Introduction
Cutaneous melanoma (CM) is the most malignant type
of skin cancers. Its incidence and overall health burden are
increasing in Western countries (1).
The predisposition to CM and the course of its development depend
on complex interactions between environmental factors, phenotype
and genotype. The principal environmental risk factor for the
pathogenesis of CM is ultraviolet radiation (2). Among the main phenotypic factors
contributing to CM are an increased number of nevi and freckles,
light skin, fair eyes and red and blonde hair colours-all
determined by genetic factors (3,4). Several
high-, medium- and low-risk genes are involved in the CM
development (5). One of the
best-known medium-risk genes is melanocortin 1 receptor gene
(MC1R) in the locus 16q24.3. MC1R has more than 100
non-synonymous single nucleotide polymorphisms (SNPs) identified so
far; many of them have a strong link with an increased risk of CM
(6,7)
and are also associated with the so-called R phenotype that is
characterized by red hair and fair skin (8). In addition, other SNPs close to the
MC1R gene have also been associated with the R phenotype and
the CM risk-these include rs258322, rs4785763 and rs8059973
(9,10). MC1R is activated by
α-melanocyte-stimulating hormone (α-MSH) that initiates an
intracellular signal cascade leading to the production of the
photoprotective pigment melanin (11). α-MSH is produced in the
post-translational processing of the pro-opiomelanocortin (POMC).
It has been demonstrated that after UV irradiation the POMC
promoter is activated by one of the major tumour suppressor
proteins TP53 (12). Almost half of
the human cancers harbour TP53 mutations (13). Furthermore, the TP53 impact on
cancer is not limited to somatic mutations and may manifest through
germline variants (14). The
rs1042522 (p.Pro72Arg, c.215C>G) is located in a proline-rich
domain of TP53, which has an important role in the
TP53-mediated apoptosis (15).
Results from CM association studies show conflicting roles for
rs1042522. Associations have been found with both Pro (16,17) and
Arg (18,19) alleles. TP53 is negatively regulated
by an auto-regulatory feedback loop with E3 ubiquitin ligase MDM2.
The MDM2 intron 1 comprises an alternative promoter P2 for
the MDM2 transcription, which is induced by TP53 (20). The rs2279744 (c.14+309T>G) is
located within this promoter. It has been shown that the G allele
of rs2279744 increases the affinity for the transcription
activation factor Sp1 thus leading to an increased MDM2
expression. This in turn causes TP53 inhibition, which might
promote tumour formation (21).
Association studies for CM have demonstrated a relationship between
rs2279744's GG genotype and the CM risk depending on age, sex
(22–24) and Breslow thickness (25), although results are not consistent.
It means that MC1R, TP53 and MDM2 are interconnected
in a shared signalling pathway regulating pigmentation.
In this study, we aim to investigate the joint
effect of the SNPs in the MC1R gene and its vicinity and
rs1042522 in TP53 and rs227974 in MDM2 in the context
of the CM risk prediction. In addition, we explore the associations
of these SNPs with pigmentation traits and tumour characteristics,
in particular ulceration and Breslow thickness, for their
predictive power of disease progression.
Materials and methods
Study population
We conducted this study using DNA samples and data
on individuals from the Latvian Genome Data Base (LGDB), a
government-funded biobank (described in (26). These data have been previously
explored to assess the variation within the MC1R gene and to
study particular SNPs on the chromosome 16 (27). In total, 479 samples were selected
for this study, including 224 unrelated healthy volunteers and 255
CM patients with histopathologically confirmed CM (ICD-10 diagnosis
code C43). An additional criterion for the inclusion of individuals
in this study was a completed questionnaire (self-reported) about
their pigmentation characteristics: Hair colour, eye colour, skin
type, freckles in childhood and adulthood, and nevi. Data about
tumour characteristics, such as Breslow thickness and ulceration,
were obtained from the medical records of CM patients. All
individuals incorporated in the study have European ancestry and
represent Latvian population, which is known to be genetically
homogeneous (28). Written, informed
consent was acquired from all LGDB participants.
Genotyping
The entire coding region of the MC1R gene was
sequenced and SNPs on chromosome 16 (rs258322, rs4785763, and
rs8059973) were genotyped as described in (27). The TP53 gene exon 4, which
contains rs1042522, was amplified using primers
5′-ATCTACAGTCCCCCTTGCGC-3′ and 5′-GCAACTGACCGTGCAAGTCA-3′ (18). The intronic promoter region of the
MDM2 gene that contains rs2279744 was amplified using
primers 5′-CGGGAGTTCAGGGTAAAGGT-3′ and 5′-AGCAAGTCGGTGCTTACCTG-3′
(21). All primers have been
synthesised at the Metabion International AG, Martinsried,
Germany. Polymerase chain reactions were performed in a 25 µl
reaction volume containing 25 ng of template DNA, 1× Taq buffer, 5%
dimethyl sulfoxide, 1.5 mM magnesium chloride, 0.24 mM dNTPs, 0.4
µM of each primer and 1.25 U TaqDNA Polymerase (Thermo
Scientific Molecular Biology). The cycling conditions were as
follows: An initial denaturation at 95°C for ten minutes; 35 cycles
of denaturation at 95°C for 30 sec, annealing at 62°C and 55°C (for
TP53 and MDM2, respectively) for 30 sec and extension
at 72°C for one minute followed by a final extension at 72°C for
seven minutes. The sequencing was done in both directions with the
primers that were used for amplification. ABI PRISM BigDye
Terminator cycle sequencing kit (Applied Biosystems) was
applied in the following conditions: 25 cycles at 94°C for 30 sec,
then at 53°C for 15 sec, and at 60°C for four minutes. Initial
analysis was carried out on an ABI PRISM 3100 Genetic analyser
according to manufacturer's instructions (Applied
Biosystems). Sequence analysis was performed and confirmed
manually using the Vector NTI (Life Technologies).
Statistical analyses
The associations between demographic
characteristics, pigmentation traits (hair colour, eye colour, skin
type, freckles in childhood, freckles in adulthood and nevi) and CM
were assessed either by the Chi-squared test or Fisher's exact
test. The age distributions for cases and controls were compared by
the Mann-Whitney test. The minor allele frequency (MAF) of a SNP
was estimated from all controls having the genotype information for
this particular SNP (224 controls for the MC1R gene SNPs,
203 for rs258322, 205 for rs4785763, 217 for rs1042522 and 215 for
rs2279744). In subsequent analyses we included only the SNPs that
had ≥4% MAF and at least one homozygote of two minor alleles in
controls and also did not significantly deviate from the
Hardy-Weinberg equilibrium. Genotyping failed for rs1042522 in one
case and for rs2279744 in another case. These two samples were
excluded from further analyses. Hence, 253 CM cases and 200
controls having genotype information for all selected SNPs were
included in the analyses.
