Introduction
Prostate cancer is the most prevalent cancer in men
in Western societies (1), but only a
small subset is highly aggressive and needs extensive treatment
(2,3). Predictive preoperative prognostic
parameters are limited to Gleason score and tumour extent on
biopsies, prostate-specific antigen (PSA) serum level and clinical
stage. Thus it is hoped that additional biomarkers can be
identified which will improve the prediction of an aggressive
tumour course.
The E26 transformation-specific (ETS) family of
transcription factors has been named after its evolutionarily
conserved DNA-binding domain (4).
ETS factors play important roles in many human tumour types. In
prostate cancer, the ETS family member ERG is fused to the TMPRSS2
serine protease in approximately 50% of cases (5–7). Another
ETS factor with relevance in prostate cancer is SAM pointed
domain-containing Ets transcription factor (SPDEF). SPDEF is
physiologically expressed in normal tissues of the prostate
(8), breast (9), ovary (10), lung (10), brain (10) and gastrointestinal tract (11). Studies on breast (9,12–14),
prostate (13,15), ovarian (16) and colon cancers (17) have described frequent dysregulation
of SPDEF with conflicting results. In prostate cancer some authors
see SPDEF as an oncogenic driver (8,13) while
others claim a tumour metastasis suppressor role for SPDEF
(15,18–22).
To better understand the role of SPDEF in prostate
cancer, we took advantage of our large prostate cancer tissue
microarray (TMA) to study expression of SPDEF with the monoclonal
antibody MAB9916 clone 4A5 by immunohistochemistry in more than
12,000 prostate cancer samples, and then compared SPDEF expression
with relevant clinical and pathological parameters in a patient
cohort which was castration-sensitive or naïve to androgenic
deprivation therapy.
Materials and methods
Patients
Radical prostatectomy specimens were available from
12,427 patients, undergoing surgery between 1992 and 2012 at the
Department of Urology and the Prostate Cancer Center Martini Clinic
at the University Medical Center Hamburg-Eppendorf. All prostate
specimens were analysed according to a standard procedure,
including a complete embedding of the entire prostate for
histological analysis (23).
Histopathological data was retrieved from the patient files,
including tumour stage, Gleason grade, nodal stage and status of
the resection margin. In addition to the classical Gleason
categories, ‘quantitative’ Gleason grading was performed by
estimating the percentage of Gleason 4 patterns as previously
described (24). Follow-up data were
available for a total of 11,152 patients with a median follow-up of
60 months (range: 1 to 241 months; Table
I). Prostate specific antigen (PSA) serum values were measured
following surgery and PSA recurrence was defined as a postoperative
serum PSA of at least 0.2 ng/ml and increasing at subsequent
measurements. The TMA manufacturing process was described earlier
in detail (25). In short, for each
patient, a 0.6 mm diameter core was taken from a representative
tissue block. The tissues were distributed among 27 TMA blocks,
each containing 144 to 522 tumour samples. For internal controls,
each TMA block also contained various control tissues, including
normal prostate tissue. The use of leftover archived diagnostic
tissues for manufacturing of tissue microarrays and their analysis
for research purposes, as well as patient data analysis, has been
approved by local laws (HmbKHG, §12a) and by the local ethics
committee (Ethics Commission Hamburg, WF-049/09). The present study
has been carried out in compliance with the Helsinki
Declaration.
 | Table I.Pathological and clinical data of the
arrayed prostate cancer samples. |
Table I.
Pathological and clinical data of the
arrayed prostate cancer samples.
| Variables | Study cohort on
TMAa, n | Biochemical
relapse, n (%) |
|---|
| Follow-up | 11,152 | 2,769 (24.8) |
| Mean/median
(months) | 64.4/60.0 | – |
| Age (years) |
|
|
|
≤50 | 323 | 81 (25.1) |
|
51–59 | 2,696 | 705 (26.1) |
|
60–69 | 6,528 | 1,610 (24.7) |
|
≥70 | 1,498 | 370 (24.7) |
| Pretreatment PSA
(ng/ml) |
|
|
|
<4 | 1,585 | 242 (15.3) |
|
4–10 | 7,480 | 1,355 (18.1) |
|
>10–20 | 2,412 | 737 (30.6) |
|
>20 | 812 | 397 (48.9) |
| pT stage (AJCC
2002) |
|
|
|
pT2 | 8,187 | 1,095 (13.4) |
|
pT3a | 2,660 | 817 (30.7) |
|
pT3b | 1,465 | 796 (54.3) |
|
pT4 | 63 | 51 (81.0) |
| Gleason grade |
|
|
|
≤3+3 | 2,297 | 230 (10.0) |
|
3+4 | 6,679 | 1,240 (18.6) |
| 3+4
Tertiary 5 | 433 | 115 (26.6) |
|
4+3 | 1,210 | 576 (47.6) |
| 4+3
Tertiary 5 | 646 | 317 (49.1) |
|
≥4+4 | 416 | 348 (83.7) |
| pN stage |
|
|
|
pN0 | 6,970 | 1,636 (23.5) |
|
pN+ | 693 | 393 (56.7) |
| Surgical
margin |
|
|
|
Negative | 9,990 | 1,848 (18.5) |
|
Positive | 2,211 | 853 (38.6) |
Immunohistochemistry
The freshly cut TMA sections were collectively
immunostained in a single run. Slides were deparaffinized and
exposed to heat-induced antigen retrieval (121°C, 5 min, pH 7,8
Tris-EDTA-citrate buffer). Primary SPDEF antibody (mouse monoclonal
antibody, MAB9916, clone 4A5; Abnova Germany, dilution 1:4050) was
applied (37°C, 60 min) and bound antibody was then visualized using
the EnVision kit (Dako, Glostrup, Denmark) according to the
manufacturer's directions. The SPDEF staining was typically nuclear
and slightly cytoplasmic in prostate cancer and usually negative in
benign prostate tissue. The staining intensity (0, 1+, 2+, and 3+)
and the fraction of positive tumour cells were separately recorded
for each tissue spot. A final score was assigned as described
(26): Negative scores had a
complete absence of staining; weak scores had a staining intensity
of 1+ in ≤70 % of the tumour cells or a staining intensity of 2+ in
≤30% of the tumour cells; moderate scores had a staining intensity
of 1+ in >70% of tumour cells, a staining intensity of 2+ in
>30% but in ≤70% of the tumour cells, or a staining intensity of
3+ in ≤30% of the tumour cells; and strong scores had a staining
intensity of 2+ in >70% of the tumour cells or a staining
intensity of 3+ in >30% of the tumour cells. Ki-67 labelling
index (Ki67-LI) data were taken from a previous publication
(27).
FISH
Details of the method were previously published for
ERG (26), 5q21 (CHD1) (28), 6q15 (MAP3K7) (29), PTEN (10q23) (30), and 3p13 (FOXP1) (31).
