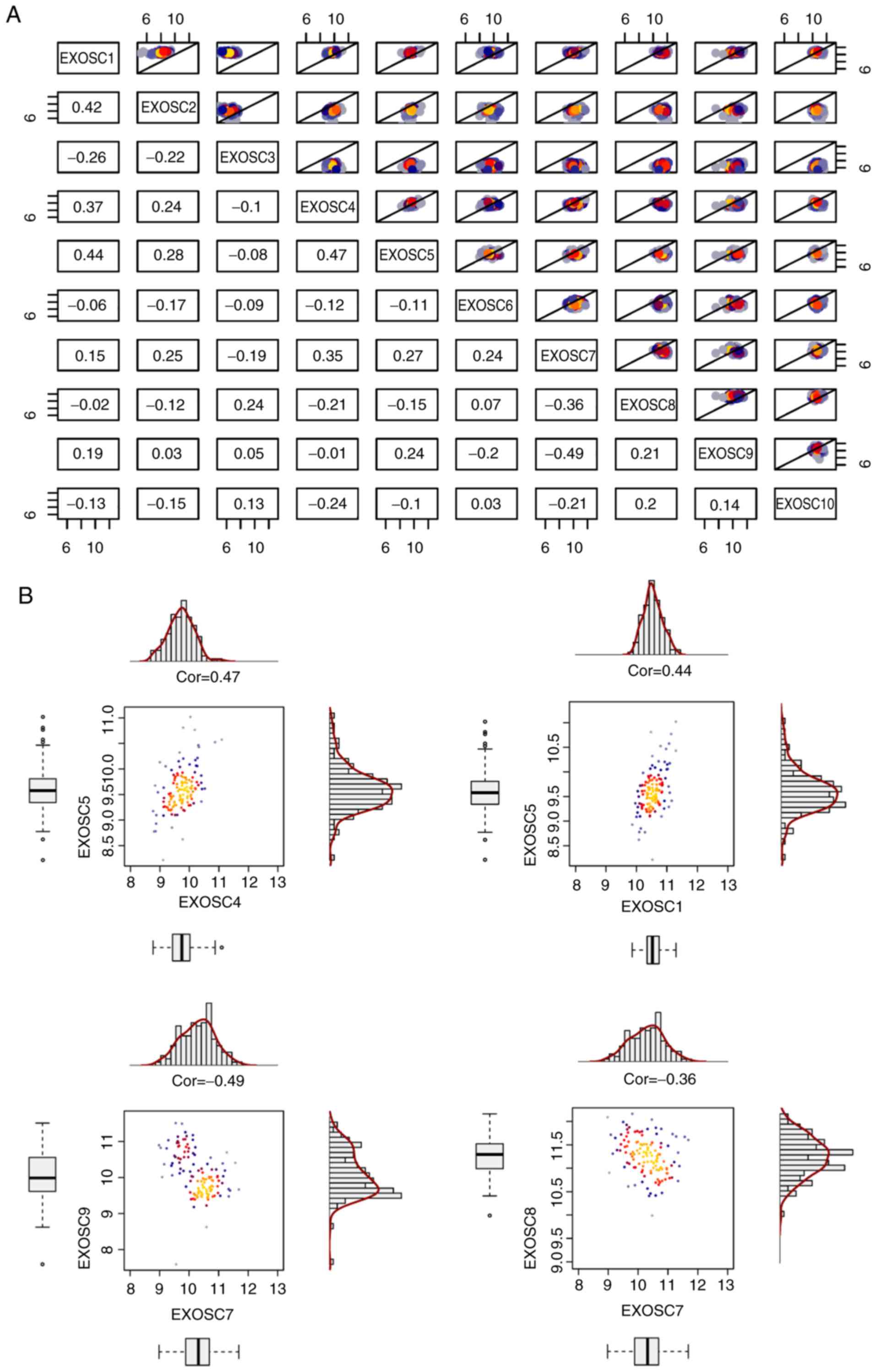 | Figure 3.Correlation plot of the expression
levels of 10 EXOSC genes in mantle cell lymphoma. (A) Correlation
plot of 10 EXOSC genes. The Pearson correlation coefficient is
presented. The X-axis and Y-axis represent gene expression levels
(log2). The colors included in the upper right section of the plot
represent dot density. Grey and blue represent mean low dot density
and red and yellow represent high dot density. (B) Correlations
between EXOSC4 and EXOSC5 (upper left), EXOSC1 and EXOSC5 (upper
right), EXOSC7 and EXOSC9 (lower left), EXOSC7 and EXOSC8 (lower
right) genes were calculated. EXOSC, exosome complex; cor, Pearson
correlation coefficient; EXOSC4, exosome component 4; EXOSC5,
exosome component 5; EXOSC1, exosome component 1; EXOSC7, exosome
component 7; EXOSC9, exosome component 9; EXOSC8, exosome component
8. |
EXOSC gene expression predicts
survival in MCL
Cosine correlation similarity was adopted to perform
unsupervised clustering of the 10 EXOSC genes in the 123 patients
with MCL (Fig. 4A). The results
indicated that the 10 EXOSC genes were significantly clustered into
2 different groups (EXOSC2 and EXOSC3 in one group, and the other 8
EXOSC genes in a second group). Furthermore, all of the ‘harmful
genes’, with the exception of EXOSC2, were in the same group.