First, univariate association analyses were carried
out using logistic regression models with and without cofactors
(age and sex). Throughout all analyses an additive model of the
contribution of alleles was assumed. Models were fitted using the
function glm in R environment. The model without cofactors
was log(P(Y=1)/(1-P(Y=1)))=µ+β·X+ε where Y was melanoma status, X
was the genotype vector for all individuals, µ was a constant
intercept, β measured the effect of the genotype upon melanoma
status and ε was the vector of error terms.
The model with cofactors was log(P(Y=1)/(1-P(Y=1)))
= µ+γ·G+α·A/100+β·X+ε where G was the sex vector and γ the effect
of sex and A was the vector of ages in years and α the effect of
age; other variables as defined above.
The significance of an association between a SNP and
CM was measured by the Wald test that was applied to the SNP term
with α=0.05. A permutation test was carried out to check whether
the observed level of association was significant (Supplementary
Material).
Multivariate models with and without cofactors (age
and sex) were built by stepwise regression using the function
stepAIC from the R package MASS. The AIC criterion
was applied to assess the significance of a model improvement after
either adding or removing of a predictor. A generalised linear
model was used throughout all multivariate analyses. A multivariate
model without cofactors was of the form
log(P(Y=1)/(1-P(Y=1)))=µ+β1·X1+···+βk·Xk+ε
where Xk was the vector of genotypes for SNP k and
βk was the effect of SNP k. A multivariate model with
cofactors had the form log(P(Y=1)/(1-P(Y=1))) =
µ+γ·G+α·A/100+β1·X1+···+βk·Xk+ε
with notation as specified above.
In addition, for each SNP (rs2228479, rs1805007,
rs1110400, rs1805008, rs258322, rs4785763), three multivariate
models were built that incorporated one of these SNPs and either
one or both of the rs1042522 and rs2279744. These models were:
log(P(Y=1)/(1-P(Y=1)))=µ+β1·X1+βs·Xs+ε;
log(P(Y=1)/(1-P(Y=1)))=µ+β2·X2+βs·Xs+ε;
log(P(Y=1)/(1-P(Y=1)))=µ+β1·X1+β2·X2+βs·Xs+ε.
Here, X1 was the vector of the rs1042522
genotypes and β1 measured their effect, X2
was the vector of the rs2279744 genotypes having the effect
β2, and Xs and βs were the
genotypes and the effect of the chosen SNP.
The associations of SNPs with pigmentation traits
were tested as well. In these analyses, we used either only
controls or solely CM cases. The strength of each association was
evaluated by Fisher's exact test and empirical P-values were
obtained by a permutation test (see Supplementary Material). In
order to understand simultaneous effects of individual SNPs upon
pigmentation traits and the CM status we used ordinal regression
approach. We modelled a genotype as an ordinal outcome and included
both the CM status and a pigmentation trait as predictors in a
regression model according to the method described in (29). Each model was fitted by the function
clm from the R package ordinal. The significance of
each likelihood ratio obtained from a comparison of two models,
with and without the CM status and a pigmentation trait, was
assessed using a permutation test (Supplementary Material).
The associations between SNPs and tumour Breslow
thickness and ulceration were explored using the patients that had
the information on the studied tumour characteristic-195 cases for
Breslow thickness and 147 cases for ulceration (68 with and 79
without ulceration). A linear regression model was used to describe
the association between a SNP and the logarithm of Breslow
thickness (exploratory analyses showed that the logarithm of
Breslow thickness roughly corresponds to a normal distribution).
The model was log(R)=µ+β·X+ε where R was a vector of the Breslow
thicknesses and other terms were defined as for univariate models
of genotype and CM associations.
The impact of a SNP on Breslow thickness was
assessed by the t-test that was applied to the SNP term. The models
were fitted by the function lm in R environment.
A generalised linear model was used to relate a SNP
to ulceration: log(P(U=1)/(1-P(U=1)=µ+β·X+ε where P(U=1) was the
probability of ulceration and other terms were as defined for
univariate models of genotype and CM associations. The significance
of an association was determined by the Wald test applied to the
genotype term. For Breslow thickness, age was chosen as a cofactor,
while for ulceration both age and sex were incorporated in models.
The two models with cofactors were
log(R)=µ+α·A/100+β·X+ε
log(P(U=1)/(1-P(U=1)=µ+γ·G+α·A/100+β·X+ε
with the notation as above. Empirical P-values
obtained from a permutation test were reported for the associations
with Breslow thickness and ulceration (Supplementary Material).
Results
Demographic and pigmentation
characteristics of the study population
Associations between the demographic and
pigmentation characteristics of the study participants (253 cases
and 200 controls) and CM are summarised in Table I. CM patients and controls did not
differ in terms of sex (P=0.149), while the control group was
younger than patients (Mann-Whitney test P=1.15×10−7).
The strongest association with an increased CM risk was observed
for skin type (P=5.74×10−9), followed by nevi
(P=7.46×10−5) and freckles, both in adulthood and
childhood (P=0.001 and 0.01 respectively).
 | Table I.Associations between demographic
data, pigmentation characteristics and cutaneous melanoma status
within the study cohort. |
Table I.
Associations between demographic
data, pigmentation characteristics and cutaneous melanoma status
within the study cohort.