Cell culture and Western blotting
DU-145 (prostate cancer) cells were obtained from
the Leibniz-Institute DSMZ-German collection of microorganisms and
cell cultures (no.: ACC 261, Human) and grown in DMEM (Dulbecco's
Modified Eagles Medium), enriched with 10% heat-inactivated and
filtered foetal bovine serum, 1% penicillin-streptomycin, 1% NEAA,
1% pyruvate. The cells were incubated in 75 cm2 flasks
at 37°C and 5% CO2. For electrophoresis and blotting
about 10×106 cells were harvested after being washed
once with 1× PBS in 150 µl lysis buffer. The cells were scratched
with a cell scraper and transferred into a precooled 1.5 ml
Eppendorf tube, rested 30 min on ice, and were centrifuged at
14,000 rpm for 5 min at 4°C. The amount of protein was measured
with the Quibit fluorometer at 550 nm. Four-times Laemmli-buffer
(BioRad) and β-mercaptoethanol (1:10 dilution) were added to the
DU-145 lysate sample and water control, heated at 95°C for 5 min,
and loaded together with the size marker Precision Plus Protein™
dual colour standard (BioRad) to a 4–15% polyacrylamide gel. The
gel was run for 30 min at 180 V in a mini-protean Tetra cell system
in 1× Tris-glycin-SDS buffer (BioRAD). The proteins were
transferred at 2A in 7 min to a polyvinylidene fluoride membrane,
washed with PBS-Tween 20× (PBS + 0.5% Tween 20), and incubated for
1 h with blocking buffer (5% milk powder in PBS-Tween 20, filtered
and boiled). MAB9916 clone 4A5 (1:1,400 in blocking buffer) was
added overnight at 4°C, washed 3× 15 min with PBS-Tween and
incubated with secondary antibody (peroxidase goat anti mouse,
diluted 1:1,000, Dianova) for 40 min at room temperature on a
shaker. The membrane was developed with enhanced chemiluminescence
substrate (Bio Rad) and recorded on a ChemiDoc™ imaging system.
Statistical analysis
Contingency tables and the χ2-test were
performed to search for associations between molecular parameters
and tumour phenotype. Ki67 labelling data were tested by ANOVA.
Kaplan-Meier curves were tested by log-rank for differences between
groups and Cox proportional hazards regression analysis was
performed to test for independence and significance between
pathological, molecular and clinical variables. Separate models
were calculated with different sets of parameters, according to
their availability before or after the prostatectomy. Statistical
calculations were done with JMP 10 (SAS Institute Inc., NC,
USA).
Results
SPDEF-staining
A total of 9403 (76%) of tumour samples were
interpretable in our TMA analysis. Reasons for non-informative
cases (3024, 24%) included lack of tissue samples or absence of
unequivocal cancer tissue in the TMA spot.
SPDEF expression in normal and
cancerous prostate tissues
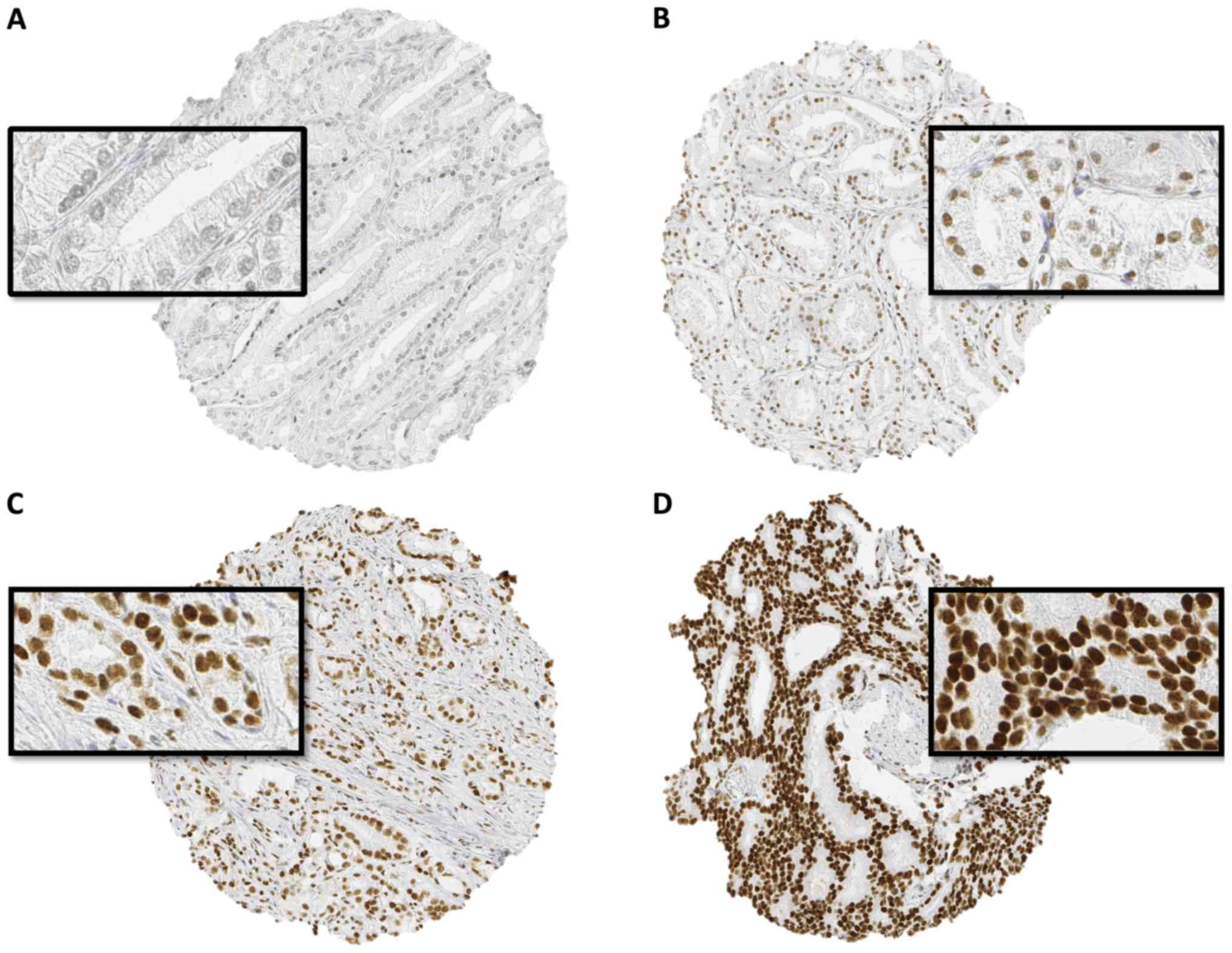
Normal prostate tissue only rarely showed weak
nuclear staining, which was limited to epithelial cells. In cancer,
nuclear SPDEF immunostaining was found in the majority (7522 of
9403; 80%) of interpretable prostate cancers and was considered
weak in 26%, moderate in 40% and strong in 14% of cases.
Representative images of SPDEF staining are given in Fig. 1.