Therefore, the EXOSC genes were considered to be associated with
the survival of MCL and also exhibited a characteristic expression
profile. Furthermore, using a fuzzy clustering of the 10 EXOSC
genes, it was possible to stratify the 123 patients with MCL into
two groups (R, ggplot2; Fig. 4B).
The EXO.index was then calculated as the ratio of the expression of
‘harmful genes’ and ‘favorable genes’. The EXO.index was
significantly associated with the survival of MCL
(P=1.73×10−7; log-rank test; Fig. 4C). The EXO.index revealed the unequal
expression levels between the ‘favorable genes’ and the ‘harmful
genes’ among the EXOSC genes. The patients with MCL in the
EXO.index-high group exhibited poorer survival rates compared with
the EXO.index-low group.
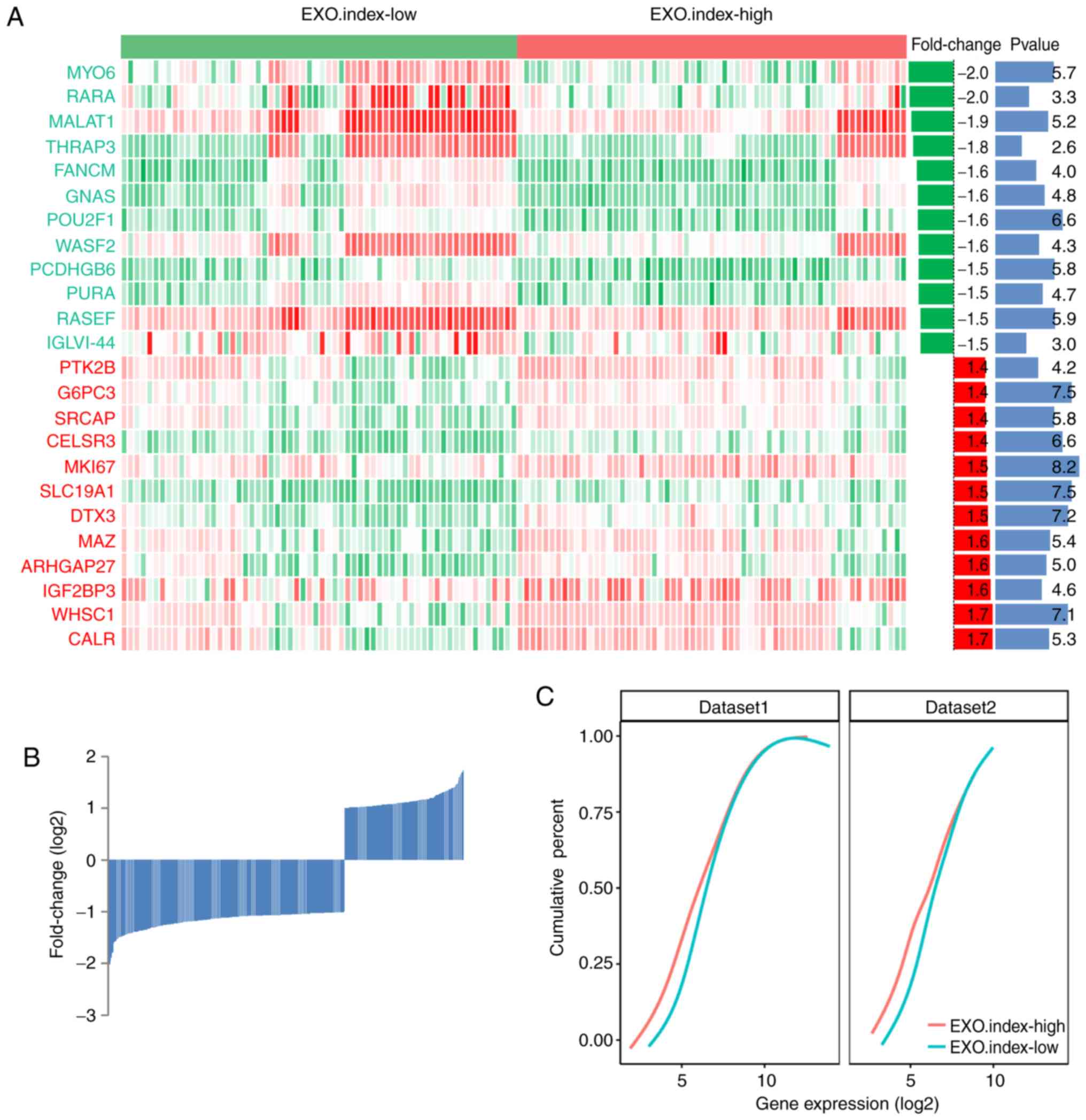
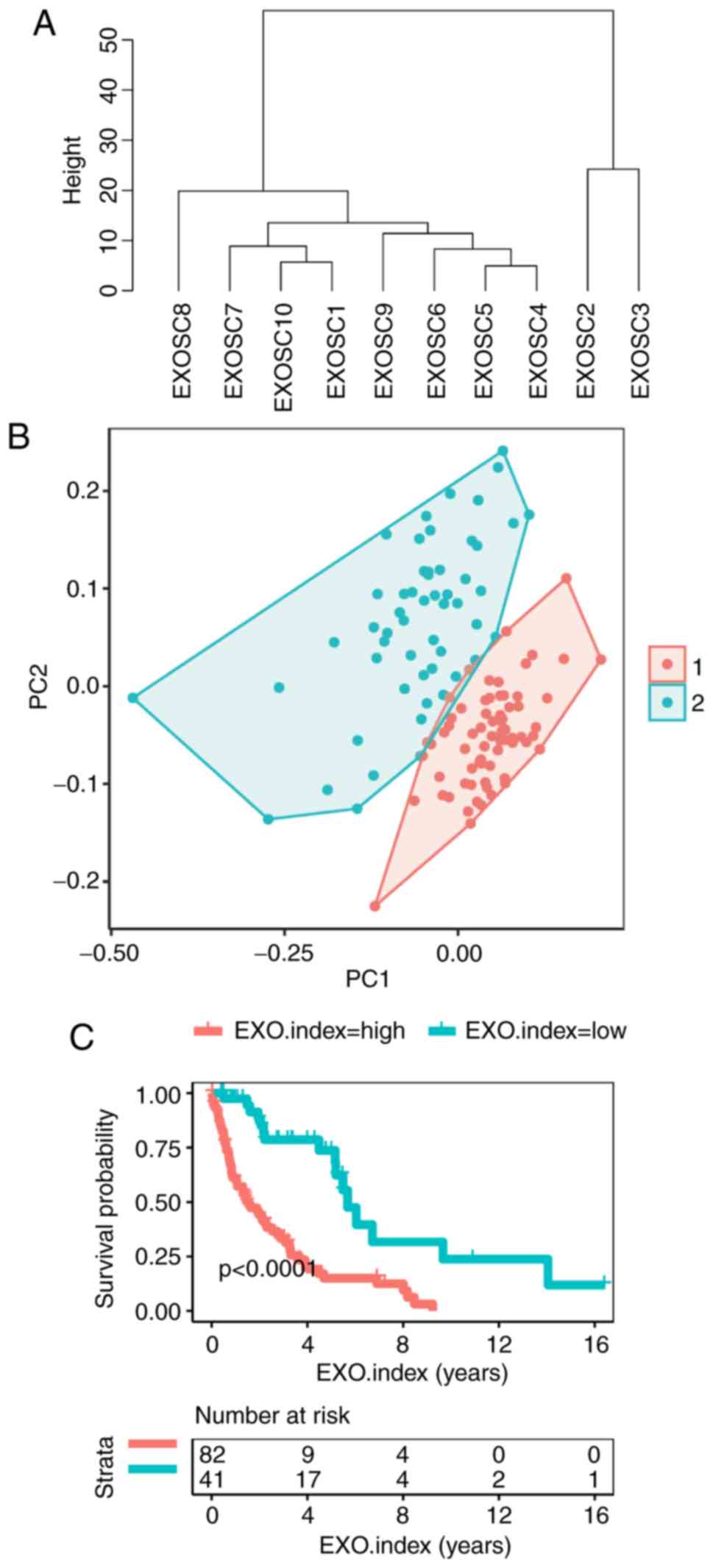 | Figure 4.A total of 10 EXOSC genes were used
as classifiers for the 123 patients with MCL. (A) Unsupervised
clustering of the expression of 10 EXOSC genes for the 123 patients
with MCL. The cluster of EXOSC genes demonstrated cosine
correlation similarity. (B) Fuzzy clustering of the 123 patients
with MCL based on the expression of the 10 EXOSC genes. (C)
Kaplan-Meier curves for overall survival of 123 patients with MCL
based on the EXO.index (P=1.73×10−7). The log-rank test
was used to compare Kaplan-Meier curves. EXOSC, exosome complex;
EXO.index, exosome complex index; MCL, mantle cell lymphoma;
EXOSC5, exosome component 5; EXOSC1, exosome component 1; EXOSC4,
exosome component 4; EXOSC2, exosome component 2; EXOSC7, exosome
component 7; EXOSC3, exosome component 3; EXOSC8, exosome component
8; EXOSC6, exosome component 6; EXOSC9, exosome component 9;