|
| Controls
(n=200) | Melanoma patients
(n=253) |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | (%) | n | (%) | P-value |
|---|
| Sex |
|
|
|
|
|
|
Female | 150 | (75.0) | 173 | (68.4) | 0.149a |
|
Male | 50 | (25.0) | 80 | (31.6) |
|
| Age | 47.5±17.5 | 56.4±15.0 |
1.15×10−7c |
| Hair colour |
|
|
|
|
|
|
Red | 5 | (2.5) | 16 | (6.3) | 0.070b,d |
|
Fair | 72 | (36.0) | 126 | (49.8) |
|
|
Brown | 109 | (54.5) | 96 | (37.9) |
|
|
Black | 11 | (5.5) | 9 | (3.6) |
|
| nd | 3 | (1.5) | 6 | (2.4) |
|
| Skin type |
|
|
|
|
|
| I | 7 | (3.5) | 28 | (11.0) |
5.74×10−9b |
| II | 28 | (14.0) | 72 | (28.5) |
|
|
III | 144 | (72.0) | 107 | (42.3) |
|
| IV | 19 | (9.5) | 41 | (16.2) |
|
| nd | 2 | (1.0) | 5 | (2.0) |
|
| Eye colour |
|
|
|
|
|
|
Blue | 62 | (31.0) | 104 | (41.1) | 0.605e |
|
Grey | 50 | (25.0) | 37 | (14.6) |
|
|
Green | 25 | (12.5) | 30 | (11.9) |
|
|
Brown | 29 | (14.5) | 30 | (11.9) |
|
|
Other | 29 | (14.5) | 47 | (18.5) |
|
| nd | 5 | (2.5) | 5 | (2.0) |
|
| Freckles in
childhood |
|
|
|
|
|
| Very
many/many | 7 | (3.5) | 21 | (8.3) |
0.010b |
|
Some | 7 | (3.5) | 7 | (2.8) |
|
|
Few | 22 | (11.0) | 37 | (14.6) |
|
| Very
few | 52 | (26.0) | 86 | (34.0) |
|
|
None | 105 | (52.5) | 96 | (37.9) |
|
| nd | 7 | (3.5) | 6 | (2.4) |
|
| Freckles in
adulthood |
|
|
|
|
|
| Very
many/many | 8 | (4.0) | 16 | (6.3) |
0.001b |
|
Some | 7 | (3.5) | 6 | (2.4) |
|
|
Few | 18 | (9.0) | 31 | (12.3) |
|
| Very
few | 46 | (23.0) | 79 | (31.2) |
|
|
None | 119 | (59.5) | 89 | (35.2) |
|
| nd | 2 | (1.0) | 32 | (12.6) |
|
| Nevi |
|
|
|
|
|
|
Many | 25 | (12.5) | 71 | (28.0) |
7.46×10−5a |
|
Some | 69 | (34.5) | 70 | (27.7) |
|
|
Few | 93 | (46.5) | 83 | (32.8) |
|
|
None | 11 | (5.5) | 23 | (9.1) |
|
| nd | 2 | (1.0) | 6 | (2.4) |
|
Genotyping results
Both SNPs genotyped within this study (rs1042522
(Pro72Arg, c.215C>G) in TP53 and rs2279744
(c.14+309T>G) in MDM2) reached 4% MAF, had at least one
homozygote of two minor alleles among controls and were in
Hardy-Weinberg equilibrium (data not shown). Thus both SNPs were
included in further analyses and were studied together with the six
previously selected SNPs on chromosome 16, namely rs2228479
(p.Val92Met, c.274G>A), rs1805007 (p.Arg151Cys, c.451C>T),
rs1110400 (p.Ile155Thr, c.464T>C) and rs1805008 (p.Arg160Trp,
c.478C>T) in the MC1R gene, as well as rs258322
(c.160+171A>G) in the CDK10 gene, and rs4785763
(n.1682A>C) in the AFG3L1P pseudogene. Among the SNPs
that passed the minor allele frequency threshold there were none
with several alternative alleles. Other MC1R SNPs found in
our cohort did not pass the inclusion criteria for further analyses
(4% MAF and at least one homozygote of two minor alleles among
controls). These included 13 non-synonymous SNPs, rs1805005
(Val60Leu, c.178G>T), rs777024553 (Ser83Leu, c.248C>T),
rs1805006 (Asp84Glu, c.252C>A), rs34540312 (Gly89Arg,
c.265G>C), rs34158934 (Thr95Met, c.284C>T), rs200616835
(Asp121Glu, c.363C>G), rs11547464 (Arg142His, c.425G>A),
rs885479 (Arg163Gln, c.488G>A), rs762096175 (Val165Ile,
c.493G>A), rs780875127 (c.495_496insGG), rs530102853 (Asp184His,
c.550G>C), rs774680166 (Val188Ile, c.562G>A), rs200000734
(Arg213Trp, c.637C>T), as well as seven synonymous SNPs,
rs201429598 (Cys133=, c.399C>T), rs201827012 (Arg151=,
c.453C>G), rs374959395 (Ala166=, c.498G>A), rs146544450
(Gln233=, c.699G>A), rs375813196 (Cys273=, c.819C>T),
rs2228478 (Thr314=, c.942A>G), rs151318945 (Ser316=,
c.948C>T). The MAFs of these variants are shown in the
supplementary Table SI.
SNP associations with CM
Among the newly genotyped SNPs, only the rs1042522
in TP53 showed a borderline significant association with CM
in the univariate analysis (P=0.065). However, this association was
not significant according to a permutation test and became weaker
after adjustment for age and sex (Table
II). Notable associations with CM were displayed by the
rs1805007 within MC1R (P=0.009), as well as rs258322 and
rs4785763 (P=0.012 and P=0.021, respectively). A permutation test
confirmed the associations for rs1805007 and rs258322 as moderately
significant (empirical P=0.051 and P=0.065, respectively). Results
were similar after the inclusion of age and sex as cofactors and
revealed a more convincing association with the CM status for
rs4785763 (P=0.012) (Table II).
 | Table II.Univariate association analyses of
individual SNPs and cutaneous melanoma risk. |
Table II.
Univariate association analyses of
individual SNPs and cutaneous melanoma risk.
|
|
|
|
|
|
|
|
| After adjustment
for sex and age |
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Gene | SNP | AF 1000 MGenomes
(EUR) % | MAF controls % (n
genotyped) | MAF patients % (n
genotyped) |
P-valuea | OR (95% CI) | Permutation
P-value | P-value | OR (95% CI) | Permutation
P-value |
|---|
| MC1R | rs2228479 | 6.9 | 8.9 (224) | 12.0 (255) | 0.166 | 1.36
(0.88–2.11) | 0.749 | 0.148 | 1.40
(0.89–2.21) | 0.686 |
|
| p.Val92Met
c.274G>A |
|
|
|
|
|
|
|
|
|
| MC1R | rs1805007 | 7.2 | 4.0 (224) | 8.0 (255) | 0.009 | 2.26
(1.23–4.16) | 0.051 | 0.009 | 2.30
(1.23–4.32) | 0.056 |
|
| p.Arg151Cys
c.451C>T |
|
|
|
|
|
|
|
|
|
| MC1R | rs1110400 | 0.8 | 4.0 (224) | 2.7 (255) | 0.314 | 0.69
(0.34–1.42) | 0.932 | 0.317 | 0.69
(0.33–1.44) | 0.944 |
|
| p.Ile155Thr
c.464T>C |
|
|
|
|
|
|
|
|
|
| MC1R | rs1805008 | 6.2 | 10.9 (224) | 12.9 (255) | 0.420 | 1.18
(0.79–1.76) | 0.982 | 0.347 | 1.22
(0.81–1.85) | 0.962 |
|
| p.Arg160Trp
c.478C>T |
|
|
|
|
|
|
|
|
|
| CDK10 | rs258322
c.160+171A>G | 9.8 | 6.7 (203) | 11.8 (255) | 0.012 | 1.84
(1.15–2.97) | 0.065 | 0.014 | 1.85
(1.13–3.01) | 0.090 |
| AFG3L1P | rs4785763
n.1682A>C | 29.9 | 34.4 (205) | 41.6 (255) | 0.021 | 1.40
(1.05–1.86) | 0.133 | 0.012 | 1.46
(1.09–1.97) | 0.079 |
| TP53 | rs1042522 | 71.5b | 65.7 (217) | 70.9 (254) | 0.065 | 1.29
(0.98–1.70) | 0.376 | 0.132 | 1.25
(0.94–1.66) | 0.652 |
|
| p.Pro72Arg
c.215C>G |
|
|
|
|
|
|
|
|
|
| MDM2 | rs2279744
c.14+309T>G | 35.5 | 28.6 (215) | 32.5 (254) | 0.267 | 1.18
(0.88–1.57) | 0.902 | 0.127 | 1.26
(0.94–1.71) | 0.637 |
Three SNPs were selected for the multivariate model
without cofactors-rs1805007 and rs2228479 from MC1R, as well
as rs1042522 from TP53 (P-values 0.005, 0.126 and 0.035,
respectively) (Table III). This
suggests that the effects of these SNPs on the CM risk might be
rather independent and complementary to each other. After the
inclusion of cofactors (age and sex) in the model, four SNPs were
selected (Table III). The
rs1805007 in MC1R remained significant, and rs1042522 in
TP53 showed a borderline significant association. Two novel
association signals were identified-rs4785763 and rs2279744 in the
MDM2 gene. Both of them seem to have small effects,
independent from rs1805007 and rs1042522, which might also have
some interplay with age and sex (Table
III).