Association with TMPRSS2:ERG fusion
status and ERG protein expression
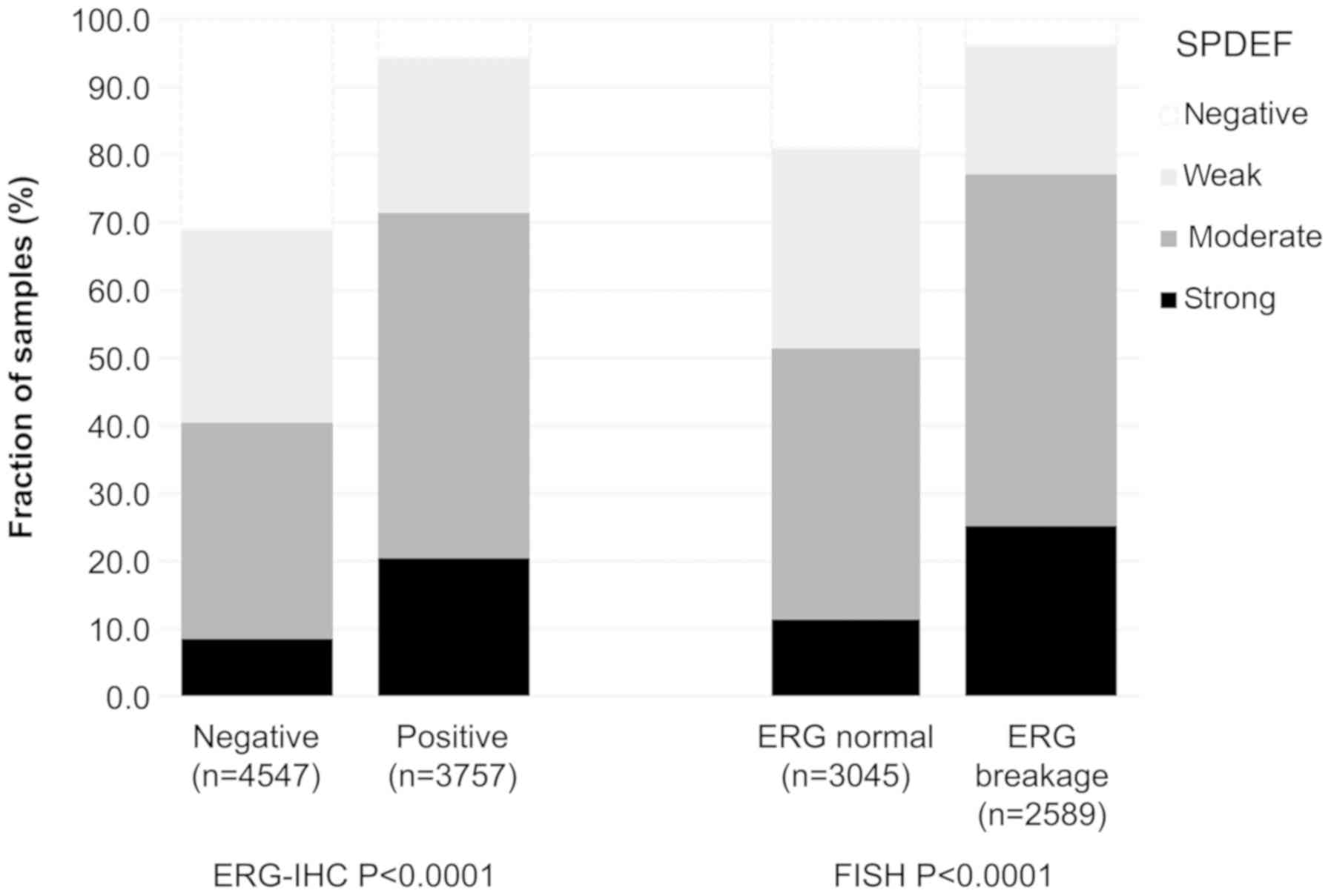
Data on TMPRSS2:ERG fusion status previously
obtained by FISH were available from 5634 and by
immunohistochemistry from 8304 tumours with evaluable SPDEF
immunostaining [18,19]. Data on both ERG FISH and IHC were
available from 5421 cancers, and concordant results (ERG IHC
positive and break by FISH or ERG IHC negative and missing break by
FISH) were found in 5171 of 5421 (95.4%) cancers. The SPDEF
staining score was associated with TMPRSS2:ERG rearrangement
and ERG positivity. For example, SPDEF immunostaining was seen in
94 and 96% of cancers with TMPRSS2:ERG fusion detected by
IHC and FISH. In contrast, only 69% of cancers without ERG staining
and 81% of cancers without ERG rearrangements were SPDEF positive
(P<0.0001 each; Fig. 2).
Associations with tumour
phenotype
High-level SPDEF immunostaining was associated with
advanced pT stage, higher Gleason grade, lymph node metastasis
(P<0.0001 each) and positive surgical margin (P=0.0105; Table II). The associations with pT stage
and high Gleason grade held true for both the ERG negative and ERG
positive cancer cohorts (P<0.0001 each; Tables III and IV).
 | Table II.Association between SPDEF expression
and prostate cancer phenotype. |
Table II.
Association between SPDEF expression
and prostate cancer phenotype.
|
|
| SPDEF (%) |
|
|---|
|
|
|
|
|
|---|
| Parameters | Number, n | Negative | Weak | Moderate | Strong | P-value |
|---|
| All cancer
samples | 9,403 | 20.0 | 26.4 | 40.1 | 13.5 |
|
| Tumour
stagea |
|
|
|
|
| <0.0001 |
|
pT2 | 5,987 | 22.9 | 27.9 | 37.7 | 11.5 |
|
|
pT3a | 2,148 | 16.2 | 24.2 | 42.1 | 17.6 |
|
|
pT3b | 1,179 | 12.6 | 23.7 | 48.1 | 15.6 |
|
|
pT4 | 53 | 13.2 | 9.4 | 50.9 | 26.4 |
|
| Gleason
gradea |
|
|
|
|
| <0.0001 |
|
≤3+3 | 2,126 | 28.9 | 32.1 | 31.7 | 7.2 |
|
|
3+4 | 5,302 | 18.8 | 26.0 | 41.5 | 13.8 |
|
|
4+3 | 1,468 | 13.8 | 20.9 | 45.5 | 19.8 |
|
|
≥4+4 | 460 | 12.4 | 23.0 | 45.4 | 19.1 |
|
| Lymph node
metastasisa |
|
|
|
|
| <0.0001 |
| N0 | 5,384 | 17.6 | 24.6 | 42.6 | 15.2 |
|
| N+ | 560 | 13.0 | 18.6 | 48.6 | 19.8 |
|
| Preoperative PSA
level (ng/ml)a |
|
|
|
|
| 0.0394 |
|
<4 | 1,132 | 16.5 | 27.6 | 43.2 | 12.7 |
|
|
4–10 | 5,602 | 20.1 | 26.6 | 39.8 | 13.4 |
|
|
>10–20 | 1,885 | 21.3 | 25.7 | 38.6 | 14.5 |
|
|
>20 | 676 | 21.0 | 24.1 | 41.6 | 13.3 |
|
| Surgical
margina |
|
|
|
|
| 0.0105 |
|
Negative | 7,419 | 20.4 | 26.9 | 39.6 | 13.1 |
|
|
Positive | 1,811 | 18.2 | 25.1 | 41.9 | 14.9 |
|
 | Table III.Association between SPDEF expression
and prostate cancer phenotype in the ERG negative subset. |
Table III.
Association between SPDEF expression
and prostate cancer phenotype in the ERG negative subset.
|
|
| SPDEF (%) |
|
|---|
|
|
|
|
|
|---|
| Parameters | Number, n | Negative | Weak | Moderate | Strong | P-value |
|---|
| All cancer
samples | 4,547 | 31.0 | 28.5 | 32.2 | 8.4 |
|
| Tumour stage |
|
|
|
|
| <0.0001 |
|
pT2 | 3,015 | 33.8 | 29.5 | 30.0 | 6.6 |
|
|
pT3a | 928 | 28.8 | 27.4 | 33.0 | 10.9 |
|
|
pT3b | 567 | 19.2 | 26.3 | 40.9 | 13.6 |
|
|
pT4 | 24 | 25.0 | 12.5 | 54.2 | 8.3 |
|
| Gleason grade |
|
|
|
|
| <0.0001 |
|
≤3+3 | 953 | 44.5 | 31.2 | 21.5 | 2.8 |
|
|
3+4 | 2,552 | 30.2 | 29.2 | 32.6 | 8.1 |
|
|
4+3 | 757 | 20.9 | 24.4 | 40.6 | 14.1 |
|
|
≥4+4 | 267 | 18.4 | 25.8 | 41.2 | 14.6 |
|
| Lymph node
metastasis |
|
|
|
|
| 0.0002 |
| N0 | 2,659 | 27.6 | 28.8 | 34.0 | 9.6 |
|
| N+ | 262 | 20.2 | 22.1 | 43.1 | 14.5 |
|
| Preoperative PSA
level (ng/ml) |
|
|
|
|
| 0.6675 |
|
<4 | 466 | 27.5 | 28.8 | 35.4 | 8.4 |
|
|
4–10 | 2,671 | 31.3 | 28.4 | 32.3 | 8.0 |
|
|
>10–20 | 998 | 31.5 | 29.0 | 30.4 | 9.2 |
|
|
>20 | 373 | 30.6 | 27.3 | 32.7 | 9.4 |
|
| Surgical
margin |
|
|
|
|
| 0.4234 |
|
Negative | 3,597 | 31.3 | 28.7 | 31.9 | 8.1 |
|
|
Positive | 870 | 29.2 | 28.5 | 32.9 | 9.4 |
|
 | Table IV.Association between SPDEF expression
and prostate cancer phenotype in the ERG positive subset. |
Table IV.