EXOSC10, exosome component 10. |
EXO.index-high group demonstrates a
lower RNA levels in MCL
The gene expression profiles of the EXO.index-high
and EXO.index-low groups of patients with MCL are demonstrated in
Fig. 5A. Overall, it was observed
that 153 genes were upregulated and 303 genes were downregulated
between the EXO.index-high and EXO. Index-low group (P<0.05;
Fig. 5B). The number of
downregulated genes in the EXO.index-high group of patients with
MCL was increased compared with the upregulated genes, indicating
that the EXO.index-high group has different ways of RNA processing
compared with the EXO.index-low group. As demonstrated in the
cumulative distribution of RNA expression of the differentially
expressed genes between the EXO.index-high vs. EXO.index-low groups
of patients with MCL, the EXO.index-high group exhibited decreased
RNA levels compared to the whole transcript profile
(P=5.14×10−3; Fig. 5C).
This result was also confirmed in a second dataset (GSE36000; n=38
samples; P=9.20×10−7; Fig.
5C).
Furthermore, the correlation coefficients of the
EXO.index with the gene expression of 10 proliferation-associated
genes, including Ki67, were determined in the 123 MCL samples
(Fig. S1). The results indicated
that EXO.index is highly correlated with the 10
proliferation-associated genes.
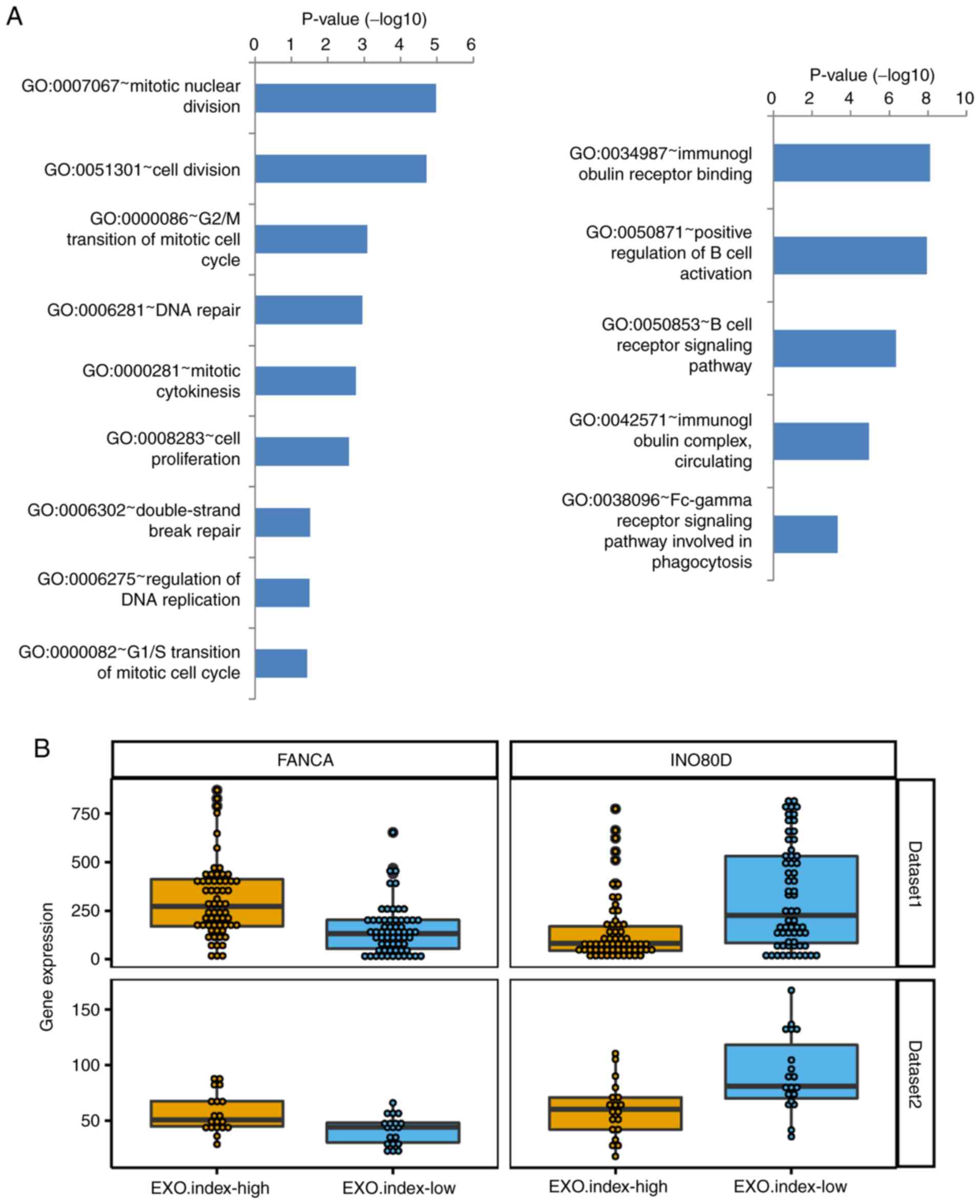
DNA repair and regulation of B-cell
pathways are significantly enriched in MCL
Enrichment analysis of the differentially expressed
genes between the EXO.index-high and the EXO.index-low group of
patients with MCL was then performed. The results of the GO pathway
analysis are summarized in Table I,
