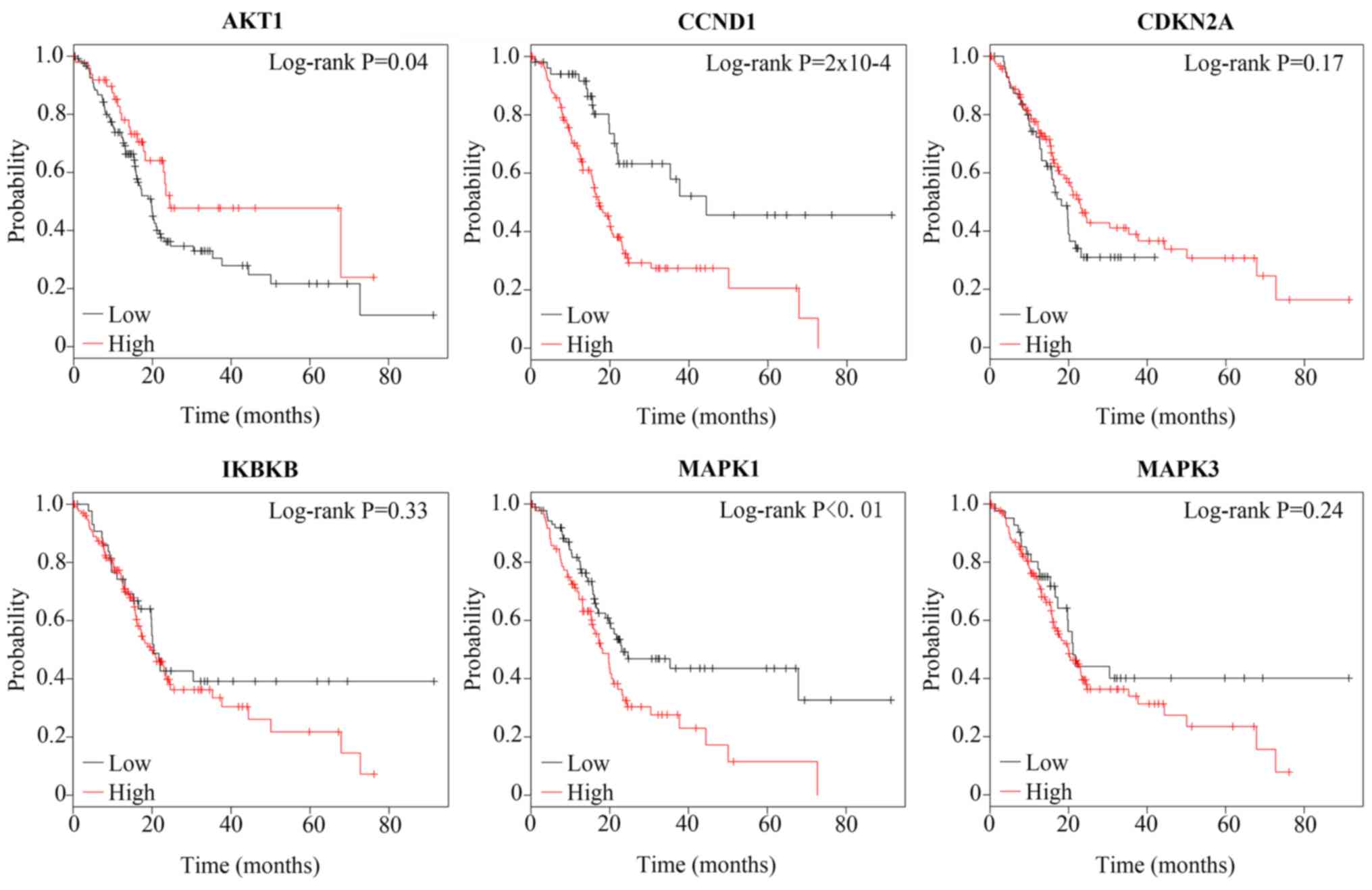
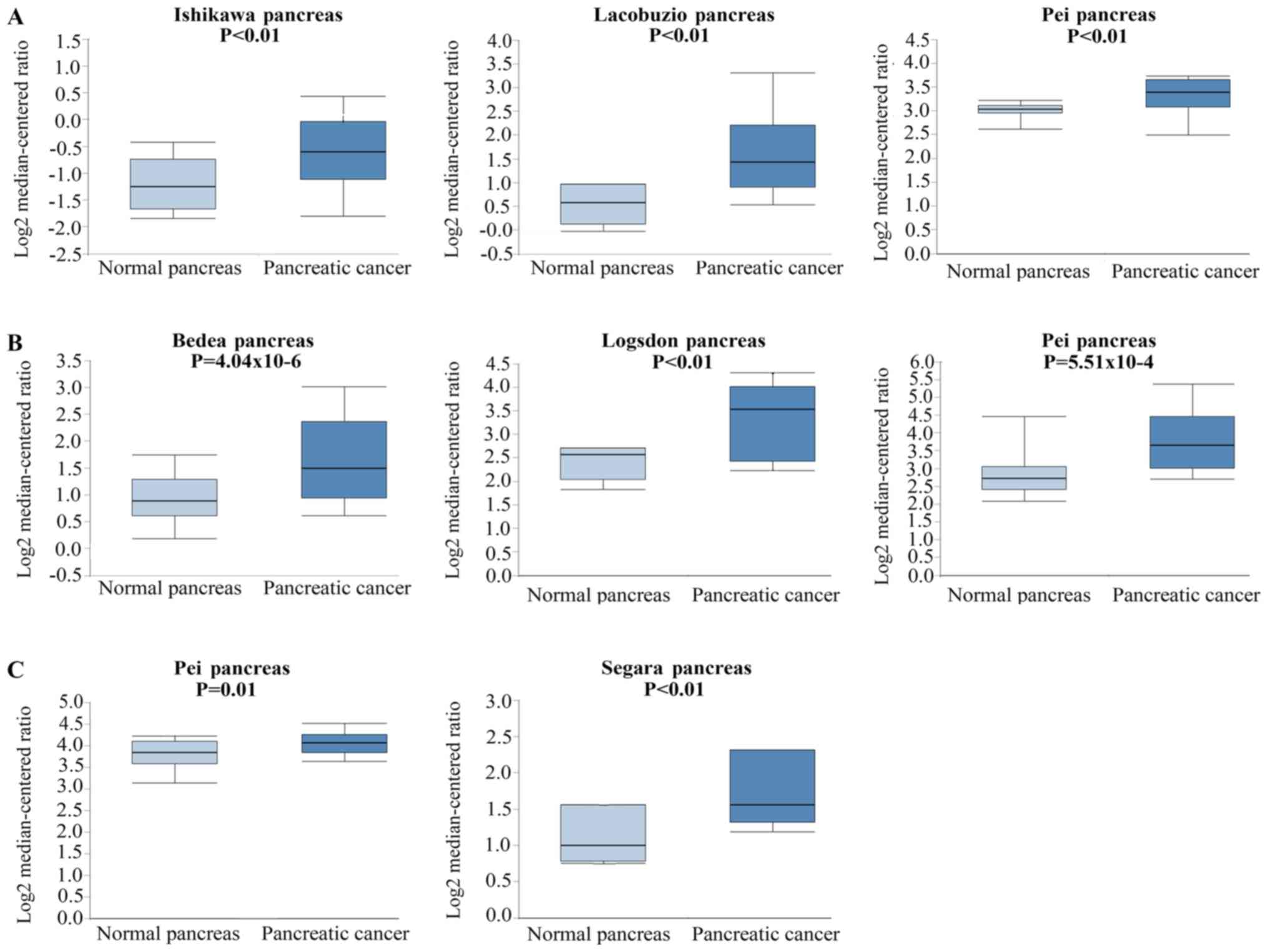
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saad AM, Turk T, Al-Husseini MJ and
Abdel-Rahman O: Trends in pancreatic adenocarcinoma incidence and
mortality in the United States in the last four decades; a
SEER-based study. BMC Cancer. 18:6882018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soignet SL, Frankel SR, Douer D, Tallman
MS, Kantarjian H, Calleja E, Stone RM, Kalaycio M, Scheinberg DA,
Steinherz P, et al: United States multicenter study of arsenic
trioxide in relapsed acute promyelocytic leukemia. J Clin Oncol.