Association between SPDEF expression
and prostate cancer phenotype in the ERG positive subset.
|
|
| SPDEF (%) |
|
|---|
|
|
|
|
|
|---|
| Parameters | Number, n | Negative | Weak | Moderate | Strong | P-value |
|---|
| All cancer
samples | 3,757 | 5.8 | 23.0 | 50.7 | 20.5 |
|
| Tumor stage |
|
|
|
|
| <0.0001 |
|
pT2 | 2,200 | 6.8 | 24.7 | 49.4 | 19.2 |
|
|
pT3a | 1,019 | 4.6 | 20.9 | 50.7 | 23.7 |
|
|
pT3b | 499 | 4.2 | 21.0 | 56.7 | 18.0 |
|
|
pT4 | 22 | 4.5 | 4.5 | 40.9 | 50.0 |
|
| Gleason grade |
|
|
|
|
| <0.0001 |
|
≤3+3 | 803 | 9.5 | 31.9 | 45.7 | 13.0 |
|
|
3+4 | 2,211 | 5.2 | 22.2 | 51.8 | 20.9 |
|
|
4+3 | 578 | 4.2 | 15.6 | 52.4 | 27.9 |
|
|
≥4+4 | 143 | 2.8 | 16.8 | 53.8 | 26.6 |
|
| Lymph node
metastasis |
|
|
|
|
| 0.4186 |
| N0 | 2,179 | 4.7 | 19.7 | 52.8 | 22.8 |
|
| N+ | 241 | 5.0 | 15.4 | 56.0 | 23.7 |
|
| Preoperative PSA
level (ng/ml) |
|
|
|
|
| 0.1717 |
|
<4 | 502 | 5.4 | 24.1 | 52.8 | 17.7 |
|
|
4–10 | 2,278 | 5.7 | 23.8 | 49.9 | 20.5 |
|
|
>10–20 | 688 | 6.4 | 20.6 | 49.7 | 23.3 |
|
|
>20 | 237 | 5.9 | 18.1 | 56.5 | 19.4 |
|
| Surgical
margin |
|
|
|
|
| 0.1312 |
|
Negative | 2,918 | 6.2 | 23.7 | 50.1 | 20.0 |
|
|
Positive | 767 | 4.7 | 21.1 | 52.7 | 21.5 |
|
Association to key genomic
deletions
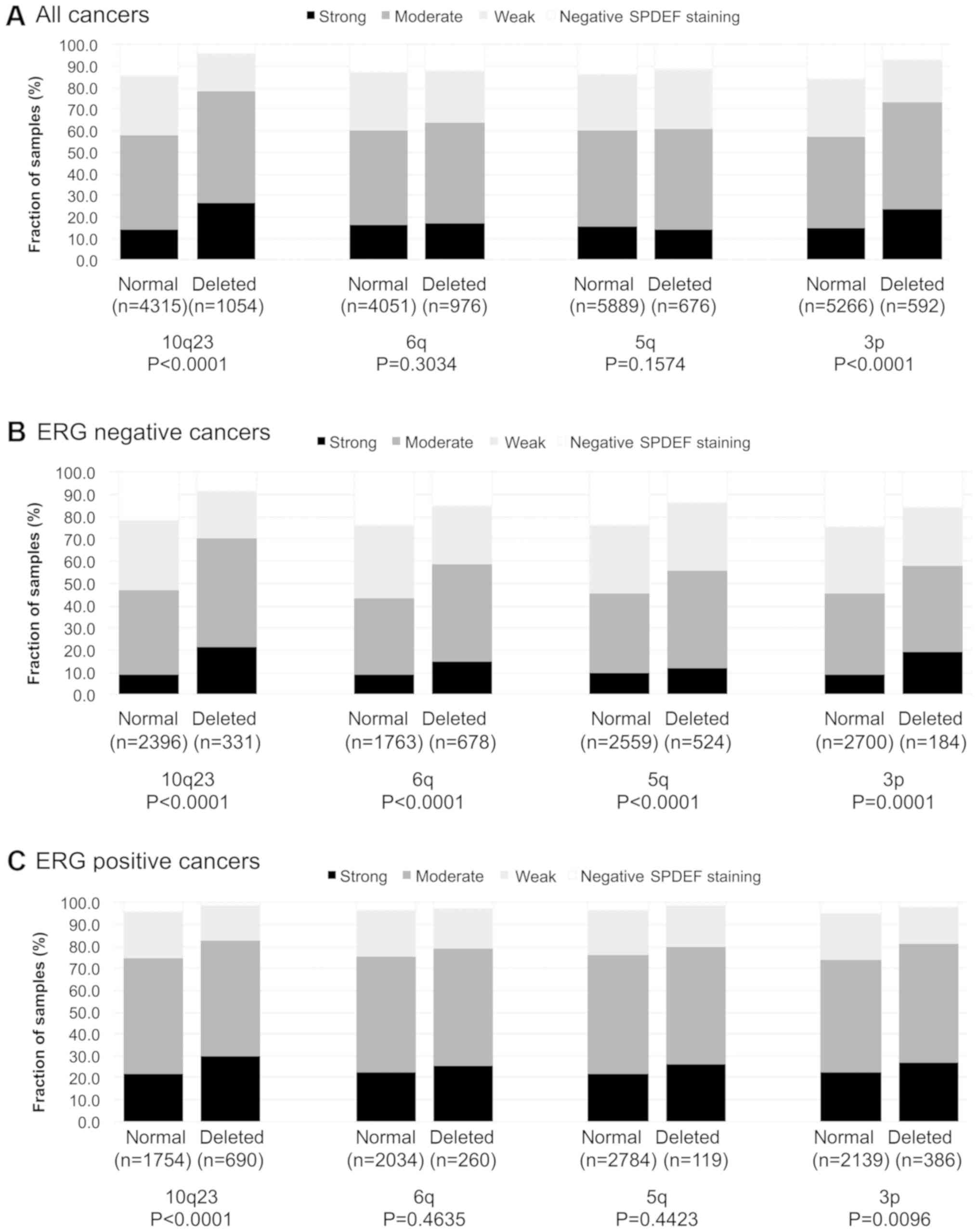
To examine whether SPDEF expression might be
associated with one or several of the most common genomic
deletions, SPDEF data were compared to pre-existing data on
PTEN (10q23), 3p13 (FOXP1), 6q15 (MAP3K7) and
5q21 (CHD1) deletions (Fig.
3A-C). Strong SPDEF expression was associated with all
analysed deletions in ERG negative cancers (P≤0.0008). In contrast,
SPDEF expression was only linked to PTEN deletions (P<0.0001)
and 3p13 deletions (P=0.0096) but not to the other genomic
deletions in ERG positive samples.
Association with Ki67-labelling index
(LI)
High levels of SPDEF immunostaining were
significantly associated with accelerated cellular proliferation as
measured by Ki67-LI (Table V;
P<0.0001). This also applied for all analysed subgroups of
tumours with identical Gleason scores (P<0.0001 for Gleason 3+3,
3+4, 4+3 and ≥4+4).
 | Table V.Association between SPDEF expression
and Ki67LI. |
Table V.