and the complete list is presented in Table SI. As indicated in Fig. 6A, ‘mitotic nuclear division’ followed
by ‘DNA repair’ was the most significantly enriched pathway among
the cell division-associated pathways, and ‘immunoglobulin receptor
binding pathway’ and ‘positive regulation of B-cell activation’
were the 2 most enriched pathways in B-cell immune-associated
pathways. Among all of the differentially expressed genes, FANCA
and INO80D were upregulated or downregulated in DNA repair pathway
(Fig. 6B). According to a previous
study, knockdown of EXOSC8 induced cell-cycle exit and promoted the
expression of several cell cycle regulatory genes that are involved
in cell-cycle arrest (33). A
similar expression pattern of these genes was also observed in the
other dataset (GSE36000; n=38 samples; Fig. 6B).
 | Table I.GO pathway analysis of differently
expressed genes. |
Table I.
GO pathway analysis of differently
expressed genes.
| Category | Term | Count | P-value | FDR |
|---|
|
GOTERM_MF_DIRECT |
GO:0034987~immunoglobulin receptor
binding | 9 |
8.0×10−09 |
3.7×10−06 |
|
GOTERM_MF_DIRECT | GO:0003823~antigen
binding | 11 |
1.1×10−05 |
4.9×10−03 |
|
GOTERM_MF_DIRECT | GO:0005524~ATP
binding | 48 |
3.3×10−05 |
1.5×10−02 |
|
GOTERM_MF_DIRECT | GO:0005515~protein
binding | 182 |
1.6×10−04 |
7.1×10−02 |
|
GOTERM_MF_DIRECT |
GO:0008017~microtubule binding | 13 |
2.5×10−04 |
1.1×10−01 |
|
GOTERM_MF_DIRECT | GO:0004386~helicase
activity | 7 |
3.4×10−03 |
7.9×10−01 |
|
GOTERM_MF_DIRECT |
GO:0003777~microtubule motor activity | 6 |
1.2×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0004252~serine-type endopeptidase
activity | 11 |
1.3×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0003682~chromatin binding | 14 |
