Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney carcinoma and the sixth and tenth most frequent type of
cancer diagnosed in men and women, respectively (1). The mortality rate of RCC reaches
140,000 cases per year worldwide, and RCC ranks as the 13th leading
cause of cancer-associated mortality (2). At present, surgical resection remains
the preferred curative therapy for RCC; however, since ~1/3 of
patients with RCC are diagnosed with advanced or metastatic RCC,
their prognosis remains unsatisfactory following curative operation
(3,4). In the past few decades, emerging
evidence demonstrated that the malignant phenotypes of tumors are
determined by the intrinsic activities of cancer cells and by the
complex interactions of various cell types in the tumor
microenvironment, in particular tumor-infiltrating immune cells
(TIICs) (5,6). Previous clinical and genomic studies
reported that RCC is an immunogenic tumor (7,8).
Furthermore, the cases of spontaneous and significant remission of
RCC are also considered to be mediated by the immune response
(9,10). In addition, immunotherapies have been
used in clinical trials of RCC alone or in combination with
targeted therapies, and have been demonstrated to prolong patients'
survival (11–13). It is therefore crucial to determine
the underlying mechanisms of tumor microenvironment infiltration
with immune cells in order to improve the immunotherapeutic
approaches to treat RCC. However, whether immune cells represent a
critical component of tumor microenvironment remains poorly
understood.
TIICs have been reported to function as tumor
promoters or suppressors in various types of cancer and to have
great prognostic value. For instance, tumor-associated macrophage
infiltration has a negative impact on prognosis in patients with
breast and bladder carcinomas, whereas CD8+ cytotoxic T
cells and CD45RO+ memory T cells, the shortest CD45 isoform which
lacks all three of A, B, and C regions, are associated with longer
overall survival (OS) and disease-free survival (DFS) (14,15).
Recent studies demonstrated that RCC is also infiltrated by
heterogeneous TIICs, including macrophages, T cells and B cells,
and that their location, density and type in RCC are associated
with prognosis (16,17). However, TIICs have largely been
characterized using tissue-based approaches, including
immunohistochemistry (IHC) and flow cytometry, which may be limited
by various factors, including the amount of tissue required, and
that the number of immune cell types may not be evaluated
simultaneously. These aforementioned approaches may therefore not
lead to comprehensive and convincing conclusions.
A novel gene-based approach named CIBERSORT has been
described; this computation method applies deconvolution of a
reference gene expression matrix and support vector regression to
estimate the fractions of various cell types, including TIICs, in
complex tissues (18,19). The diversity and landscape of TIICs
can therefore be determined by this approach. The present study
used CIBERSORT analytical tool to deconvolute gene expression data
and determine 22 immune cell types in RCC tissues, and further
investigated their clinical significance.
Materials and methods
Data acquisition from the cancer
genome atlas (TCGA) cohort
Gene expression profiles of RCC (n=895) and normal
samples (n=128), and clinical characteristics from the
corresponding patients were downloaded from the TCGA data portal in
March 2019 (20). RNA sequencing
data were normalized with the mean-variance modeling at the
observational level (voom) method (21), which converted count data to values
more similar to those resulting from microarrays. For clinical
data, only patients with RCC and complete information (n=421) were
included. Patients with RCC who had missing data for age, gender,
Fuhrman grade, Tumor-Node-Metastasis (TNM) classification, survival
time or status were excluded.
Evaluation of TIICs
CIBERSORT analytical tool is a deconvolution
algorithm that applies a set of barcode gene expression values that
corresponds to a ‘signature matrix’ of 547 genes to precisely
determine the immune cell composition in data from bulk tumor
samples of mixed cell types (18).
To quantify the proportion of 22 immune cell types in RCC tissues,
normalized gene expression datasets were prepared, and data were
uploaded to the CIBERSORT web portal (http://cibersort.stanford.edu/), with the algorithm
run and the number of permutations being set to 1,000. A total of
22 types of TIICs together with CIBERSORT metrics, including
CIBERSORT P-value, Pearson's correlation coefficient and root mean
square error, were quantified for each sample. The statistical
significance of the deconvolution consequences of all cell subsets
was reflected by the CIBERSORT P-value, which was used to filter
out deconvolution with a less significant fitting accuracy. To meet
the requirement of a CIBERSORT P≤0.05, normal samples (n=17) and
RCC samples (n=617) were selected.
IHC
Specimens from 20 patients diagnosed with RCC at the
First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) between February 2017 and October 2018 and who
received tumor resection surgery were used for IHC. All specimens
were fixed with 4% formalin at room temperature for 24 h, embedded
with paraffin and sliced into 4-µm sections and stored at −20°C.
For IHC, sections were deparaffinized with 100% xylene for 30 min
at room temperature and rehydrated with a decreasing gradient of
ethanol for 30 min at room temperature (100% for 7 min, 95% for 5
min, 85% for 5 min and 70% for 5 min). Sections were immersed in a
boiling sodium citrate-hydrochloric acid buffer solution and heated
in a microwave at 96–98°C for the antigen retrieval for 25 min.
After cooling at room temperature, 3% hydrogen peroxide was used to
block endogenous peroxidase at room temperature for 10 min.