Association between SPDEF expression
and Ki67LI.
| Gleason | SPDEF IHC | Number, n | Ki67LI, mean ±
SEM | P-value |
|---|
| Total | Negative | 1,247 | 1.44±0.07 | <0.0001 |
|
| Weak | 1,502 | 2.75±0.07 |
|
|
| Moderate | 2,163 | 3.24±0.06 |
|
|
| Strong | 689 | 3.80±0.10 |
|
| ≤3+3 | Negative | 369 | 1.25±0.10 | <0.0001 |
|
| Weak | 386 | 2.46±0.10 |
|
|
| Moderate | 374 | 2.65±0.10 |
|
|
| Strong | 80 | 3.16±0.22 |
|
| 3+4 | Negative | 647 | 1.43±0.09 | <0.0001 |
|
| Weak | 801 | 2.50±0.08 |
|
|
| Moderate | 1,240 | 3.10±0.06 |
|
|
| Strong | 409 | 3.43±0.11 |
|
| 4+3 | Negative | 97 | 1.41±0.33 | <0.0001 |
|
| Weak | 134 | 4.01±0.28 |
|
|
| Moderate | 218 | 3.47±0.22 |
|
|
| Strong | 98 | 4.23±0.33 |
|
| ≥4+4 | Negative | 39 | 2.05±0.74 | <0.0001 |
|
| Weak | 60 | 3.85±0.60 |
|
|
| Moderate | 98 | 4.95±0.47 |
|
|
| Strong | 41 | 7.73±0.72 |
|
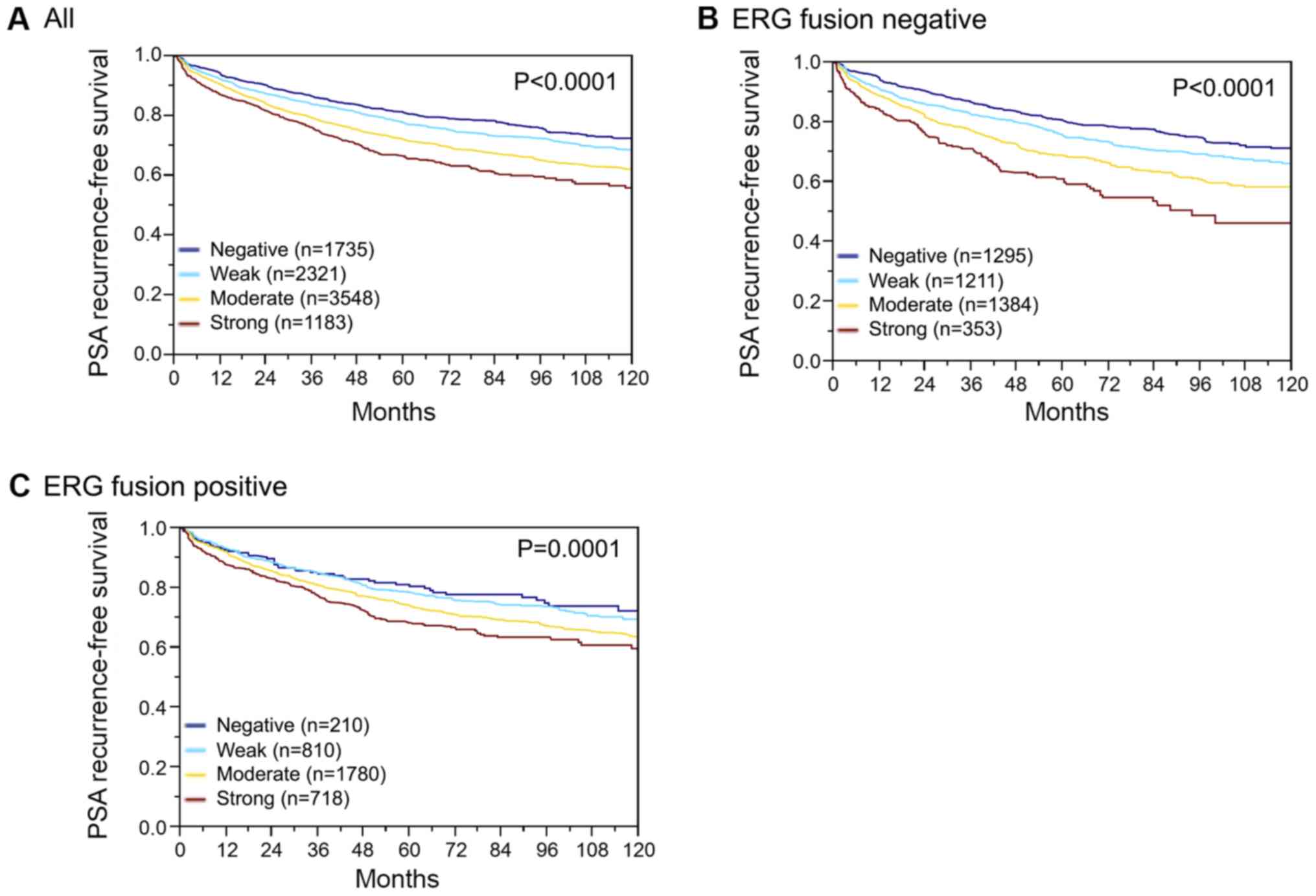
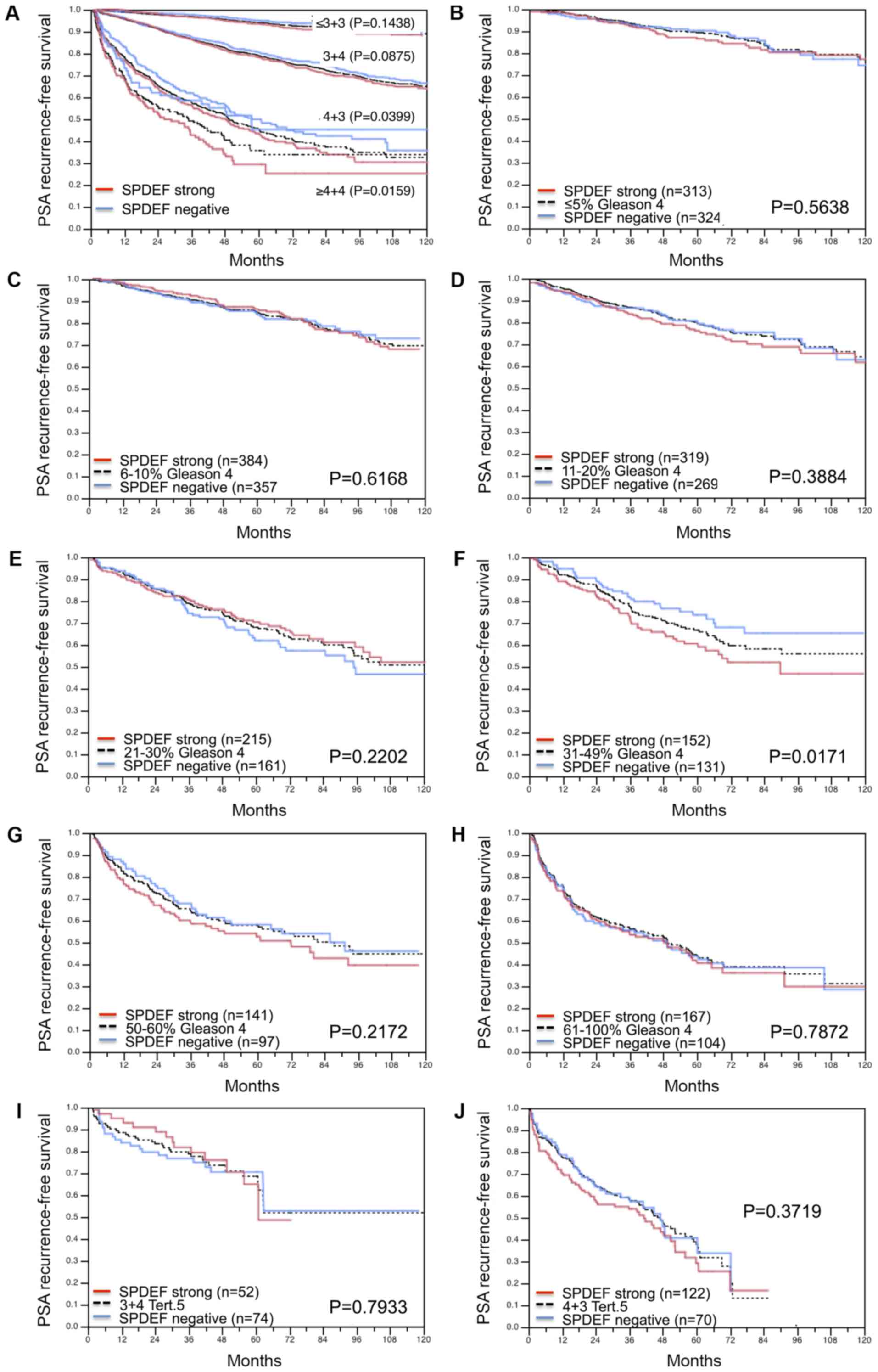
Association with PSA recurrence
Follow-up data were available for 8787 patients with
interpretable SPDEF immunostaining on the TMA. A highly significant
association was seen between PSA recurrence and elevated SPDEF
expression (P<0.0001; Fig. 4A).