 | Table III.SNPs associated with cutaneous
melanoma selected by stepwise regression. |
Table III.
SNPs associated with cutaneous
melanoma selected by stepwise regression.
| A, Regression model
without cofactors |
|---|
|
|---|
| Gene | SNP | P-value | OR | 95% CI |
|---|
| MC1R | rs1805007 | 0.005 | 2.43 | 1.31–4.50 |
| TP53 | rs1042522 | 0.035 | 1.35 | 1.02–1.78 |
| MC1R | rs2228479 | 0.126 | 1.41 | 0.91–2.19 |
|
| B, Regression
model with cofactors (age and sex) |
|
| Gene | SNP | P-value | OR | 95% CI |
|
| MC1R | rs1805007 | 0.036 | 2.03 | 1.05–3.93 |
| TP53 | rs1042522 | 0.083 | 1.32 | 0.96–1.80 |
| AFG3L1P | rs4785763 | 0.079 | 1.30 | 0.97–1.74 |
| MDM2 | rs2279744 | 0.133 | 1.26 | 0.93–1.72 |
In the latter multivariate model age emerged as a
significant risk factor for CM with P=1.44×10−8
(OR=34.64, 95% CI=10.17–118.00) and sex also differentiated the
risk with a borderline significance (P=0.050, OR=1.56, 95%
CI=1.00–2.42).
To understand the interactions between rs1042522 and
rs2279744 and their impact on the CM risk in combination with
MC1R and other chromosome 16 SNPs, further multivariate
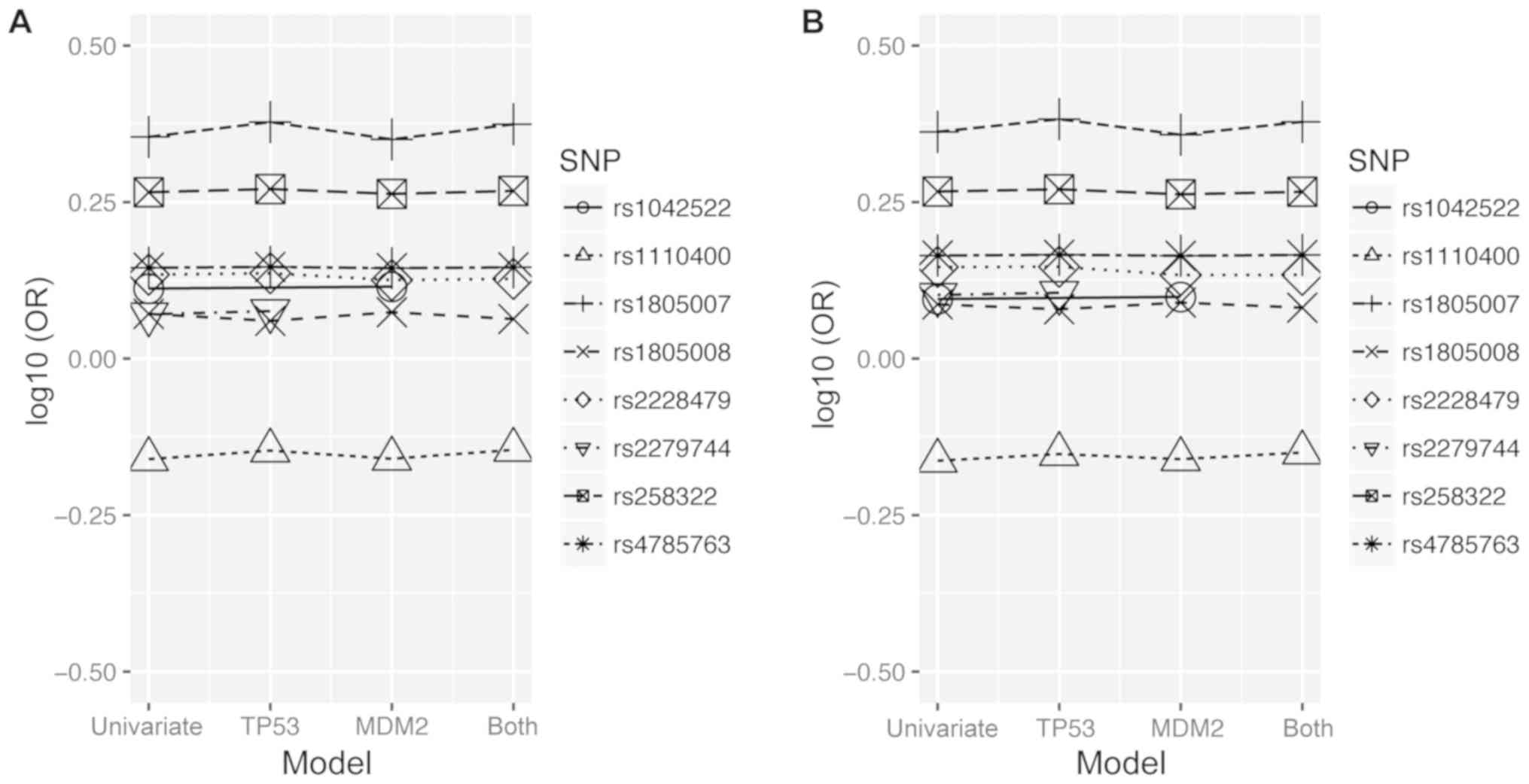
models were fitted. Fig. 1 shows the
changes in the logarithm of the odds ratio (OR) for each SNP
depending on the context of a model. The highest impact on the CM
risk was consistently displayed by rs1805007 within MC1R
having log10(OR)=0.35; its effect became more prominent when it was
accompanied by the rs1042522 of TP53: log10(OR)=0.37
(Fig. 1A). This trend was not
significantly altered by the presence of cofactors, age and sex
(Fig. 1B). Other types of potential
interactions are unlikely due to flat log10(OR) trends depicted in
Fig. 1.