Subsequently, sections were washed three times with TBS and blocked
with normal goat serum at room temperature for 15 min. Sections
were incubated with a monoclonal antibody against forkhead box P3
(FOXP3; 1:50; cat. no. 98377; Cell Signaling Technology, Inc.)
overnight at 4°C. Sections were then washed three times with TBS
and incubated with a goat anti-rabbit immunoglobulin G secondary
antibody at 37°C for 30 min. Sections were washed three times with
TBS and incubated with streptavidin-biotin-conjugated horseradish
peroxidase substrate at 37°C for another 30 min. The 3% hydrogen
peroxide, normal goat serum, secondary antibody, and
streptavidin-biotin-conjugated horseradish peroxidase substrate
were included in an IHC kit (cat. no. SP9001; ZSGB-BIO). Signal was
visualized with diaminobenzidine (ZSGB-BIO), and sections were
counterstained with hematoxylin at room temperature for
approximately 10 sec. Sections were analyzed as previously
described (19). Briefly, Treg
density was determined according to the percentage of positive
cells normalized to the total number of cells under a light
microscope, at ×400 magnification.
Statistical analysis
Statistical analyses were performed with SPSS 22.0
software (IBM Corp.), GraphPad Prism 5.0 software (GraphPad
Software, Inc.) and R software (version 3.5.2; R Foundation).
Kaplan-Meier analysis was used to determine survival curves that
reflect the association between immune cells and corresponding
clinical follow-ups, which were evaluated by the log-rank test. The
median value of the proportion of each cell type was used for
Kaplan-Meier analysis and modeled as continuous variable to obtain
interpretable hazard ratios (HRs). Kruskal-Wallis test was used to
analyze the association between TIICs and clinical information
(Fuhrman grade and TNM classification). Univariate and multivariate
Cox regression analyses were used to estimate the prognostic
significance of the inferred TIICs. Two tailed Student's t-test was
used to examine the statistical relevance between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Profile of immune infiltration in RCC
and normal samples
According to the CIBERSORT algorithm, the 22 immune
cell types infiltrated in RCC samples were investigated and their
proportions were visualized. The results revealed the profile of
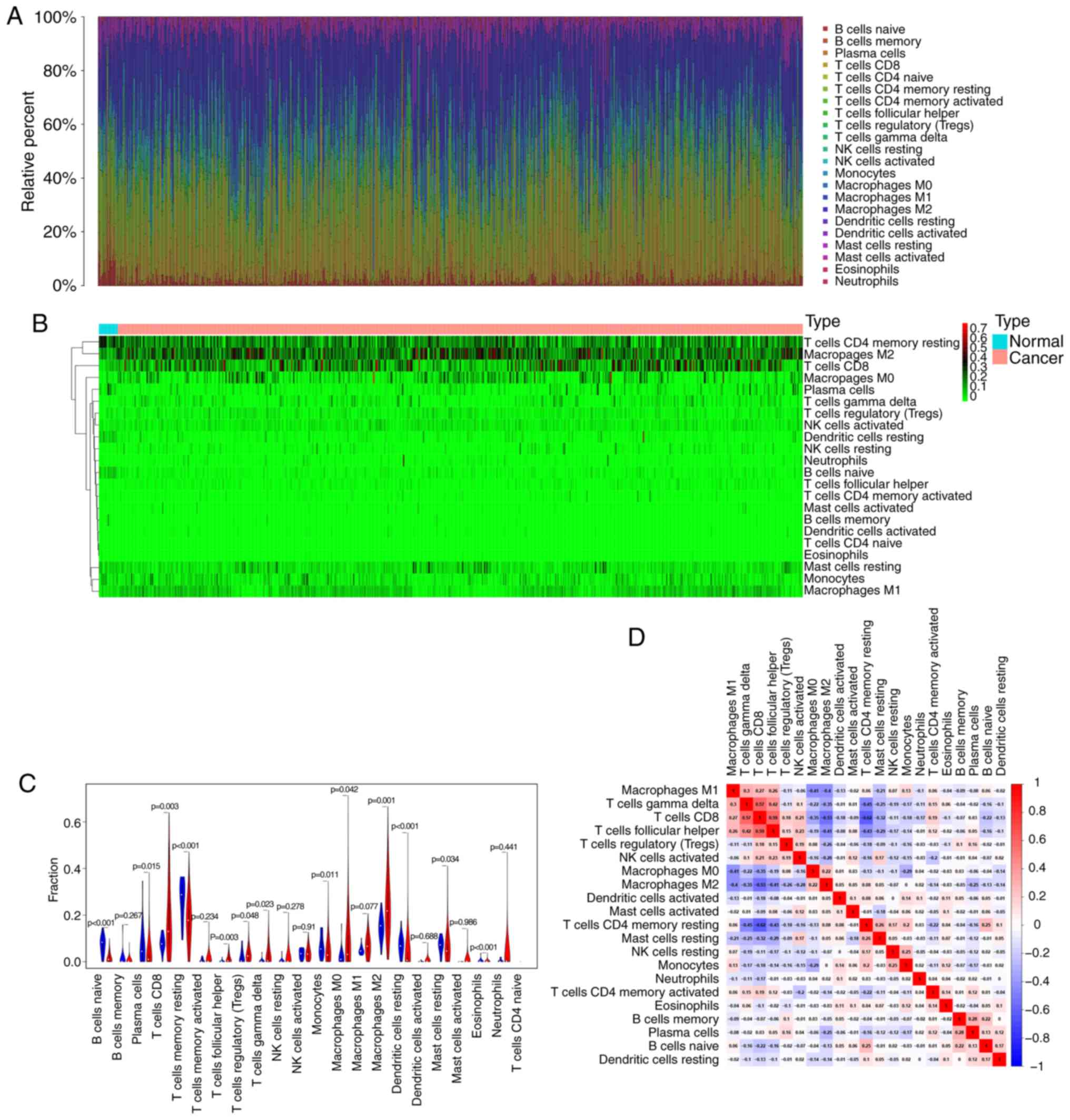
immune infiltration in RCC samples. The bar plot demonstrated the
results from 17 normal samples and 617 RCC samples. The proportions
of immune cells were variable in each sample (Fig. 1A). In addition, unsupervised
hierarchical clustering was used to identify the subpopulations of
immune cells. The normal and tumor samples were divided into two
discrete groups, and the proportions of TIICs varied between these
groups (Fig. 1B). These results
suggested that the variation of TIICs proportions may be an
essential characteristic that could describe the individual
differences. As presented in Fig.