Subset analysis further revealed that the prognostic impact was
stronger in the subgroup of ERG negative (P<0.0001; Fig. 4B) than in ERG positive cancers
(P=0.0001; Fig. 4C). Further subset
analyses of cancers with identical classical and quantitative
Gleason grade revealed a prognostic impact of SPDEF immunostaining
beyond classical Gleason grade for the 4+3 and ≥4+4 score groups
(Fig. 5A) and the quantitative
Gleason grade group 31–49% Gleason 4 (Fig. 5B-J).
Multivariate analysis
To evaluate the clinical relevance of SPDEF
expression in different scenarios, four different types of
multivariable analyses were performed, as previously described
(32). In brief, scenario 1 included
postoperative parameters (pathological tumour stage, pathological
lymph node status (pN), surgical margin status, preoperative PSA
value, pathological Gleason grade). In scenario 2, all
postoperatively available parameters with exception of nodal status
were included. This takes into account that the indication and
extent of lymph node dissection is not standardized in the surgical
therapy of prostate cancer. Two additional scenarios had the
purpose to model the preoperative situation: Scenario 3 included
preoperative PSA, clinical tumour stage (cT stage) and Gleason
grade obtained on the prostatectomy specimen. Since postoperative
determination of a tumour's Gleason grade is of higher quality than
the preoperatively determined Gleason grade [subject to sampling
errors and consequently under-grading in more than one third of the
cases (33)], another multivariable
analysis was added. In scenario 4, the preoperative Gleason grade
obtained on the original biopsy was combined with preoperative PSA,
cT stage and SPDEF expression. In all four scenarios, the
multivariate analysis demonstrated that SPDEF expression provides
independent prognostic information in the subset of ERG negative
cancers and also in the non-stratified cohort of all cancers. In
the ERG positive subset, only the preoperative model 4 showed a
significant prognostic effect (P=0.0024, Table VI). The overall univariate Cox
proportional hazard ratio for strong versus negative SPDEF
expression was weak (1.99, 95% CI 1.72–2.20, Table SI).
 | Table VI.Multivariate analysis of Cox
proportional hazards for biochemical RPE in various models and ERG
subsets. |
Table VI.
Multivariate analysis of Cox
proportional hazards for biochemical RPE in various models and ERG
subsets.
|
|
|
| P-value of
χ2 test |
|---|
|
|
|
|
|
|---|
| ERG subset | Model | Number, n | Preoperative
PSA-Level | pT stage | cT stage | RPE Gleason | Gleason Biopsy | Nodal stage | R status | SPDEF
expression |
|---|
| Total | 1 | 5,203 | <0.0001 | <0.0001 | – | <0.0001 | – | <0.0001 | 0.0006 | 0.0074 |
|
| 2 | 8,265 | <0.0001 | <0.0001 | – | <0.0001 | – | – | <0.0001 | 0.0020 |
|
| 3 | 8,131 | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | <0.0001 |
|
| 4 | 8,021 | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | <0.0001 |
| Negative | 1 | 2,564 | 0.0002 | <0.0001 | – | <0.0001 | – | <0.0001 | 0.1299 | 0.0013 |
|
| 2 | 3,986 | <0.0001 | <0.0001 | – | <0.0001 | – | – | 0.0014 | <0.0001 |
|
| 3 | 3,942 | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | <0.0001 |
|
| 4 | 3,891 | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | <0.0001 |
| Positive | 1 | 2,116 | 0.0013 | <0.0001 | – | <0.0001 | – | 0.0041 | 0.0113 | 0.4037 |
|
| 2 | 3,307 | <0.0001 | <0.0001 | – | <0.0001 | – | – | 0.0002 | 0.6837 |
|
| 3 | 3,230 | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | 0.4474 |
|
| 4 | 3,182 | <0.0001 | – | 0.0031 | – | <0.0001 | – | – | 0.0024 |
Discussion
The results of our study identify high SPDEF
expression as an independent predictor of poor prognosis in
prostate cancers. The increased SPDEF staining in the analysed
prostate cancers, compared to the negative to weak SPDEF expression
in normal prostate tissue, suggests for role of SPDEF in prostate
cancer development. These observations fit well with a recent study
by Situ et al (34)
describing low or absent SPDEF staining in normal prostate tissue
but strong up regulation in cancer. Sood et al (13) also described a slightly increasing
rate of SPDEF positivity from normal (27%) to high-grade prostatic
intraepithelial neoplasia (33%) and invasive prostate cancer (40%).
Within our 9,403 interpretable prostate carcinomas, elevated SPDEF
expression levels were strikingly linked to advanced tumour stage,
high Gleason grade, rapid cell proliferation and early PSA
recurrence. These findings fit well with a role for SPDEF as a
potential oncogenic driver in prostate cancer. Other authors had
also reported increasing SPDEF expression from intermediate to high
Gleason grade in prostate cancer (13,34). A
link between high SPDEF expression and cancer development and
progression was also found in breast (12,35,36) and
ovarian (16,37) cancers. It is of note that others have
reported loss of SPDEF expression during tumour progression, and
found reduced levels of SPDEF in prostate cancer as compared to
normal epithelium in cohorts of 40 and 73 patients (15,20).
Similar findings were also reported for breast (14), bladder (38) and colon (17) cancer. Such discrepancies are most
likely due to differences in the reagents used, and possibly also
due to patient cohort selection issues. We are confident in our
reagents used. The monoclonal antibody (MAB9916 clone 4A5 from
Abnova) used in this study was tested by Western blot, where it
showed a single band of the predicted molecular mass of 37.5 kDa
(Fig. S1) (8).