19:3852–3860. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mathews V, George B, Chendamarai E,
Lakshmi KM, Desire S, Balasubramanian P, Viswabandya A, Thirugnanam
R, Abraham A, Shaji RV, et al: Single-agent arsenic trioxide in the
treatment of newly diagnosed acute promyelocytic leukemia:
Long-term follow-up data. J Clin Oncol. 28:3866–3871. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghavamzadeh A, Alimoghaddam K, Rostami S,
Ghaffari SH, Jahani M, Iravani M, Mousavi SA, Bahar B and Jalili M:
Phase II study of single-agent arsenic trioxide for the front-line
therapy of acute promyelocytic leukemia. J Clin Oncol.
29:2753–2757. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lo-Coco F, Avvisati G, Vignetti M, Thiede
C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona
E, et al: Retinoic acid and arsenic trioxide for acute
promyelocytic leukemia. N Engl J Med. 369:111–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Burnett AK, Russell NH, Hills RK, Bowen D,
Kell J, Knapper S, Morgan YG, Lok J, Grech A, Jones G, et al:
Arsenic trioxide and all-trans retinoic acid treatment for acute
promyelocytic leukaemia in all risk groups (AML17): Results of a
randomised, controlled, phase 3 trial. Lancet Oncol. 16:1295–1305.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seftel MD, Barnett MJ, Couban S, Leber B,
Storring J, Assaily W, Fuerth B, Christofides A and Schuh AC: A
Canadian consensus on the management of newly diagnosed and
relapsed acute promyelocytic leukemia in adults. Curr Oncol.
21:234–250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L, Liu L, Zhan S, Chen L, Wang Y,
Zhang Y, Du J, Wu Y and Gu L: Arsenic trioxide suppressed migration
and angiogenesis by targeting FOXO3a in gastric cancer cells. Int J
Mol Sci. 19(pii): E37392018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhong L, Xu F and Chen F: Arsenic trioxide
induces the apoptosis and decreases NF-KB expression in lymphoma
cell lines. Oncol Lett. 16:6267–6274. 2018.PubMed/NCBI
|
|
12
|
Mao MH, Huang HB, Zhang XL, Li K, Liu YL
and Wang P: Additive antitumor effect of arsenic trioxide combined
with intravesical bacillus Calmette-Guerin immunotherapy against
bladder cancer through blockade of the IER3/Nrf2 pathway. Biomed
Pharmacother. 107:1093–1103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du S, Liu K, Gao P, Li Z and Zheng J:
Differential anticancer activities of arsenic trioxide on head and
neck cancer cells with different human papillomavirus status. Life
Sci. 212:182–193. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo D, Zhang X, Du R, Gao W, Luo N, Zhao
S, Li Y, Chen R, Wang H, Bao Y, et al: Low dosage of arsenic
trioxide (As2O3) inhibits angiogenesis in
epithelial ovarian cancer without cell apoptosis. J Biol Inorg
Chem. 23:939–947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao JK, Wang LX, Long B, Ye XT, Su JN, Yin
XY, Zhou XX and Wang ZW: Arsenic Trioxide inhibits cell growth and
invasion via Down-Regulation of Skp2 in pancreatic cancer cells.
Asian Pac J Cancer Prev. 16:3805–3810. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang YA, You ZH and Chen X: A systematic
prediction of drug-target interactions using molecular fingerprints
and protein sequences. Curr Protein Pept Sci. 19:468–478. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36:D901–D906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kuhn M, von Mering C, Campillos M, Jensen
LJ and Bork P: STITCH: Interaction networks of chemicals and
proteins. Nucleic Acids Res. 36:D684–D688. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bindea G, Galon J and Mlecnik B: CluePedia
Cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Biankin AV, Waddell N, Kassahn KS, Gingras
MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J,
et al: Pancreatic cancer genomes reveal aberrations in axon
guidance pathway genes. Nature. 491:399–405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bailey P, Chang DK, Nones K, Johns AL,
Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC,
et al: Genomic analyses identify molecular subtypes of pancreatic
cancer. Nature. 531:47–52. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sanchez-Vega F, Mina M, Armenia J, Chatila
WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia
S, et al: Oncogenic signaling pathways in the cancer genome atlas.