1C, the differences in each immune cell between normal and RCC
samples were further analyzed. The results demonstrated that 13
immune cells were significantly different between the two sample
types. In particular, CD8 T cells, follicular helper T cells,
Tregs, γδ T cells, and M0 and M2 macrophages were more present in
RCC samples than in normal samples, whereas B cells naïve, plasma
cells, CD4 memory resting T cells, monocytes, resting dendritic
cells, resting mast cells and the eosinophil fraction were lower in
RCC samples compared with those in normal samples. These TIICs may
therefore be involved in the development of RCC and may be
associated with the prognosis of patients with RCC. Furthermore,
the results from the correlation matrix demonstrated that
follicular helper T cells had the strongest positive correlation
with CD8 T cells, whereas CD4 memory resting T cells had the
strongest negative correlation with CD8 T cells, and that the
proportions of other TIICs subpopulations presented a weak to
moderate correlation (Fig. 1D).
Taken together, these findings suggested that the deviations of the
RCC immuno-landscape from normal tissue may have significant
clinical implications.
Prognostic value of TIICs in RCC
samples
The correlation between prognosis of patients with
RCC and TIIC subpopulations was further investigated. Following
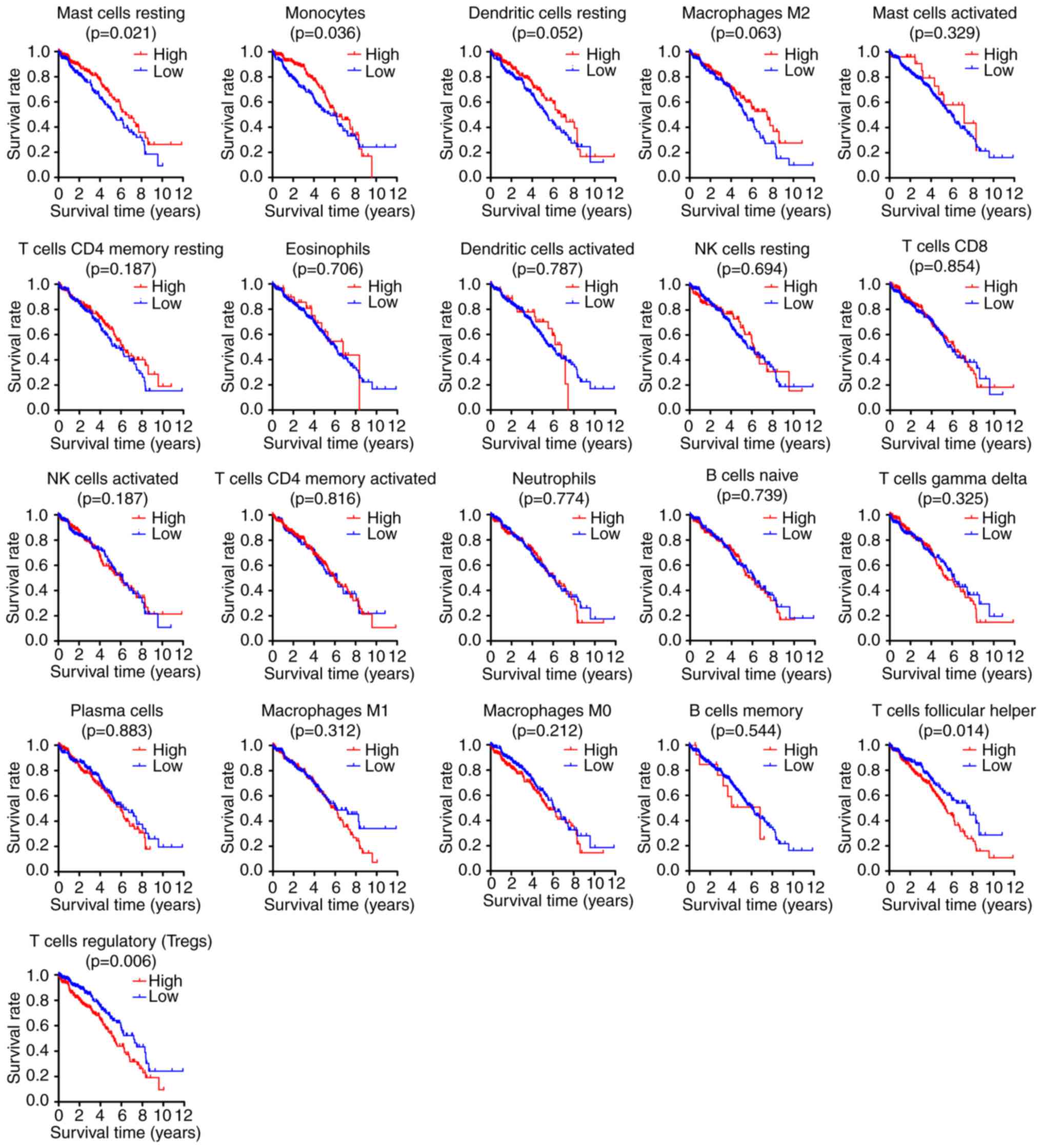
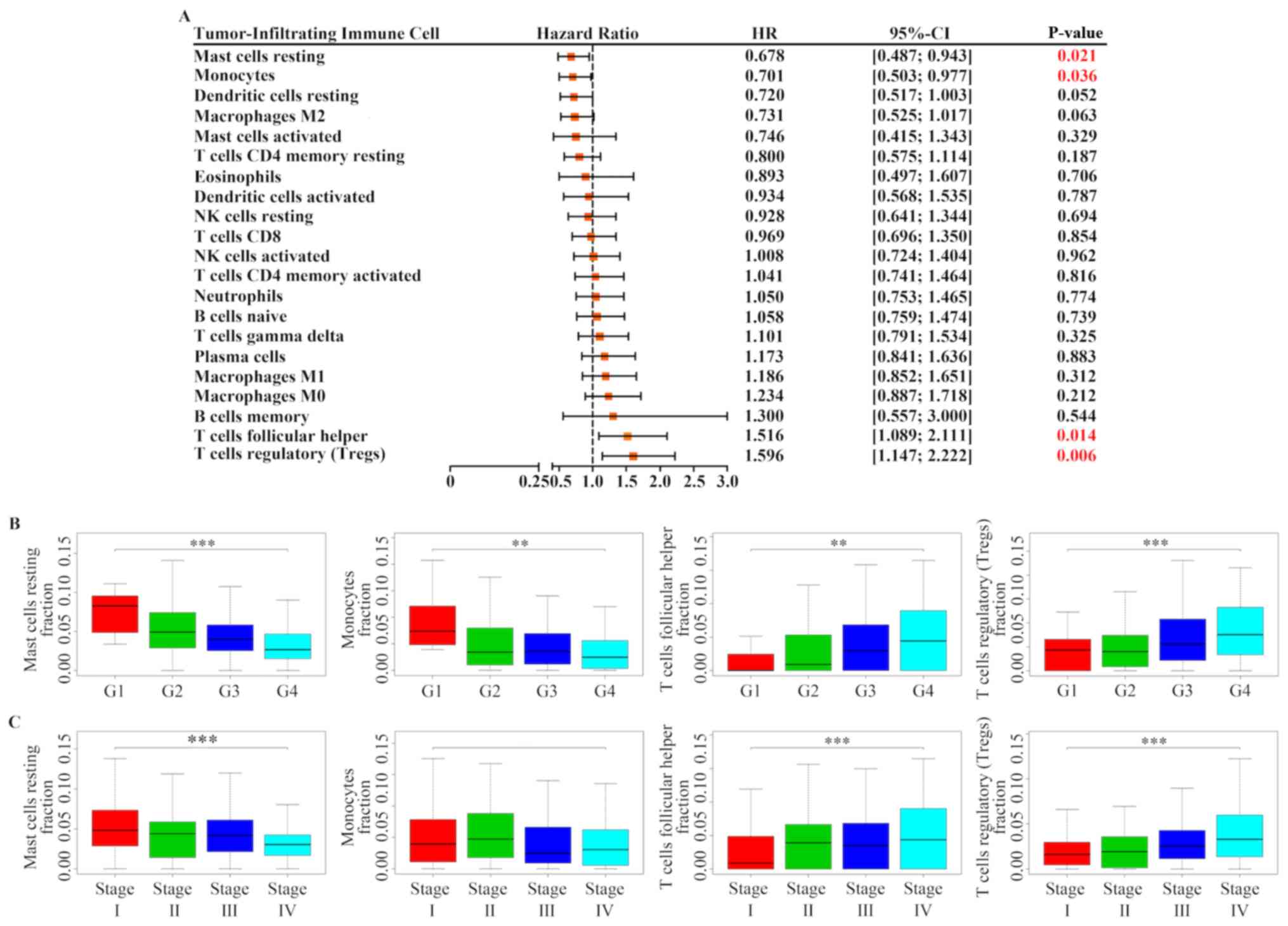
Kaplan-Meier analysis and log-rank test, the results demonstrated
that increased proportion of resting mast cells [HR=0.678; 95%
confidence interval (CI), 0.487–0.943; P=0.021] and monocytes
(HR=0.701; 95% CI, 0.503–0.977; P=0.036) was associated with a
favorable outcome, whereas a greater proportion of follicular
helper T cells (HR=1.516; 95% CI, 1.089–2.111; P=0.014) and Tregs
(HR=1.596; 95% CI, 1.147–2.222; P=0.006) was associated with poor
prognosis (Figs. 2 and 3A). In addition, the association between
these four immune cell types and patients' clinical information was
determined. As presented in Fig. 3B and
C, Tregs and follicular helper T cells were positively
associated with Fuhrman grade and TNM classification, whereas
resting mast cells were negatively associated with Fuhrman grade
and TNM classification. Furthermore, monocytes were also negatively
associated with Fuhrman grade, but was not associated with TNM
classification.