The prognostic effect of SPDEF expression was seen
in both ERG positive and ERG negative cancers, but was stronger in
the latter group. It is well known that aberrant expression of the
ERG transcription factor leads to dysregulation of at least 1,600
genes in affected prostate cancer cells, and that these changes may
impact the role of various prognostic molecular features. In
earlier studies on the same set of tumours, prognostic factors that
were restricted to either ERG positive (39,40) or
ERG negative cancers (29,41) were often found. Given the different
average SPDEF expression levels between ERG positive and ERG
negative cancers, it is possible that our immunohistochemistry
protocol was better suited to distinguish expression differences in
cancers with somewhat lower expression levels, such as in ERG
negative cancers, than in tumours with higher expression, such as
in ERG positive cancers. Most chromosomal deletions are decidedly
more frequent in either ERG positive (3p, PTEN) or ERG negative
(5q, 6q) cancers (29,31,39,42).
Because SPDEF is also associated with ERG status, a positive
association with 3p and PTEN deletions and an inverse association
with 5q and 6q deletions was expected in unselected prostate cancer
cohorts. The association of high SPDEF expression with the cell
proliferation marker Ki-67 fits well with the role of SPDEF as an
oncogenic activator and with previous reports (8,13).
Our data are restricted to naïve prostate cancer
patients, who had not received androgen deprivation therapy (ADT).
Under ADT and in patients with castration resistant prostate cancer
after ADT as well as in transgenic mouse models and prostate cancer
cell lines it has been shown that SPDEF suppresses tumour
metastasis (15,18–20,22).
While ADT reduces SPDEF expression and cell proliferation, it
relieves repression of TGFB1, CCL2, and MMP9 key drivers of
metastasis. This provides an example of how a therapy which blocks
growth of the primary tumour, may paradoxically promote metastasis
(43).
The data of this study suggest that the SPDEF
protein level may constitute a clinically useful marker in prostate
cancer. SPDEF expression exerted a prognostic impact that was
independent of established prognostic parameters. It is of note
that the most critical clinical need in prostate cancer is not
finding prognostic markers that are independent of established
parameters. Most of all, parameters are needed that are more
reproducible and reliable than the established ones. The Gleason
grade, the strongest established prognostic parameter, suffers from
very substantial interobserver variability reaching up to 40% in
individual biopsies (44). Based on
the data from this study, we assume that SPDEF expression
measurement has potential to become part of a future
multiparametric prognostic test for prostate cancer prognosis
assessment.
In summary, these data show that SPDEF is a weak to
moderate prognostic parameter in prostate cancer, especially in the
ERG negative subset. Our data are consistent with a particularly
strong up regulation of SPDEF in response to accelerated cell
proliferation in the subset of PTEN deficient and genetically
instable prostate cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Wilfried Fehrle
(Department of Pathology, University Medical Center
Hamburg-Eppendorf, Hamburg, Germany) for help in revision of the
manuscript, and are grateful to Mrs. Sünje Seekamp and Mrs. Inge
Brandt (Department of Pathology, University Medical Center
Hamburg-Eppendorf, Hamburg, Germany) for excellent technical
assistance.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JM, FB, RS and GS designed the study, and drafted
the manuscript. TS, MG, JRI, HHe and HHu participated in study
design. KS, CB, CG, AH, VR and SW performed IHC analysis and
scoring. FJ, CMK, TM, NCB, FL, FV, ML, CF and SM participated in
pathology data analysis. CH-M, NCB and RS performed statistical
analysis. SB, KM, and DH participated in data interpretation, and
helped to draft the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the Ärztekammer Hamburg
approved the study protocol (approval no. WF-049/09). According to
local laws (HmbKHG §12a), patient informed consent was not
required. Patient records/information were anonymized and
de-identified prior to analysis. All procedures were performed in
compliance with the principles outlined in the Helsinki
Declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ERG
|
ETS-related gene
|
|
ETS
|
E26 transformation-specific
|
|
SPDEF
|
SAM pointed domain-containing Ets
transcription factor
|
|
TMA
|
tissue microarray
|
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
UICC
|
International Union Against Cancer
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thompson IM Jr and Tangen CM: Prostate
cancer-uncertainty and a way forward. N Engl J Med. 367:270–271.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wilt TJ, Brawer MK, Jones KM, Barry MJ,
Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, et al:
Radical prostatectomy versus observation for localized prostate
cancer. N Engl J Med. 367:203–213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharrocks AD: The ETS-domain transcription
factor family. Nat Rev Mol Cell Biol. 2:827–837. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clark JP and Cooper CS: ETS gene fusions
in prostate cancer. Nat Rev Urol. 6:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar-Sinha C, Tomlins SA and Chinnaiyan
AM: Recurrent gene fusions in prostate cancer. Nat Rev Cancer.
8:497–511. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomlins SA, Bjartell A, Chinnaiyan AM,
Jenster G, Nam RK, Rubin MA and Schalken JA: ETS gene fusions in
prostate cancer: From discovery to daily clinical practice. Eur
Urol. 56:275–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oettgen P, Finger E, Sun Z, Akbarali Y,
Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, et al:
PDEF, a novel prostate epithelium-specific ets transcription
factor, interacts with the androgen receptor and activates
prostate-specific antigen gene expression. J Biol Chem.
275:1216–1225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghadersohi A and Sood AK: Prostate
epithelium-derived Ets transcription factor mRNA is overexpressed
in human breast tumors and is a candidate breast tumor marker and a
breast tumor antigen. Clin Cancer Res. 7:2731–2738. 2001.PubMed/NCBI
|
|
10
|
Ghadersohi A, Odunsi K, Lele S, Collins Y,
Greco WR, Winston J, Liang P and Sood AK: Prostate derived Ets
transcription factor shows better tumor-association than other
cancer-associated molecules. Oncol Rep. 11:453–458. 2004.PubMed/NCBI
|
|
11
|
Horst D, Gu X, Bhasin M, Yang Q, Verzi M,
Lin D, Joseph M, Zhang X, Chen W, Li YP, et al: Requirement of the
epithelium-specific Ets transcription factor Spdef for mucous gland
cell function in the gastric antrum. J Biol Chem. 285:35047–35055.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gunawardane RN, Sgroi DC, Wrobel CN, Koh
E, Daley GQ and Brugge JS: Novel role for PDEF in epithelial cell
migration and invasion. Cancer Res. 65:11572–11580. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sood AK, Saxena R, Groth J, Desouki MM,
Cheewakriangkrai C, Rodabaugh KJ, Kasyapa CS and Geradts J:
Expression characteristics of prostate-derived Ets factor support a
role in breast and prostate cancer progression. Hum Pathol.
38:1628–1638. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feldman RJ, Sementchenko VI, Gayed M,
Fraig MM and Watson DK: Pdef expression in human breast cancer is
correlated with invasive potential and altered gene expression.
Cancer Res. 63:4626–4631. 2003.PubMed/NCBI
|
|
15
|
Johnson TR, Koul S, Kumar B, Khandrika L,
Venezia S, Maroni PD, Meacham RB and Koul HK: Loss of PDEF, a
prostate-derived Ets factor is associated with aggressive phenotype
of prostate cancer: Regulation of MMP 9 by PDEF. Mol Cancer.
9:1482010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rodabaugh KJ, Mhawech-Fauceglia P, Groth
J, Lele S and Sood AK: Prostate-derived Ets factor is overexpressed
in serous epithelial ovarian tumors. Int J Gynecol Pathol.
26:10–15. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moussa O, Turner DP, Feldman RJ,
Sementchenko VI, McCarragher BD, Desouki MM, Fraig M and Watson DK:
PDEF is a negative regulator of colon cancer cell growth and
migration. J Cell Biochem. 108:1389–1398. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu X, Zerbini LF, Out HH, Bhasin M, Yang
Q, Joseph MG, Grall F, Onatunde T, Correa RG and Libermann TA:
Reduced PDEF expression increases invasion and expression of
mesenchymal genes in prostate cancer cells. Cancer Res.