Associations between SNPs,
pigmentation traits and melanoma
We searched for associations between individual SNPs
and pigmentation traits within the set of controls first. We found
associations between rs1805007 in the MC1R gene and red hair
colour (empirical P=0.040) as well as with skin types I and II
(empirical P=0.049). In addition, the rs2279744 from MDM2
was associated with brown eye colour (empirical P=0.015) (data not
shown).
Next, associations were identified simultaneously
for each pigmentation trait and the CM status. Three different SNPs
emerged as significant from this analysis (Table IV). The MC1R rs1805007 was
associated with CM and with red hair as well with the skin types I
and II and the presence of freckles in childhood. The rs1805008 in
MC1R was strongly associated with red hair and also related
to the presence of freckles in childhood. However, this SNP did not
exhibit any prominent relationship with CM, which implies that it
primarily determines the pigmentation traits. Furthermore,
rs2279744 from MDM2 displayed an association with brown eye
colour and no convincing association with CM (Table IV).
 | Table IV.SNPs exhibiting significant
associations with CM and pigmentation traits. |
Table IV.
SNPs exhibiting significant
associations with CM and pigmentation traits.
|
|
|
|
|
| Association with
pigmentation | Association with
CM | Overall model |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Gene | SNP | Pigmentation
trait | Reference
phenotype | Alternative
phenotype | OR (95%
CI)a | OR (95% CI) | P-value | Permutation
P-value |
|---|
| MC1R | rs1805007 | Hair colour | Other | Red | 3.56
(1.34–9.44) | 2.34
(1.22–4.46) | 0.0007 | 0.01 |
|
|
| Skin type | III, IV | I, II | 2.24
(1.24–4.07) | 2.07
(1.07–4.01) | 0.0003 | 0.004 |
|
|
| Freckles in
childhood | None or few | Some or many | 1.76
(0.92–3.34) | 2.54
(1.31–4.91) | 0.002 | 0.017 |
| MC1R | rs1805008 | Hair colour | Other | Red | 4.46
(1.76–11.28) | 1.08
(0.68–1.70) | 0.008 | 0.058 |
|
|
| Freckles in
childhood | None or few | Some or many | 2.69
(1.62–4.50) | 1.08
(0.68–1.71) | 0.0007 | 0.006 |
| MDM2 | rs2279744 | Eye colour | Other | Brown | 2.35
(1.37–4.02) | 1.30
(0.87–1.94) | 0.003 | 0.027 |
Associations between SNPs and tumour
characteristics
We also looked at the connections between the eight
selected SNPs and tumour ulceration and Breslow thickness. The only
SNP that showed an association with ulceration was rs4785763-its
alternative allele displayed a protective effect (P=0.038, OR=0.58,
95% CI=0.35–0.97). The effect became less pronounced after the
adjustment for age and sex (P=0.125, OR=0.66, 95% CI=0.38–1.12).
However, according to a permutation test this association was not
significant. Associations with Breslow thickness were exhibited by
two MC1R SNPs-rs2228479 and rs1110400. These associations
remained consistent in models with and without cofactors. The
rs1110400 was associated with thicker tumours (P=0.035, OR=2.49,
95% CI=1.07–5.77 and P=0.027, OR=2.60, 95% CI=1.12–6.00 for models
without and with cofactors, respectively), and rs2228479 was
related to thinner tumours (P=0.036, OR=0.69, 95% CI=0.48–0.97 and
P=0.037, OR=0.69, 95% CI=0.49–0.98 for models without and with
cofactors, respectively). However, after applying a permutation
test, these associations became weaker and lost their
significance.
Discussion
In this paper, we explored the associations between
CM and SNPs in the TP53 (rs1042522) and the MDM2
(rs2279744) genes alongside the SNPs in the MC1R gene and
other chromosome 16 SNPs previously shown to modify the CM risk in
the population of Latvia. The strongest association with CM was
consistently displayed by the rs1805007 of MC1R. The
association of this SNP with CM has been discovered in numerous
studies of various populations across the world with overall
OR=1.80 (95% CI=1.58–2.06) (30).
This OR is slightly lower than the one observed in our study
(OR=2.26, 95% CI=1.23–4.16). However, ORs similar to ours were
obtained for the geographically close Swedish population (OR=2.32,
95% CI=1.77–3.05) (31) as well as
for Dutch (OR=2.5, 95% CI=1.4–4.5) (32), French (OR=2.35, 95% CI=1.78–3.11,
P=5.10×10−10) (33), and
Spanish (OR=2.71, 95% CI=1.63–4.52, P=0.00013) (31) populations.
A moderately significant association was found for
another chromosome 16 SNP, rs258322. However, this SNP has been
shown to be linked with the MC1R SNPs, rs1805007, rs1805008
and rs1805009 (p.Asp294His, c.880G>C), all of them associated
with an increased CM risk (10,27,34,35).
Previously, we have shown that rs4785763 is associated with CM in
Latvian population as well (27).
Here, we confirm this by the multivariate model that includes age
and sex. One of the limitations of the current study is the small
number of individuals, which might hinder discovering SNPs with
minor effects on CM. However, multivariate models can be more
sensitive in elucidating various association signals and can reveal
weaker associations more clearly after conditioning on a few
strongly associated SNPs. This might also be the reason why,
according to a univariate analysis, the rs1042522 within
TP53 seems to be associated with CM only moderately, but is
included in a multivariate model. Moreover, rs1042522 emerged as
significant even after rs1805007 had entered the model. These
observations suggest that rs1042522 has a small and independent
effect on the CM risk with respect to rs1805007.
