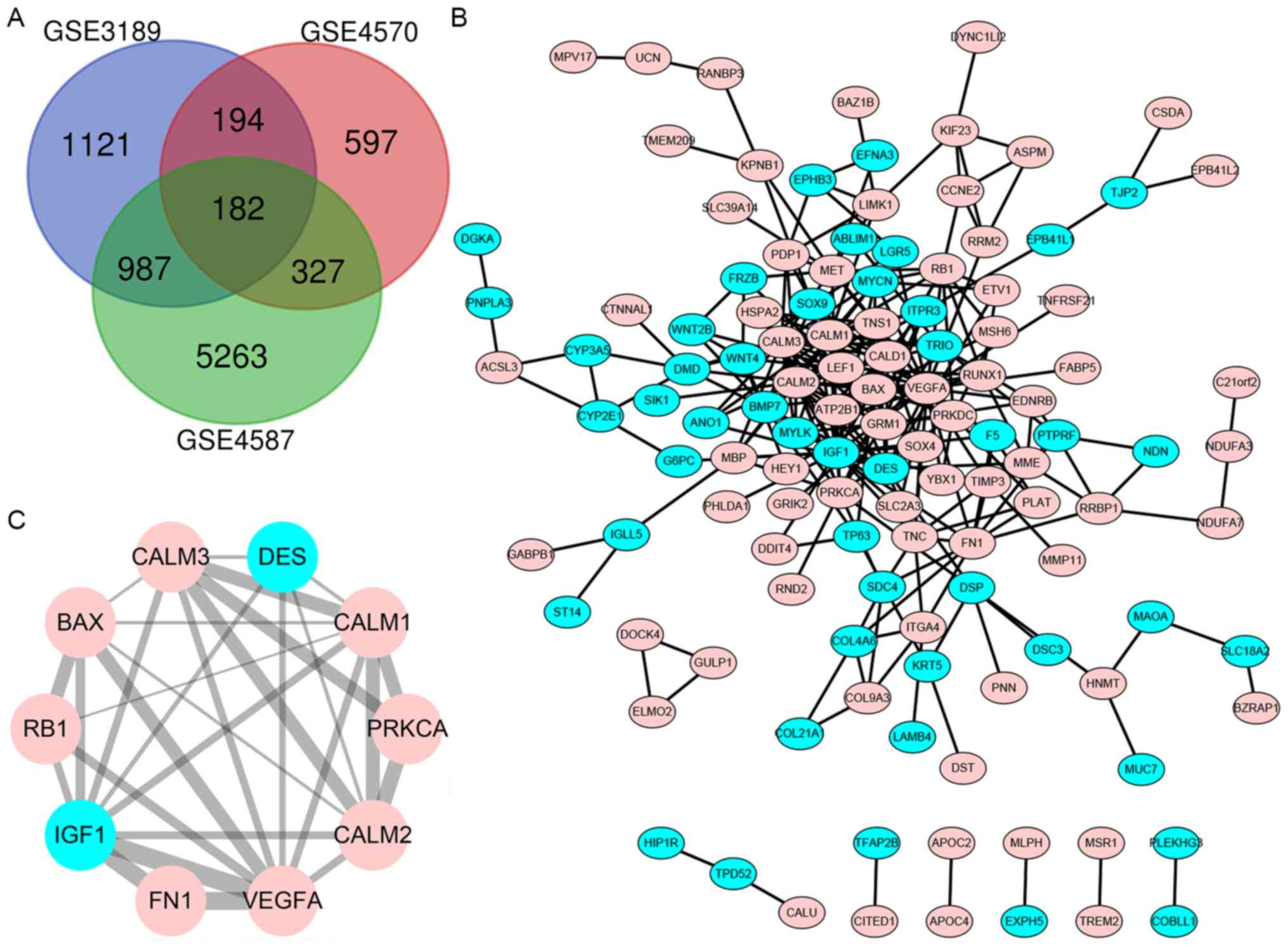