Cox regression model for OS
To determine whether inferred TIICs were independent
risk factors for the prognosis of patients with RCC, Cox regression
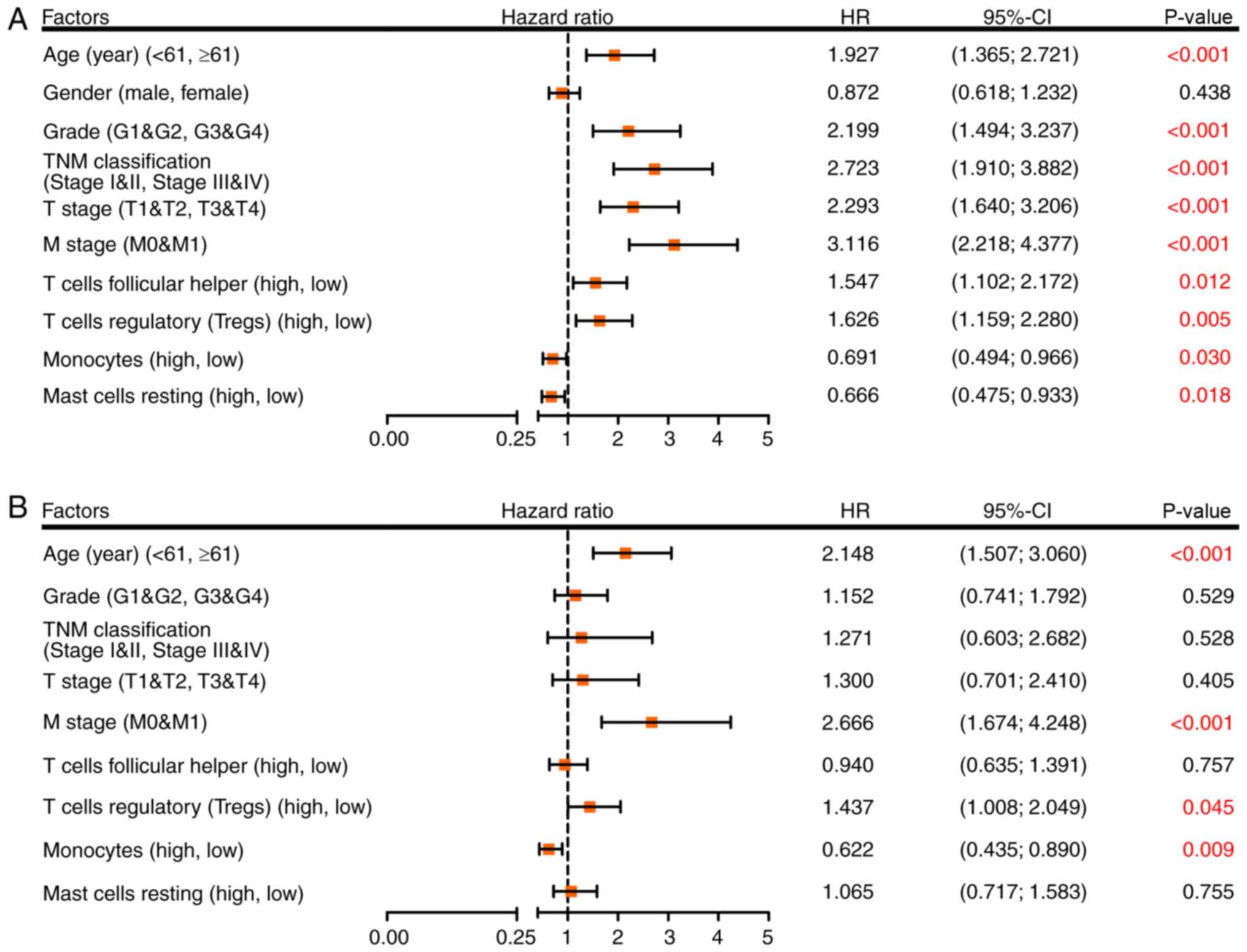
analysis was used. Univariate analysis demonstrated that age,
grade, TNM classification, T stage, M stage, follicular helper T
cells, Tregs, monocytes and resting mast cells were significant
risk factors for RCC outcome (Fig.
4A). These factors were then incorporated into a multivariate
analysis, and the results demonstrated that age ≥61 years
(HR=2.148; 95% CI, 1.507–3.060; P<0.001), M1 stage (HR=2.666;
95% CI, 1.674–4.248; P<0.001), a high proportion of Tregs
(HR=1.437; 95% CI, 1.008–2.049; P=0.045) and a low proportion of
monocytes (HR=0.622; 95% CI, 0.435–0.890; P=0.009) were separately
associated with an increased risk of mortality (Fig. 4B).