Interestingly enough, we found an association
between the CM risk and rs1042522's Arg allele, which is the most
frequent allele of this SNP in our cohort. Indeed, the Arg allele
is more prevalent in individuals having light skin and living in
higher latitudes while the ancestral Pro allele is more widespread
in populations with darker skin that live closer to equator, most
probably due to evolutionary selection (36). The association of rs1042522 with CM
has been reported for both alleles in literature. For example, Shen
et al (2003) showed an association between CM and the
Arg/Arg genotype in the US population (OR=1.43, 95% CI=1.02–2.02)
(18). This association was
especially strong in individuals older than 50 years (OR=2.32, 95%
CI=1.39–3.88). Later Li et al (2008) confirmed this
association in a larger US-based study (OR=1.28, 95% CI=1.05–1.55)
(19). The Arg/Arg homozygote has
also been associated with CM in a Brazilian population (OR=1.76,
95% CI=1.09–2.83, P=0.020) (37) and
a small effect attributable to the Arg allele was also identified
when analysing specific genotype subgroups (Arg/Pro vs. Pro/Pro)
(38). Several studies have shown an
association between CM and the Pro allele. Such an association has
been found in German (OR=2.49, 95% CI=1.30–4.75, P=0.006) (39) and Greek populations (OR=3.17, 95%
CI=1.03–9.78) (17). There are also
studies that have not found associations between any of the
rs1042522 alleles and the CM risk-a Dutch population study
(40), US Nurses' Health Study
(16), a Scottish study (41) and Italian population studies
(25). To our knowledge, there is
only one study in which the relationships between rs1042522 and the
MC1R gene SNPs have been analysed. The authors of this study
found that the association with the CM risk became stronger for the
Pro/Pro genotype in the absence of such MC1R SNPs as
rs1805007, rs1805008 and rs1805009, which are related to red hair
(OR=2.99, 95% CI=1.02–8.78) (17).
More recently, it has also been shown that mutations in TP53
gene are associated with faster progression and poorer overall
survival as well as with weaker response to the anti-CTLA-4 therapy
in melanoma (42). These
observations indicate a possible role of the TP53 gene in
predicting the outcome of a therapy.
Interestingly, rs2228479 within MC1R
displayed a small effect on CM in a multivariate model, suggesting
it might have an independent impact on the CM risk. However, after
the cofactors, age and sex, were included in the model, the effect
of rs2228479 disappeared. Previous studies of the involvement of
this SNP in the modification of the CM risk report contrasting
results. A meta-analysis did not find an association of rs2228479
with CM (43). Subsequent
meta-analyses revealed that rs2228479 is associated with CM and has
a small OR (1.08–1.32) (7,30). Hence the effect of this SNP on the CM
development is still controversial.
Another SNP that entered a multivariate model was
rs2279744 from MDM2. Moreover, rs2279744 was incorporated
only in the model with cofactors suggesting some interplay between
this SNP and age and/or sex. Previous studies also show some
evidence for an age- and sex-dependent effect of rs2279744 on CM,
although their results are not consistent. A couple of studies have
demonstrated an association between rs2279744's minor allele
homozygote GG and the CM risk, especially for younger women or
women with a hereditary CM (22,24). In
contrast, another study showed that women with the GG genotype
might actually be at lower risk of developing CM at a young age
(23). Most studies do not tend to
find a convincing association between this SNP in the MDM2
gene and the CM risk (25,44–46).
As expected, we identified associations between CM
and skin type, nevi, freckles and hair colour. Notably we have a
larger proportion of dark skinned individuals with skin types III
and IV in the control group than in the case group. The former
proportion within controls is unexpected in the light-skinned
population of Latvia. This might be explained by the fact that
pigmentation traits were self-reported. It also means that it was
important to include skin type as a cofactor when assessing
associations between genotypes and melanoma. We have done thus when
looking at the associations between SNPs, pigmentation traits and
melanoma. The results revealed that one of the analysed SNPs was
associated with both skin type and melanoma; for other variants
skin type was not relevant and did not seem to introduce any bias.
The lowest estimate of the OR for rs1805007 and melanoma (OR=2.07)
might be more precise than other estimates of ours because it takes
into account skin type.
One of the major pigmentation regulators is
MC1R. Previously several MC1R SNPs have been
associated both with CM and red hair or fair skin, e.g.
rs1805006 (p.Asp84Glu, c.252C>A), rs11547464 (p.Arg142His,
c.425G>A), rs1805007, rs1805008, and rs1805009 (43). We also found an association between
the most strongly CM-associated SNP in our study, rs1805007, and
red hair and fair skin as well as with the presence of freckles in
childhood. Yet another MC1R SNP, rs1805008, was associated
with red hair only. The associations of this SNP were consistent
throughout all, univariate and multivariate, analyses performed
with this study. This result is somewhat different to other
studies. It seems that in our cohort the increased melanoma risk
for individuals carrying the rs1805008 alternative allele is
attributable to the presence of freckles and the red hair
phenotype. In addition, rs2279744 in MDM2 displayed an
association with brown eye colour. To our knowledge, only one study
has looked at the association between rs2279744 and eye colour in
CM patients. In that study, genotypes with G allele (TG and GG)
were found more often in patients with dark eyes (45), which is similar to our results.
Several studies have shown that the presence of
MC1R SNPs is associated with tumour thickness-associations
have been demonstrated both with thicker (47–49) and
thinner tumours (50). Similarly,
rs2279744 from MDM2 has been shown to increase the
MDM2 expression (21), which
in turn has been linked to the CM thickness (51). The GG genotype of rs2279744 turned
out to be significantly associated with CM in patients having
tumours thicker than 0.75 mm (25).
In the latter study, the association between Breslow thickness and
rs1042522 in TP53 was also assessed and not found (25). In this study, we were not able to
show a convincing association between any of the SNPs analysed and
Breslow thickness, as none of them could be validated by a
permutation test. Associations between MC1R SNPs, in
particular the so-called ‘R’ variants, and ulceration have also
been described (49). However, we
were not able to confirm these in our cohort. The rs4785763 from
chromosome 16 initially showed an association with ulceration but
did not withstand a permutation test. So far no associations have
been found between rs2279744 in MDM2 or rs1042522 in
TP53 and ulceration (24),
and our results confirmed this. Larger cohorts of individuals with
less missing data on tumour features would be necessary to draw
definite conclusions about such associations.
To conclude, we have demonstrated that rs1042522
within TP53 has an independent effect on the CM risk, which
complements the effect of the strongly associated rs1805007
residing in MC1R. Both of these SNPs needs to be taken into
account in melanoma risk calculation. The rs2279744 in MDM2
is associated with eye colour and has a small, if any, effects
towards CM in Latvian population.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The current study was supported by TRANSCAN via The
State Education Development Agency, Republic of Latvia (grant no.
GENMEL Z/15/1285 - PRL15/15).
Availability of data and materials
The datasets used and/or analysed in the present
study are available from the corresponding author upon a reasonable
request.
Authors' contributions
AO performed genotyping and data analysis. DR
performed statistical analyses and interpreted results. DP
conceived and designed the current study, and interpreted the data.
All authors wrote, edited and approved the final manuscript.
Ethics approval and consent to
participate
The Central Medical Ethics Committee of Latvia
approved the protocols for sample collection (approval nos.
A-3/2006 and A-7/2007), which were part of the project ‘Creation of
Genome Data Base of Latvian population’ (protocol no.