Cell. 173:321–337.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Witkiewicz AK, McMillan EA, Balaji U, Baek
G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et
al: Whole-exome sequencing of pancreatic cancer defines genetic
diversity and therapeutic targets. Nat Commun. 6:67442015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lanczky A, Nagy A, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ishikawa M, Yoshida K, Yamashita Y, Ota J,
Takada S, Kisanuki H, Koinuma K, Choi YL, Kaneda R, Iwao T, et al:
Experimental trial for diagnosis of pancreatic ductal carcinoma
based on gene expression profiles of pancreatic ductal cells.
Cancer Sci. 96:387–393. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iacobuzio-Donahue CA, Maitra A, Olsen M,
Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq
R, et al: Exploration of global gene expression patterns in
pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol.
162:1151–1162. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
35
|
Logsdon CD, Simeone DM, Binkley C,
Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R and Hanash
S: Molecular profiling of pancreatic adenocarcinoma and chronic
pancreatitis identifies multiple genes differentially regulated in
pancreatic cancer. Cancer Res. 63:2649–2657. 2003.PubMed/NCBI
|
|
36
|
Segara D, Biankin AV, Kench JG, Langusch
CC, Dawson AC, Skalicky DA, Gotley DC, Coleman MJ, Sutherland RL
and Henshall SM: Expression of HOXB2, a retinoic acid signaling
target in pancreatic cancer and pancreatic intraepithelial
neoplasia. Clin Cancer Res. 11:3587–3596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chari ST: Detecting early pancreatic
cancer: Problems and prospects. Semin Oncol. 34:284–294. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han JB, Sang F, Chang JJ, Hua YQ, Shi WD,
Tang LH and Liu LM: Arsenic trioxide inhibits viability of
pancreatic cancer stem cells in culture and in a xenograft model
via binding to SHH-Gli. Onco Targets Ther. 6:1129–1138. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lang M, Wang X, Wang H, Dong J, Lan C, Hao
J, Huang C, Li X, Yu M, Yang Y, et al: Arsenic trioxide plus PX-478
achieves effective treatment in pancreatic ductal adenocarcinoma.
Cancer Lett. 378:87–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Michl P and Downward J: Mechanisms of
disease: PI3K/AKT signaling in gastrointestinal cancers. Z
Gastroenterol. 43:1133–1139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yan L, Wang Y, Wang ZZ, Rong YT, Chen LL,
Li Q, Liu T, Chen YH, Li YD, Huang ZH and Peng J: Cell motility and
spreading promoted by CEACAM6 through cyclin D1/CDK4 in human
pancreatic carcinoma. Oncol Rep. 35:418–426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
McWilliams RR, Wieben ED, Chaffee KG,
Antwi SO, Raskin L, Olopade OI, Li D, Highsmith WE Jr, Colon-Otero
G, Khanna LG, et al: CDKN2A Germline rare coding variants and risk
of pancreatic cancer in minority populations. Cancer Epidemiol
Biomarkers Prev. 27:1364–1370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liptay S, Weber CK, Ludwig L, Wagner M,
Adler G and Schmid RM: Mitogenic and antiapoptotic role of
constitutive NF-kappaB/Rel activity in pancreatic cancer. Int J
Cancer. 105:735–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fujioka S, Sclabas GM, Schmidt C,
Frederick WA, Dong QG, Abbruzzese JL, Evans DB, Baker C and Chiao
PJ: Function of nuclear factor kappaB in pancreatic cancer
metastasis. Clin Cancer Res. 9:346–354. 2003.PubMed/NCBI
|
|
45
|
Hu Y, Yang H, Lu XQ, Xu F, Li J and Qian
J: ARHI suppresses pancreatic cancer by regulating MAPK/ERK 1/2
pathway. Pancreas. 44:342–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hu B, Shi C, Jiang HX and Qin SY:
Identification of novel therapeutic target genes and pathway in
pancreatic cancer by integrative analysis. Medicine (Baltimore).
96:e82612017. View Article : Google Scholar : PubMed/NCBI
|