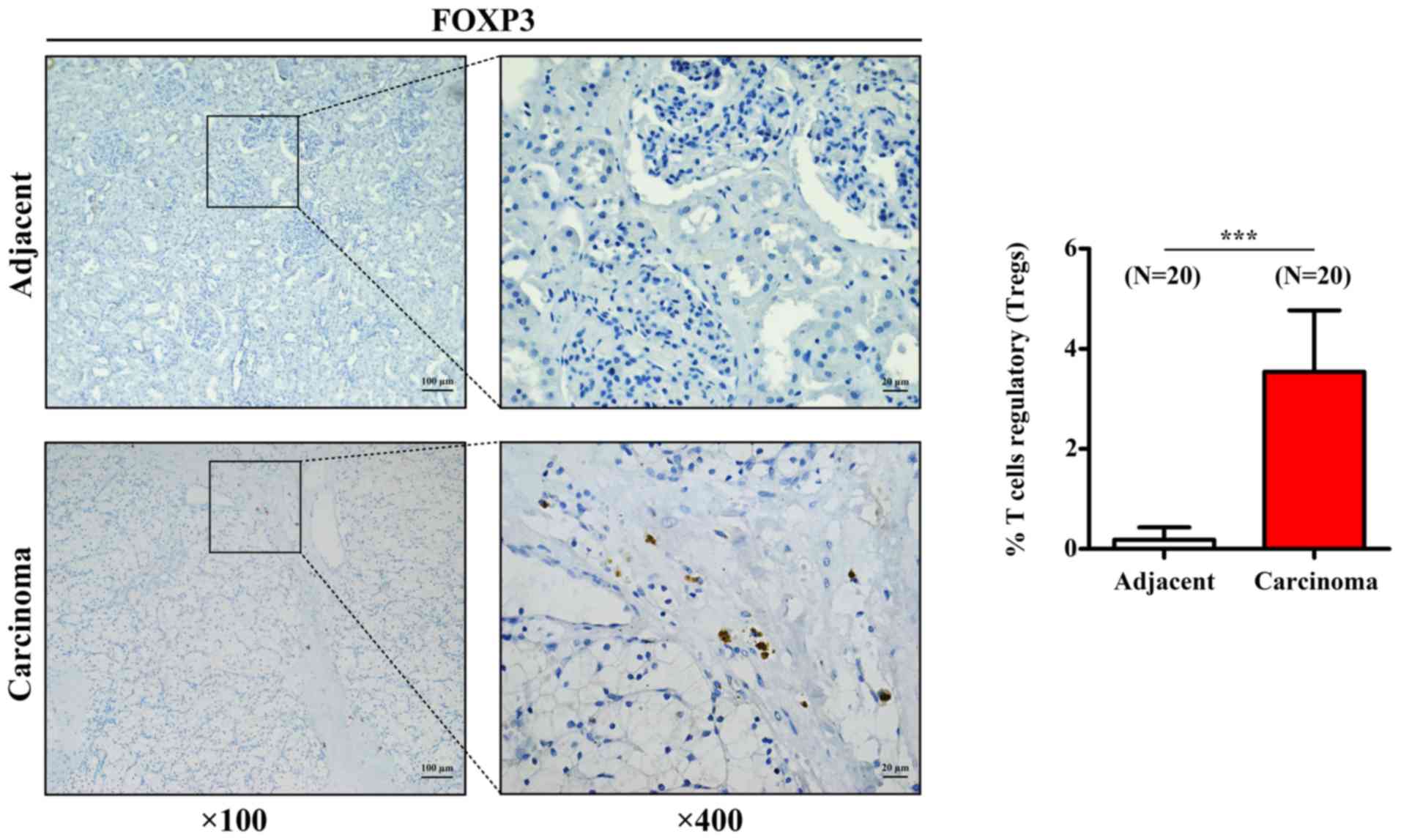
Tregs infiltration in RCC samples
To further confirm the aforementioned results
acquired for Tregs, the expression of the transcription factor
FOXP3, which is the characteristic marker of Tregs, was assessed by
IHC in 20 human RCC tissues and 20 paired adjacent tissues. As
presented in Fig. 5, Treg density
was significantly higher in RCC tissues compared with that in
adjacent tissues, which was similar to the results from CIBERSORT
analysis.
Discussion
Tumors are no longer considered as bulks of
malignant cancer cells, but as a complex tumor microenvironments
where other cell subpopulations corrupted by malignant neoplastic
cells are recruited in order to constitute a self-sufficient
biological structure (22). The
tumor-related microenvironment comprises various cell populations,
including immune cells, cancer-associated fibroblasts and
endothelial cells (23). In
addition, numerous chemokines, cytokines, proangiogenic mediators,
growth factors and proteins of the extracellular matrix are
involved in the interaction between these subpopulations of cells,
such as immune cells, cancer-associated fibroblasts and endothelial
cells, and cancer cells (24). The
predominant cells recruited to and activated in the tumor
microenvironment are immune cells (24). The immune system has been reported to
possess cancer-inhibiting and cancer-promoting roles in the tumor
microenvironment, and is associated with the process of cancer
development (25–27). In RCC, a high variety of infiltrating
immune cells has been reported. These immune cells can activate the
immune response and are associated with clinical outcome. For
example, a high level of CD8+ T lymphocytes is
associated with prolonged OS in patients with RCC (28).
In the present study, the results from the
deconvolution of bulk gene expression data from TCGA cohort allowed
the comprehensive analysis of the different immune cell
infiltrations in RCC samples. Great differences in the immune cell
composition between RCC and normal tissues were observed. Since CD4
naïve T cells were not infiltrated in the majority of samples,
correlation analysis and subsequent survival analysis could not be
performed. Furthermore, the association between these
immune-infiltrated cells and the clinical outcome of patients with
RCC was investigated. The results demonstrated that a greater
proportion of resting mast cells and monocytes was associated with
a favorable prognosis, whereas a high level of follicular helper T
cells and Tregs was associated with poor prognosis. In addition,
following multivariate Cox regression analysis, Tregs and monocytes
were determined as independent risk factors for the outcome of
patients with RCC.
Tregs are adaptive immune cells that can suppress an
excessive immune response and maintain self-tolerance and
homeostasis (29). In cancer, Tregs
can suppress the antitumor response, leading to tumor immune escape
(30). A previous meta-analysis
demonstrated that a high level of infiltration of FOXP3+
Tregs in the majority of solid tumors is significantly associated
with poor prognosis of patients with cancer, including renal cancer
(31). In patients with RCC, it has
been reported that an increased number of Tregs in peripheral blood
and tumors is negatively correlated with OS (32,33). A
high proportion of infiltrated Tregs in RCC may therefore indicate
a worse outcome, which is similar to the CIBERSORT findings from
the present study. Furthermore, the present study demonstrated that
the proportion of Tregs was associated with tumor grade and TNM
classification, which was also the case for other cancer types
(34,35). In addition, the results from IHC
demonstrated that FOXP3 expression in Tregs was increased in RCC
samples compared with that in adjacent tumors. Taken together,
these findings suggested that Tregs may represent a high-risk
factor for worse outcome in patients with RCC, and that decreasing
Treg infiltration may be considered as a potential therapy in
RCC.