01-29/2016-1-1).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Whiteman DC, Green AC and Olsen CM: The
growing burden of invasive melanoma: Projections of incidence rates
and numbers of new cases in six susceptible populations through
2031. J Invest Dermatol. 136:1161–1171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gilchrest BA, Eller MS, Geller AC and Yaar
M: The pathogenesis of melanoma induced by ultraviolet radiation. N
Engl J Med. 340:1341–1348. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gandini S, Sera F, Cattaruzza MS, Pasquini
P, Abeni D, Boyle P and Melchi CF: Meta-analysis of risk factors
for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer.
41:28–44. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gandini S, Sera F, Cattaruzza MS, Pasquini
P, Zanetti R, Masini C, Boyle P and Melchi CF: Meta-analysis of
risk factors for cutaneous melanoma: III. Family history, actinic
damage and phenotypic factors. Eur J Cancer. 41:2040–2059. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Read J, Wadt K and Hayward NK: Melanoma
genetics. J Med Genet. 53:1–14. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pérez Oliva AB, Fernéndez LP, Detorre C,
Herráiz C, Martínez-Escribano JA, Benítez J, Lozano Teruel JA,
García-Borrón JC, Jiménez-Cervantes C and Ribas G: Identification
and functional analysis of novel variants of the human melanocortin
1 receptor found in melanoma patients. Hum Mutat. 30:811–822. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Williams PF, Olsen CM, Hayward NK and
Whiteman DC: Melanocortin 1 receptor and risk of cutaneous
melanoma: A meta-analysis and estimates of population burden. Int J
Cancer. 129:1730–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duffy DL, Box NF, Chen W, Palmer JS,
Montgomery GW, James MR, Hayward NK, Martin NG and Sturm RA:
Interactive effects of MC1R and OCA2 on melanoma risk phenotypes.
Hum Mol Genet. 13:447–461. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bishop DT, Demenais F, Iles MM, Harland M,
Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E,
et al: Genome-wide association study identifies three loci
associated with melanoma risk. Nat Genet. 41:920–925. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han J, Kraft P, Nan H, Guo Q, Chen C,
Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, et al: A
genome-wide association study identifies novel alleles associated
with hair color and skin pigmentation. PLoS Genet. 4:e10000742008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hunt G, Kyne S, Wakamatsu K, Ito S and
Thody AJ: Nle4DPhe7 alpha-melanocyte-stimulating hormone increases
the eumelanin: Phaeomelanin ratio in cultured human melanocytes. J
Invest Dermatol. 104:83–85. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui R, Widlund HR, Feige E, Lin JY,
Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter
SR and Fisher DE: Central role of p53 in the suntan response and
pathologic hyperpigmentation. Cell. 128:853–864. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stracquadanio G, Wang X, Wallace MD,
Grawenda AM, Zhang P, Hewitt J, Zeron-Medina J, Castro-Giner F,
Tomlinson IP, Goding CR, et al: The importance of p53 pathway
genetics in inherited and somatic cancer genomes. Nat Rev Cancer.
16:251–265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sakamuro D, Sabbatini P, White E and
Prendergast GC: The polyproline region of p53 is required to
activate apoptosis but not growth arrest. Oncogene. 15:887–898.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han J, Cox DG, Colditz GA and Hunter DJ:
The p53 codon 72 polymorphism, sunburns, and risk of skin cancer in
US caucasian women. Mol Carcinog. 45:694–700. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stefanaki I, Stratigos AJ, Dimisianos G,
Nikolaou V, Papadopoulos O, Polydorou D, Gogas H, Tsoutsos D,
Panagiotou P, Kanavakis E, et al: P53 codon 72 Pro homozygosity
increases the risk of cutaneous melanoma in individuals with dark
skin complexion and among noncarriers of melanocortin 1 receptor
red hair variants. Br J Dermatol. 156:357–362. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen H, Liu Z, Strom SS, Spitz MR, Lee JE,
Gershenwald JE, Ross MI, Mansfield PF, Duvic M, Ananthaswamy HN and
Wei Q: P53 codon 72 arg homozygotes are associated with an
increased risk of cutaneous melanoma. J Invest Dermatol.
121:1510–1514. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Chen K, Liu Z, Wang LE, Gershenwald
JE, Lee JE, Prieto VG, Duvic M, Grimm EA and Wei Q: Polymorphisms
of TP53 Arg72Pro, but not p73 G4C14>A4TA4 and p21 Ser31Arg,
contribute to risk of cutaneous melanoma. J Invest Dermatol.
128:1585–1588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zauberman A, Flusberg D, Haupt Y, Barak Y
and Oren M: A functional p53-responsive intronic promoter is
contained within the human mdm2 gene. Nucleic Acids Res.
23:2584–2592. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bond GL, Hu W, Bond EE, Robins H, Lutzker
SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al: A
single nucleotide polymorphism in the MDM2 promoter attenuates the
p53 tumor suppressor pathway and accelerates tumor formation in
humans. Cell. 119:591–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thunell LK, Bivik C, Wäster P, Fredrikson
M, Stjernström A, Synnerstad I, Rosdahl I and Enerbäck C: MDM2
SNP309 promoter polymorphism confers risk for hereditary melanoma.
Melanoma Res. 24:190–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cotignola J, Chou JF, Roy P, Mitra N,
Busam K, Halpern AC and Orlow I: Investigation of the effect of
MDM2 SNP309 and TP53 Arg72Pro polymorphisms on the age of onset of
cutaneous melanoma. J Invest Dermatol. 132:1471–1478. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Firoz EF, Warycha M, Zakrzewski J, Pollens
D, Wang G, Shapiro R, Berman R, Pavlick A, Manga P, Ostrer H, et
al: Association of MDM2 SNP309, age of onset, and gender in
cutaneous melanoma. Clin Cancer Res. 15:2573–2580. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Capasso M, Ayala F, Avvisati RA, Russo R,
Gambale A, Mozzillo N, Ascierto PA and Iolascon A: MDM2 SNP309 and
p53 arg72Pro in cutaneous melanoma: Association between SNP309 GG
genotype and tumor breslow thickness. J Hum Genet. 55:518–524.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rovite V, Wolff-Sagi Y, Zaharenko L,
Nikitina-Zake L, Grens E and Klovins J: Genome database of the
latvian population (LGDB): Design, goals, and primary results. J
Epidemiol. 28:353–360. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ozola A, Ruklisa D and Pjanova D:
Association of the 16q24.3 region gene variants rs1805007 and
rs4785763 with heightened risk of melanoma in latvian population.