1.8×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0008253~5′-nucleotidase activity | 3 |
2.3×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT | GO:0003677~DNA
binding | 40 |
2.9×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0003690~double-stranded DNA
binding | 5 |
5.0×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT | GO:0042393~histone
binding | 6 |
5.9×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT | GO:0004674~protein
serine/threonine kinase activity | 12 |
6.0×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT | GO:0098641~cadherin
binding involved in cell-cell adhesion | 10 |
6.2×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0035662~Toll-like receptor 4
binding | 2 |
6.7×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT | GO:0051015~actin
filament binding | 6 |
7.7×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0050544~arachidonic acid binding | 2 |
8.3×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT |
GO:0008026~ATP-dependent helicase
activity | 3 |
8.8×10−02 | 1.0×10° |
|
GOTERM_MF_DIRECT | GO:0004672~protein
kinase activity | 11 |
9.0×10−02 | 1.0×10° |
Discussion
In eukaryotic cells, the exosome complex exists in
the cytoplasm, nucleus and particularly the nucleolus. In the
nucleus, the exosome is required to correct processing of several
small RNA molecules, including rRNA, small nuclear (sn)RNA and
small nucleolar RNA (34). In the
nucleolus, the exosome is involved in the processing of the 5.8S
rRNA and several snRNAs (34–36). In
the cytoplasm, the exosome has a role in degrading mRNA (9,37,38). It
has been demonstrated that mutations in EXOSC3 and EXOSC8 are
associated with human neurological diseases (27,39).