Monocytes circulate in peripheral blood, migrate
into tumor tissue and differentiate into different macrophage types
in the tumor microenvironment (36).
In humans, monocytes are heterogeneous and divided into three
subsets, including classical, intermediate and non-classical,
according to their expression of CD14 and CD16 (37). It is commonly considered that
classical monocytes essentially differentiate into M1 macrophages,
which contribute to the promotion of inflammation and tumor
suppression (38). Conversely,
intermediate and non-classical monocytes usually differentiate into
M2 macrophages, which serve immunosuppressive and tumor-promoting
roles (39). Furthermore, it has
been demonstrated that the composition of the monocyte subset is
altered in numerous diseases, including cancer (40,41).
Notably, the mutual conversion of M1 and M2 macrophage are valuable
for prognosis (42). In the present
study, monocytes were associated with positive prognosis in
patients with RCC. However, several intersections in the survival
curve of monocytes were highlighted, and the survival rate of the
patients with RCC exhibiting a high monocyte count and those
exhibiting a low monocyte count was reversed after ~8 years. These
results may be due to changes in the composition of monocyte
subsets, which may affect the prognosis of patients; however, this
hypothesis requires further investigation.
In conclusion, the present study demonstrated that
deconvolution of whole-tissue gene expression data by CIBERSORT
provided refined information on the immune cell landscape of RCC
samples. Furthermore, the different immunoprofiles between RCC and
normal tissues may represent crucial tools that could be considered
as potential targets for immunotherapies. In addition, the present
study highlighted that TIICs may have an independent prognostic
value in RCC. Coupling reliable deconvolution algorithms with
large-scale genomic data, including TCGA cohort, may have the
potential to reveal the biological and clinical implications of any
cell that infiltrates the tumor microenvironment in RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant. no. 81874092).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XG and WH designed the study. GZ and XG prepared the
figures and drafted the manuscript. GZ, LP, HY, FL, XL and XZ
performed the experiments and analyzed the data. XG and WH provided
reagents and analytic tools, and revised the manuscript. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Chongqing Medical
University, and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Capitanio U, Bensalah K, Bex A, Boorjian
SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P:
Epidemiology of renal cell carcinoma. Eur Urol. 75:74–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fisher R, Gore M and Larkin J: Current and
future systemic treatments for renal cell carcinoma. Semin Cancer
Biol. 23:38–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Climent MA, Munoz-Langa J,
Basterretxea-Badiola L and Santander-Lobera C: Systematic review
and survival meta-analysis of real world evidence on first-line
pazopanib for metastatic renal cell carcinoma. Crit Rev Oncol
Hematol. 121:45–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Candido J and Hagemann T: Cancer-related
inflammation. J Clin Immunol. 33 (Suppl 1):S79–S84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Swann JB and Smyth MJ: Immune surveillance
of tumors. J Clin Invest. 117:1137–1146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thompson RH, Dong H, Lohse CM, Leibovich
BC, Blute ML, Cheville JC and Kwon ED: PD-1 is expressed by
tumor-infiltrating immune cells and is associated with poor outcome
for patients with renal cell carcinoma. Clin Cancer Res.
13:1757–1761. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vogelzang NJ, Priest ER and Borden L:
Spontaneous regression of histologically proved pulmonary
metastases from renal cell carcinoma: A case with 5-year followup.
J Urol. 148:1247–1248. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janiszewska AD, Poletajew S and
Wasiutynski A: Spontaneous regression of renal cell carcinoma.
Contemp Oncol (Pozn). 17:123–127. 2013.PubMed/NCBI
|
|
11
|
Considine B and Hurwitz ME: Current status
and future directions of immunotherapy in renal cell carcinoma.
Curr Oncol Rep. 21:342019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Escudier B: Emerging immunotherapies for
renal cell carcinoma. Ann Oncol. 23 (Suppl 8):viii35–viii40. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amin A, Dudek AZ, Logan TF, Lance RS,
Holzbeierlein JM, Knox JJ, Master VA, Pal SK, Miller WH Jr, Karsh
LI, et al: Survival with AGS-003, an autologous dendritic
cell-based immunotherapy, in combination with sunitinib in
unfavorable risk patients with advanced renal cell carcinoma (RCC):
Phase 2 study results. J Immunother Cancer. 3:142015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bingle L, Brown NJ and Lewis CE: The role
of tumour-associated macrophages in tumour progression:
Implications for new anticancer therapies. J Pathol. 196:254–265.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fridman WH, Pages F, Sautes-Fridman C and
Galon J: The immune contexture in human tumours: Impact on clinical
outcome. Nat Rev Cancer. 12:298–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schraml P, Athelogou M, Hermanns T, Huss R
and Moch H: Specific immune cell and lymphatic vessel signatures
identified by image analysis in renal cancer. Mod Pathol.
32:1042–1052. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liss MA, Chen Y, Rodriguez R, Pruthi D,
Johnson-Pais T, Wang H, Mansour A and Kaushik D: Immunogenic
heterogeneity of renal cell carcinoma with venous tumor thrombus.
Urology. 124:168–173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rohr-Udilova N, Klinglmuller F,
Schulte-Hermann R, Stift J, Herac M, Salzmann M, Finotello F,
Timelthaler G, Oberhuber G, Pinter M, et al: Deviations of the
immune cell landscape between healthy liver and hepatocellular
carcinoma. Sci Rep. 8:62202018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei L, Jin Z, Yang S, Xu Y, Zhu Y and Ji
Y: TCGA-assembler 2: Software pipeline for retrieval and processing
of TCGA/CPTAC data. Bioinformatics. 34:1615–1617. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Statistics for Biology and Health Springer New
York; New York, NY: pp. 397–420. 2005, View Article : Google Scholar
|
|
22
|
Peltanova B, Raudenska M and Masarik M:
Effect of tumor microenvironment on pathogenesis of the head and
neck squamous cell carcinoma: A systematic review. Mol Cancer.