Meta Gene. 18:87–92. 2018. View Article : Google Scholar
|
|
28
|
Pliss L, Timša L, Rootsi S, Tambets K,
Pelnena I, Zole E, Puzuka A, Sabule A, Rozane S, Lace B, et al:
Y-Chromosomal lineages of latvians in the context of the genetic
variation of the eastern-baltic region. Ann Hum Genet. 79:418–430.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Reilly PF, Hoggart CJ, Pomyen Y, Calboli
FC, Elliott P, Jarvelin MR and Coin LJ: MultiPhen: Joint model of
multiple phenotypes can increase discovery in GWAS. PLoS One.
7:e348612012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Antonopoulou K, Stefanaki I, Lill CM,
Chatzinasiou F, Kypreou KP, Karagianni F, Athanasiadis E, Spyrou
GM, Ioannidis JPA, Bertram L, et al: Updated field synopsis and
systematic meta-analyses of genetic association studies in
cutaneous melanoma: The MelGene database. J Invest Dermatol.
135:1074–1079. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gudbjartsson DF, Sulem P, Stacey SN,
Goldstein AM, Rafnar T, Sigurgeirsson B, Benediktsdottir KR,
Thorisdottir K, Ragnarsson R, Sveinsdottir SG, et al: ASIP and TYR
pigmentation variants associate with cutaneous melanoma and basal
cell carcinoma. Nat Genet. 40:886–891. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kennedy C, ter Huurne J, Berkhout M, Gruis
N, Bastiaens M, Bergman W, Willemze R and Bavinck JN: Melanocortin
1 receptor (MC1R) gene variants are associated with an increased
risk for cutaneous melanoma which is largely independent of skin
type and hair color. J Invest Dermatol. 117:294–300. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guedj M, Bourillon A, Combadières C,
Rodero M, Dieudé P, Descamps V, Dupin N, Wolkenstein P, Aegerter P,
Lebbe C, et al: Variants of the MATP/SLC45A2 gene are protective
for melanoma in the french population. Hum Mutat. 29:1154–1160.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Demenais F, Corda E, Barrett J, Iles M,
Gillanders EM, Goldstein AM, Kanetsky PA, Bakker E, Bishop DT,
Newton-Bishop JA, et al: Importance of sequencing rare variants
after a genome-wide association study (GWAS): The MC1R gene, 16q24
region and melanoma story. Genet Epidemiol. 33:757–758. 2009.
|
|
35
|
Nan H, Kraft P, Qureshi AA, Guo Q, Chen C,
Hankinson SE, Hu FB, Thomas G, Hoover RN, Chanock S, et al:
Genome-wide association study of tanning phenotype in a population
of european ancestry. J Invest Dermatol. 129:2250–2257. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Beckman G, Birgander R, Sjalander A, Saha
N, Nolmberg PA, Kivela A and Beckman L: Is p53 polymorphism
maintained by natural selection? Hum Hered. 44:266–270. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oliveira C, Rinck-Junior JA, Lourenço GJ,
Moraes AM and Lima CS: Assessment of the XPC (A2920C), XPF
(T30028C), TP53 (Arg72Pro) and GSTP1 (Ile105Val) polymorphisms in
the risk of cutaneous melanoma. J Cancer Res Clin Oncol.
139:1199–1206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Geng P, Liao Y, Ruan Z and Liang H:
Increased risk of cutaneous melanoma associated with p53 Arg72Pro
polymorphism. PLoS One. 10:e01181122015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gwosdz C, Scheckenbach K, Lieven O,
Reifenberger J, Knopf A, Bier H and Balz V: Comprehensive analysis
of the p53 status in mucosal and cutaneous melanomas. Int J Cancer.
118:577–582. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bastiaens MT, Struyk L, Tjong-A-Hung SP,
Gruis N, ter Huurne J, Westendorp RG, Vermeer BJ, Bavinck JN and
ter Schegget J: Cutaneous squamous cell carcinoma and p53 codon 72
polymorphism: A need for screening? Mol Carcinog. 30:56–61. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Povey JE, Darakhshan F, Robertson K,
Bisset Y, Mekky M, Rees J, Doherty V, Kavanagh G, Anderson N,
Campbell H, et al: DNA repair gene polymorphisms and genetic
predisposition to cutaneous melanoma. Carcinogenesis. 28:1087–1093.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiao W, Du N, Huang T, Guo J, Mo X, Yuan
T, Chen Y, Ye T, Xu C, Wang W, et al: TP53 Mutation as potential
negative predictor for response of anti-CTLA-4 therapy in
metastatic melanoma. EBioMedicine. 32:119–124. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Raimondi S, Sera F, Gandini S, Iodice S,
Caini S, Maisonneuve P and Fargnoli MC: MC1R variants, melanoma and
red hair color phenotype: A meta-analysis. Int J Cancer.
122:2753–2760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nan H, Qureshi AA, Hunter DJ and Han J: A
functional SNP in the MDM2 promoter, pigmentary phenotypes, and
risk of skin cancer. Cancer Causes Control. 20:171–179. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Oliveira C, Lourenço GJ, Rinck-Junior JA,
Cintra ML, Moraes AM and Lima CS: Association between genetic
polymorphisms in apoptosis-related genes and risk of cutaneous
melanoma in women and men. J Dermatol Sci. 74:135–141. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qin J, Cong X, Jin J, Chu Z, Gu X and Cai
Y: Association between MDM2 SNP309 and skin cancer: A meta-analysis
of case-control studies. J Dermatol Sci. 79:171–173. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Landi MT, Kanetsky PA, Tsang S, Gold B,
Munroe D, Rebbeck T, Swoyer J, Ter-Minassian M, Hedayati M,
Grossman L, et al: MC1R, ASIP, and DNA repair in sporadic and
familial melanoma in a mediterranean population. J Natl Cancer
Inst. 97:998–1007. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fargnoli MC, Spica T, Sera F, Pellacani G,
Chiarugi A, Seidenari S, Carli P, Chimenti S and Peris K: Re: MC1R,
ASIP, and DNA repair in sporadic and familial melanoma in a
mediterranean population. J Natl Cancer Inst. 98:144–145. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Taylor NJ, Busam KJ, From L, Groben PA,
Anton-Culver H, Cust AE, Begg CB, Dwyer T, Gallagher RP, Gruber SB,
et al: Inherited variation at MC1R and histological characteristics
of primary melanoma. PLoS One. 10:e01199202015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Davies JR, Randerson-Moor J, Kukalizch K,
Harland M, Kumar R, Madhusudan S, Nagore E, Hansson J, Höiom V,
Ghiorzo P, et al: Inherited variants in the MC1R gene and survival
from cutaneous melanoma: A BioGenoMEL study. Pigment Cell Melanoma
Res. 25:384–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rajabi P, Karimian P and Heidarpour M: The
relationship between MDM2 expression and tumor thickness and
invasion in primary cutaneous malignant melanoma. J Res Med Sci.
17:452–455. 2012.PubMed/NCBI
|