67:4219–4226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Turner DP, Findlay VJ, Moussa O,
Semenchenko VI, Watson PM, LaRue AC, Desouki MM, Fraig M and Watson
DK: Mechanisms and functional consequences of PDEF protein
expression loss during prostate cancer progression. Prostate.
71:1723–1735. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ghadersohi A, Sharma S, Zhang S, Azrak RG,
Wilding GE, Manjili MH and Li F: Prostate-derived Ets transcription
factor (PDEF) is a potential prognostic marker in patients with
prostate cancer. Prostate. 71:1178–1188. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Steffan JJ, Koul S, Meacham RB and Koul
HK: The transcription factor SPDEF suppresses prostate tumor
metastasis. J Biol Chem. 287:29968–29978. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng XH, Black M, Ustiyan V, Le T,
Fulford L, Sridharan A, Medvedovic M, Kalinichenko VV, Whitsett JA
and Kalin TV: SPDEF inhibits prostate carcinogenesis by disrupting
a positive feedback loop in regulation of the Foxm1 oncogene. PLoS
Genet. 10:e10046562014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Erbersdobler A, Fritz H, Schnöger S,
Graefen M, Hammerer P, Huland H and Henke RP: Tumour grade,
proliferation, apoptosis, microvessel density, p53, and bcl-2 in
prostate cancers: Differences between tumours located in the
transition zone and in the peripheral zone. Eur Urol. 41:40–46.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sauter G, Steurer S, Clauditz TS, Krech T,
Wittmer C, Lutz F, Lennartz M, Janssen T, Hakimi N, Simon R, et al:
Clinical utility of quantitative gleason grading in prostate
biopsies and prostatectomy specimens. Eur Urol. 69:592–598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kononen J, Bubendorf L, Kallioniemi A,
Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G
and Kallioniemi OP: Tissue microarrays for high-throughput
molecular profiling of tumor specimens. Nat Med. 4:844–847. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Minner S, Wittmer C, Graefen M, Salomon G,
Steuber T, Haese A, Huland H, Bokemeyer C, Yekebas E, Dierlamm J,
et al: High level PSMA expression is associated with early PSA
recurrence in surgically treated prostate cancer. Prostate.
71:281–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Minner S, Jessen B, Stiedenroth L, Burandt
E, Köllermann J, Mirlacher M, Erbersdobler A, Eichelberg C, Fisch
M, Brümmendorf TH, et al: Low level HER2 overexpression is
associated with rapid tumor cell proliferation and poor prognosis
in prostate cancer. Clin Cancer Res. 16:1553–1560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burkhardt L, Fuchs S, Krohn A, Masser S,
Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et
al: CHD1 is a 5q21 tumor suppressor required for ERG rearrangement
in prostate cancer. Cancer Res. 73:2795–2805. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kluth M, Hesse J, Heinl A, Krohn A,
Steurer S, Sirma H, Simon R, Mayer PS, Schumacher U, Grupp K, et
al: Genomic deletion of MAP3K7 at 6q12-22 is associated with early
PSA recurrence in prostate cancer and absence of TMPRSS2:ERG
fusions. Mod Pathol. 26:975–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Krohn A, Diedler T, Burkhardt L, Mayer PS,
De Silva C, Meyer-Kornblum M, Kötschau D, Tennstedt P, Huang J,
Gerhäuser C, et al: Genomic deletion of PTEN is associated with
tumor progression and early PSA recurrence in ERG fusion-positive
and fusion-negative prostate cancer. Am J Pathol. 181:401–412.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krohn A, Seidel A, Burkhardt L, Bachmann
F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M, et
al: Recurrent deletion of 3p13 targets multiple tumour suppressor
genes and defines a distinct subgroup of aggressive ERG
fusion-positive prostate cancers. J Pathol. 231:130–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Burdelski C, Reiswich V, Hube-Magg C,
Kluth M, Minner S, Koop C, Graefen M, Heinzer H, Tsourlakis MC,
Wittmer C, et al: Cytoplasmic accumulation of sequestosome 1 (p62)
is a predictor of biochemical recurrence, rapid tumor cell
proliferation, and genomic instability in prostate cancer. Clin
Cancer Res. 21:3471–3479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Epstein JI, Feng Z, Trock BJ and
Pierorazio PM: Upgrading and downgrading of prostate cancer from
biopsy to radical prostatectomy: Incidence and predictive factors
using the modified Gleason grading system and factoring in tertiary
grades. Eur Urol. 61:1019–1024. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Situ J, Zhang H, Lu L, Li K, Hu C and Wang
DJ: Clinical significance of PSMA, TERT and PDEF in malignant
tumors of the prostate. Eur Rev Med Pharmacol Sci. 21:3347–3352.
2017.PubMed/NCBI
|
|
35
|
Mukhopadhyay A, Khoury T, Stein L,
Shrikant P and Sood AK: Prostate derived Ets transcription factor
and Carcinoembryonic antigen related cell adhesion molecule 6
constitute a highly active oncogenic axis in breast cancer.
Oncotarget. 4:610–621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sood AK, Geradts J and Young J:
Prostate-derived Ets factor, an oncogenic driver in breast cancer.
Tumour Biol. 39:10104283176916882017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ghadersohi A, Odunsi K, Zhang S, Azrak RG,
Bundy BN, Manjili MH and Li F: Prostate-derived Ets transcription
factor as a favorable prognostic marker in ovarian cancer patients.
Int J Cancer. 123:1376–1384. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tsui KH, Lin YH, Chung LC, Chuang ST, Feng
TH, Chiang KC, Chang PL, Yeh CJ and Juang HH: Prostate-derived ets
factor represses tumorigenesis and modulates
epithelial-to-mesenchymal transition in bladder carcinoma cells.
Cancer Lett. 375:142–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Krohn A, Freudenthaler F, Harasimowicz S,
Kluth M, Fuchs S, Burkhardt L, Stahl P, C Tsourlakis M, Bauer M,
Tennstedt P, et al: Heterogeneity and chronology of PTEN deletion
and ERG fusion in prostate cancer. Mod Pathol. 27:1612–1620. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kluth M, Runte F, Barow P, Omari J,
Abdelaziz ZM, Paustian L, Steurer S, Christina Tsourlakis M, Fisch
M, Graefen M, et al: Concurrent deletion of 16q23 and PTEN is an
independent prognostic feature in prostate cancer. Int J Cancer.
137:2354–2363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kluth M, Scherzai S, Büschek F, Fraune C,
Möller K, Höflmayer D, Minner S, Göbel C, Möller-Koop C, Hinsch A,
et al: 13q deletion is linked to an adverse phenotype and poor
prognosis in prostate cancer. Genes Chromosomes Cancer. 57:504–512.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grupp K, Diebel F, Sirma H, Simon R,
Breitmeyer K, Steurer S, Hube-Magg C, Prien K, Pham T, Weigand PU,
et al: SPINK1 expression is tightly linked to 6q15- and
5q21-deleted ERG-fusion negative prostate cancers but unrelated to
PSA recurrence. Prostate. 73:1690–1698. 2013.PubMed/NCBI
|
|
43
|
Luk IY, Reehorst CM and Mariadason JM:
ELF3, ELF5, EHF and SPDEF transcription factors in tissue
homeostasis and cancer. Molecules. 23(pii): E21912018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Egevad L, Ahmad AS, Algaba F, Berney DM,
Boccon-Gibod L, Compérat E, Evans AJ, Griffiths D, Grobholz R,
Kristiansen G, et al: Standardization of Gleason grading among 337
European pathologists. Histopathology. 62:247–256. 2013. View Article : Google Scholar : PubMed/NCBI
|