However, the biological implications and prognostic significance of
EXOSC family genes in MCL remain unknown. By analyzing the
expression of EXOSC genes in MCL, the present Bioinformatics study
indicated that abnormal EXOSC gene expression may predict survival
in MCL and alter the expression levels of RNAs compared with those
in the whole transcript profile.
MCL is a rare and refractory subtype of
non-Hodgkin's lymphomas, with a median overall survival time of 5–7
years. In previous studies, 20 ‘proliferation signature’ genes
(23), a PCR-based 5-gene model
(24), the Ki67 proliferation index
(25), and the MIPI (26) were used as prognostic models for
predicting the survival of patients with MCL. Among these
prognostic models, MIPI is the most commonly used, and is applied
to stratify patients with MCL into three risk groups: Low risk;
intermediate risk; and high risk, on the basis of age, leukocyte
count, Eastern Cooperative Oncology Group performance status and
lactic dehydrogenase levels (22,40).
However, it is not universally accepted for predicting survival in
MCL. Therefore, novel biomarkers for predicting the survival of MCL
are urgently required (26). In the
present analysis, it was indicated that the expression levels of
more than one-half of the EXOSC genes (6 out of 10) were
significantly associated with survival in MCL (P<0.05; log-rank
test). Furthermore, a comprehensive EXO.index was established to
predict the survival of patients with MCL. The EXO.index exhibited
an improved performance in predicting survival compared with each
specific EXOSC protein alone (P=1.73×10−7). Furthermore,
the unequal expression of EXOSC genes may predict poorer survival
in patients with MCL.
In addition, 3 key results from the present study
were identified to support the study conclusions. Firstly, the
expression of EXOSC genes was demonstrated to be a good classifier
in MCL in the fuzzy clustering. Furthermore, the EXO.index was
determined, and the patients with MCL were stratified into
EXO.index-high and EXO.index-low groups. The EXO.index-high group
exhibited decreased RNA levels compared with the whole transcript
profile. In addition, the differentially expressed genes in the
EXO.index-high group were enriched in cell division and DNA repair
pathways, which may lead to poor survival of patients with MCL.
However, there are certain limitations to the
present study: There may be confounding factors during the survival
comparison of EXO.index-high and EXO.index-low groups for the
absent of comparison all the clinical information between each
group. As compensation, the correlation coefficient of EXO.index
with the gene expression of 10 proliferation-associated genes,
including KI67, was calculated in 123 MCL samples. The results
indicated that the EXO.index is highly correct with the 10
proliferation-associated genes.
In conclusion, the expression of EXOSC genes was
demonstrated to be a good classifier in MCL. Abnormal expression of
EXOSC genes predicts poor survival in MCL. Furthermore, the
patients with MCL in the EXO.index-high group exhibited poorer
survival rates and decreased RNA levels compared with the whole
transcript profile. The EXOSC genes were indicated to be associated
with cell division and DNA repair pathways, which may result in
poorer survival of patients with MCL.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by
National Natural Science Foundation of China (grant no. 81800195),
the Interdisciplinary Medicine Seed Fund of Peking University
(grant no. BMU2018MB004), the Beijing Natural Science Foundation
(grant nos. 7132183 and 7182178) and the China Health Promotion
Foundation (grant no. CHPF-zlkysx-001).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
HJ and XRL conceived the project. WZ, JZ and XH
analyzed the data. WZ, JZ, XH, XNL, JL, WL, PY, JW, KH, HMJ, XRL
and XZ contributed towards the interpretation of the data. All
authors wrote and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Houseley J and Tollervey D: The many
pathways of RNA degradation. Cell. 136:763–776. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilusz CJ, Wormington M and Peltz SW: The
cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol.