18:632019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Senbabaoglu Y, Gejman RS, Winer AG, Liu M,
Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A,
et al: Tumor immune microenvironment characterization in clear cell
renal cell carcinoma identifies prognostic and
immunotherapeutically relevant messenger RNA signatures. Genome
Biol. 17:2312016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bremnes RM, Al-Shibli K, Donnem T, Sirera
R, Al-Saad S, Andersen S, Stenvold H, Camps C and Busund LT: The
role of tumor-infiltrating immune cells and chronic inflammation at
the tumor site on cancer development, progression, and prognosis:
Emphasis on non-small cell lung cancer. J Thorac Oncol. 6:824–833.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ishigami S, Natsugoe S, Tokuda K, Nakajo
A, Che X, Iwashige H, Aridome K, Hokita S and Aikou T: Prognostic
value of intratumoral natural killer cells in gastric carcinoma.
Cancer. 88:577–583. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ribatti D, Ennas MG, Vacca A, Ferreli F,
Nico B, Orru S and Sirigu P: Tumor vascularity and
tryptase-positive mast cells correlate with a poor prognosis in
melanoma. Eur J Clin Invest. 33:420–425. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Visser KE, Eichten A and Coussens LM:
Paradoxical roles of the immune system during cancer development.
Nat Rev Cancer. 6:24–37. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yao J, Xi W, Zhu Y, Wang H, Hu X and Guo
J: Checkpoint molecule PD-1-assisted CD8+ T lymphocyte
count in tumor microenvironment predicts overall survival of
patients with metastatic renal cell carcinoma treated with tyrosine
kinase inhibitors. Cancer Manag Res. 10:3419–3431. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lahl K, Loddenkemper C, Drouin C, Freyer
J, Arnason J, Eberl G, Hamann A, Wagner H, Huehn J and Sparwasser
T: Selective depletion of Foxp3+ regulatory T cells
induces a scurfy-like disease. J Exp Med. 204:57–63. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sakaguchi S, Miyara M, Costantino CM and
Hafler DA: FOXP3+ regulatory T cells in the human immune
system. Nat Rev Immunol. 10:490–500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shang B and Liu Y, Jiang SJ and Liu Y:
Prognostic value of tumor-infiltrating FoxP3+ regulatory
T cells in cancers: A systematic review and meta-analysis. Sci Rep.
5:151792015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Griffiths RW, Elkord E, Gilham DE, Ramani
V, Clarke N, Stern PL and Hawkins RE: Frequency of regulatory T
cells in renal cell carcinoma patients and investigation of
correlation with survival. Cancer Immunol Immunother. 56:1743–1753.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Finke JH, Rini B, Ireland J, Rayman P,
Richmond A, Golshayan A, Wood L, Elson P, Garcia J, Dreicer R and
Bukowski R: Sunitinib reverses type-1 immune suppression and
decreases T-regulatory cells in renal cell carcinoma patients. Clin
Cancer Res. 14:6674–6682. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Papaioannou E, Sakellakis M, Melachrinou
M, Tzoracoleftherakis E, Kalofonos H and Kourea E: A Standardized
Evaluation Method for FOXP3+ Tregs and CD8+
T-cells in breast carcinoma: Association with breast carcinoma
subtypes, stage and prognosis. Anticancer Res. 39:1217–1232. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seminerio I, Descamps G, Dupont S, de
Marrez L, Laigle JA, Lechien JR, Kindt N, Journe F and Saussez S:
Infiltration of FoxP3+ regulatory T cells is a strong
and independent prognostic factor in head and neck squamous cell
carcinoma. Cancers (Basel). 11(pii): E2272019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Furth R and Cohn ZA: The origin and
kinetics of mononuclear phagocytes. J Exp Med. 128:415–435. 1968.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ziegler-Heitbrock L, Ancuta P, Crowe S,
Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph
GJ, et al: Nomenclature of monocytes and dendritic cells in blood.
Blood. 116:e74–e80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gui T, Shimokado A, Sun Y, Akasaka T and
Muragaki Y: Diverse roles of macrophages in atherosclerosis: From
inflammatory biology to biomarker discovery. Mediators Inflamm.
2012:6930832012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee HW, Choi HJ, Ha SJ, Lee KT and Kwon
YG: Recruitment of monocytes/macrophages in different tumor
microenvironments. Biochim Biophys Acta. 1835:170–179.
2013.PubMed/NCBI
|
|
40
|
Movahedi K, Laoui D, Gysemans C, Baeten M,
Stangé G, Van den Bossche J, Mack M, Pipeleers D, In't Veld P, De
Baetselier P and Van Ginderachter JA: Different tumor
microenvironments contain functionally distinct subsets of
macrophages derived from Ly6C(high) monocytes. Cancer Res.
70:5728–5739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Romano E, Kusio-Kobialka M, Foukas PG,
Baumgaertner P, Meyer C, Ballabeni P, Michielin O, Weide B, Romero
P and Speiser DE: Ipilimumab-dependent cell-mediated cytotoxicity
of regulatory T cells ex vivo by nonclassical monocytes in melanoma
patients. Proc Natl Acad Sci USA. 112:6140–6145. 2015. View Article : Google Scholar : PubMed/NCBI
|