2:237–246. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lehner B and Sanderson CM: A Protein
Interaction Framework for Human mRNA Degradation. Genome Res.
14:1315–1323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Z and Kiledjian M: Functional link
between the mammalian exosome and mRNA decapping. Cell.
107:751–762. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen CY, Gherzi R, Ong SE, Chan EL,
Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M and Karin M:
AU binding proteins recruit the exosome to degrade ARE-containing
mRNAs. Cell. 107:451–464. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schilders G, van Dijk E, Raijmakers R and
Pruijn GJ: Cell and molecular biology of the exosome: How to make
or break an RNA. Int Rev Cytol. 251:159–208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Dijk EL, Schilders G and Pruijn GJ:
Human cell growth requires a functional cytoplasmic exosome, which
is involved in various mRNA decay pathways. RNA. 13:1027–1035.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderson JR, Mukherjee D, Muthukumaraswamy
K, Moraes KC, Wilusz CJ and Wilusz J: Sequence-specific RNA binding
mediated by the RNase PH domain of components of the exosome. RNA.
12:1810–1816. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lejeune F, Li X and Maquat LE:
Nonsense-mediated mRNA decay in mammalian cells involves decapping,
deadenylating, and exonucleolytic activities. Mol Cell. 12:675–687.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma
D, Lv J, Heng J, Ding Y, Xue Y, et al: m6 A modulates
haematopoietic stem and progenitor cell specification. Nature.
549:273–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li HB, Tong J, Zhu S, Batista PJ, Duffy
EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al:
m6 A mRNA methylation controls T cell homeostasis by
targeting the IL-7/STAT5/SOCS pathways. Nature. 548:338–342. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anderson JR, Armitage JO and Weisenburger
DD: Epidemiology of the non-Hodgkin's lymphomas: Distributions of
the major subtypes differ by geographic locations. Non-hodgkin's
lymphoma classification project. Ann Oncol. 9:717–720. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Skarbnik AP and Goy AH: Mantle cell
lymphoma: State of the art. Clin Adv Hematol Oncol. 13:44–55.
2015.PubMed/NCBI
|
|
14
|
Zhou Y, Wang H, Fang W, Romaguer JE, Zhang
Y, Delasalle KB, Kwak L, Yi Q, Du XL and Wang M: Incidence trends
of mantle cell lymphoma in the United States between 1992 and 2004.
Cancer. 113:791–798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghielmini M and Zucca E: How I treat
mantle cell lymphoma. Blood. 114:1469–1476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dreyling M and Hiddemann W; European MCL
Network, : Current treatment standards and emerging strategies in
mantle cell lymphoma. Hematology Am Soc Hematol Educ Program.
542–551. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zucca E, Stein H and Coiffier B: European
lymphoma task force (ELTF): Report of the workshop on mantle cell
lymphoma (MCL). Ann Oncol. 5:507–511. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Banks PM, Chan J, Cleary ML, Delsol G, De
Wolf-Peeters C, Gatter K, Grogan TM, Harris NL, Isaacson PG, Jaffe
ES, et al: Mantle cell lymphoma. A proposal for unification of
morphologic, immunologic, and molecular data. Am J Surg Pathol.
16:637–640. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rajabi B and Sweetenham JW: Mantle cell
lymphoma: Observation to transplantation. Ther Adv Hematol.
6:37–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Herrmann A, Hoster E, Zwingers T,
Brittinger G, Engelhard M, Meusers P, Reiser M, Forstpointner R,
Metzner B, Peter N, et al: Improvement of overall survival in
advanced stage mantle cell lymphoma. J Clin Oncol. 27:511–518.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martin P, Chadburn A, Christos P, Furman
R, Ruan J, Joyce MA, Fusco E, Glynn P, Elstrom R, Niesvizky R, et
al: Intensive treatment strategies may not provide superior
outcomes in mantle cell lymphoma: Overall survival exceeding 7
years with standard therapies. Ann Oncol. 19:1327–1330. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vose JM: Mantle cell lymphoma: 2012 update
on diagnosis, risk-stratification, and clinical management. Am J
Hematol. 87:604–609. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosenwald A, Wright G, Wiestner A, Chan
WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink
HK, Smeland EB, et al: The proliferation gene expression signature
is a quantitative integrator of oncogenic events that predicts
survival in mantle cell lymphoma. Cancer Cell. 3:185–197. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hartmann E: Five-gene model to predict
survival in mantle-cell lymphoma using frozen or formalin-fixed,
paraffin-embedded tissue. J Clin Oncol. 26:4966–4972. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Determann O, Hoster E, Ott G, Wolfram
Bernd H, Loddenkemper C, Leo Hansmann M, Barth TE, Unterhalt M,
Hiddemann W, Dreyling M, et al: Ki-67 predicts outcome in
advanced-stage mantle cell lymphoma patients treated with anti-CD20
immunochemotherapy: Results from randomized trials of the European
MCL network and the German low Grade lymphoma Study Group. Blood.
111:2385–2387. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hoster E, Dreyling M, Klapper W,
Gisselbrecht C, van Hoof A, Kluin-Nelemans HC, Pfreundschuh M,
Reiser M, Metzner B, Einsele H, et al: A new prognostic index
(MIPI) for patients with advanced-stage mantle cell lymphoma.
Blood. 111:558–565. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan J, Yourshaw M, Mamsa H,
Rudnik-Schöneborn S, Menezes MP, Hong JE, Leong DW, Senderek J,
Salman MS, Chitayat D, et al: Mutations in the RNA exosome
component gene EXOSC3 cause pontocerebellar hypoplasia and spinal
motor neuron degeneration. Nat Genet. 44:704–708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Scott DW, Abrisqueta P, Wright GW, Slack
GW, Mottok A, Villa D, Jares P, Rauert-Wunderlich H, Royo C, Clot
G, et al: New molecular assay for the proliferation signature in
mantle cell lymphoma applicable to formalin-fixed paraffin-embedded
biopsies. J Clin Oncol. 35:1668–1677. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
The Gene Ontology Consortium, . The Gene
Ontology Resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mi H, Muruganujan A, Ebert D, Huang X and
Thomas PD: PANTHER version 14: More genomes, a new PANTHER GO-slim
and improvements in enrichment analysis tools. Nucleic Acids Res.
47:D419–D426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mciver SC, Kang YA, Devilbiss AW,
O'Driscoll CA, Ouellette JN, Pope NJ, Camprecios G, Chang CJ, Yang
D, Bouhassira EE, et al: The exosome complex establishes a
barricade to erythroid maturation. Blood. 124:2285–2297. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Allmang C, Kufel J, Chanfreau G, Mitchell
P, Petfalski E and Tollervey D: Functions of the exosome in rRNA,
snoRNA and snRNA synthesis. EMBO J. 18:5399–5410. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mitchell P, Petfalski E, Shevchenko A,
Mann M and Tollervey D: The Exosome: A conserved eukaryotic RNA
processing complex containing multiple 3′->5′Exoribonucleases.
Cell. 91:457–466. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schilders G, Raijmakers R, Raats JM and
Pruijn GJ: MPP6 is an exosome-associated RNA-binding protein
involved in 5.8S rRNA maturation. Nucleic Acids Res. 33:6795–6804.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wilson MA, Meaux S and van Hoof A: A
genomic screen in yeast reveals novel aspects of nonstop mRNA
metabolism. Genetics. 177:773–784. 2016. View Article : Google Scholar
|
|
38
|
Lin WJ, Duffy A and Chen CY: Localization
of AU-rich element-containing mRNA in cytoplasmic granules
containing exosome subunits. J Biol Chem. 282:19958–19968. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Boczonadi V, Müller JS, Pyle A, Munkley J,
Dor T, Quartararo J, Ferrero I, Karcagi V, Giunta M, Polvikoski T,
et al: EXOSC8 mutations alter mRNA metabolism and cause
hypomyelination with spinal muscular atrophy and cerebellar
hypoplasia. Nat Commun. 5:42872014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Eskelund CW, Dahl C, Hansen JW, Westman M,
Kolstad A, Pedersen LB, Montano-Almendras CP, Husby S, Freiburghaus
C, Ek S, et al: TP53 mutations identify younger mantle cell
lymphoma patients who do not benefit from intensive
chemoimmunotherapy. Blood. 130:1903–1910. 2017. View Article : Google Scholar : PubMed/NCBI
|