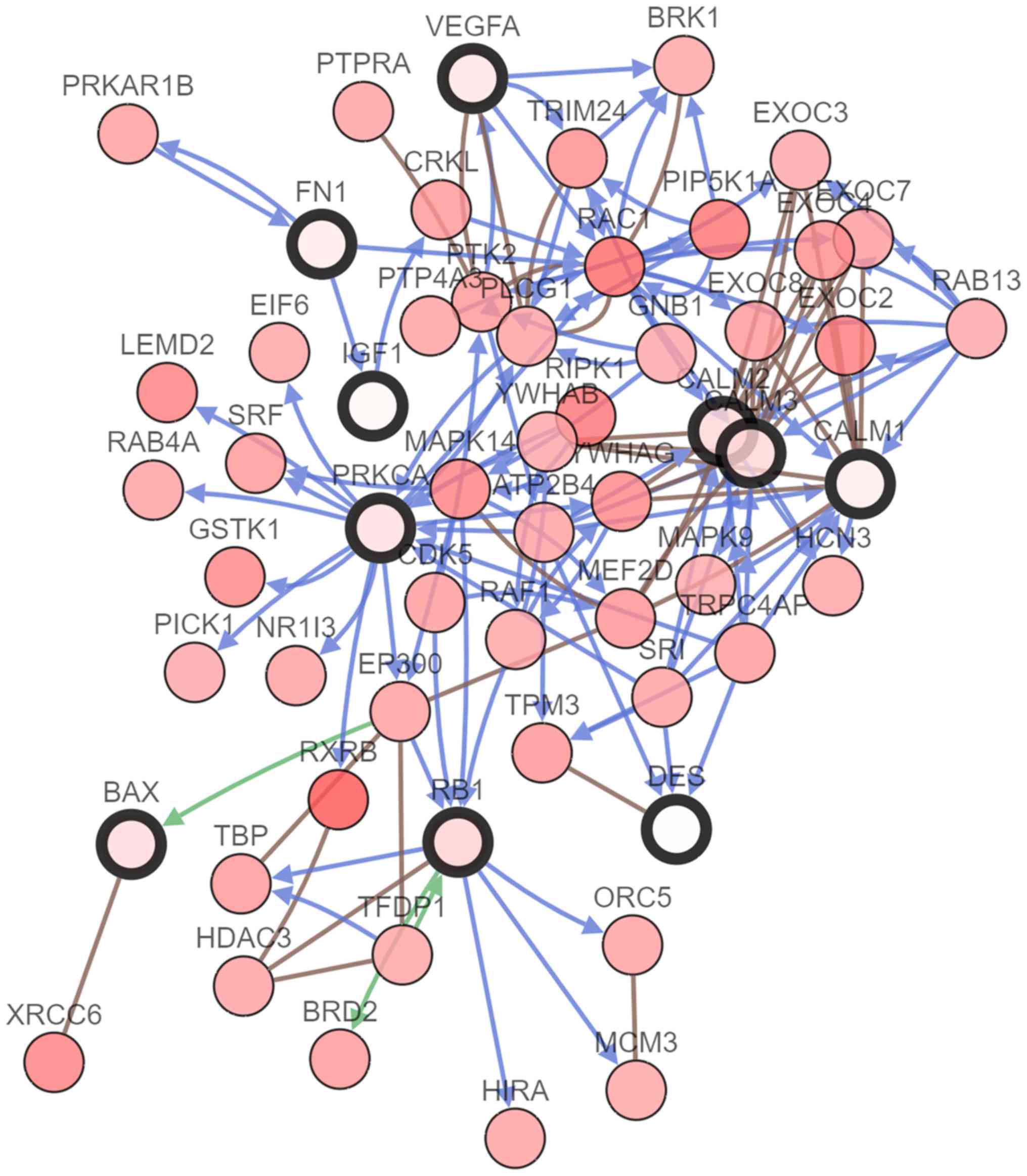
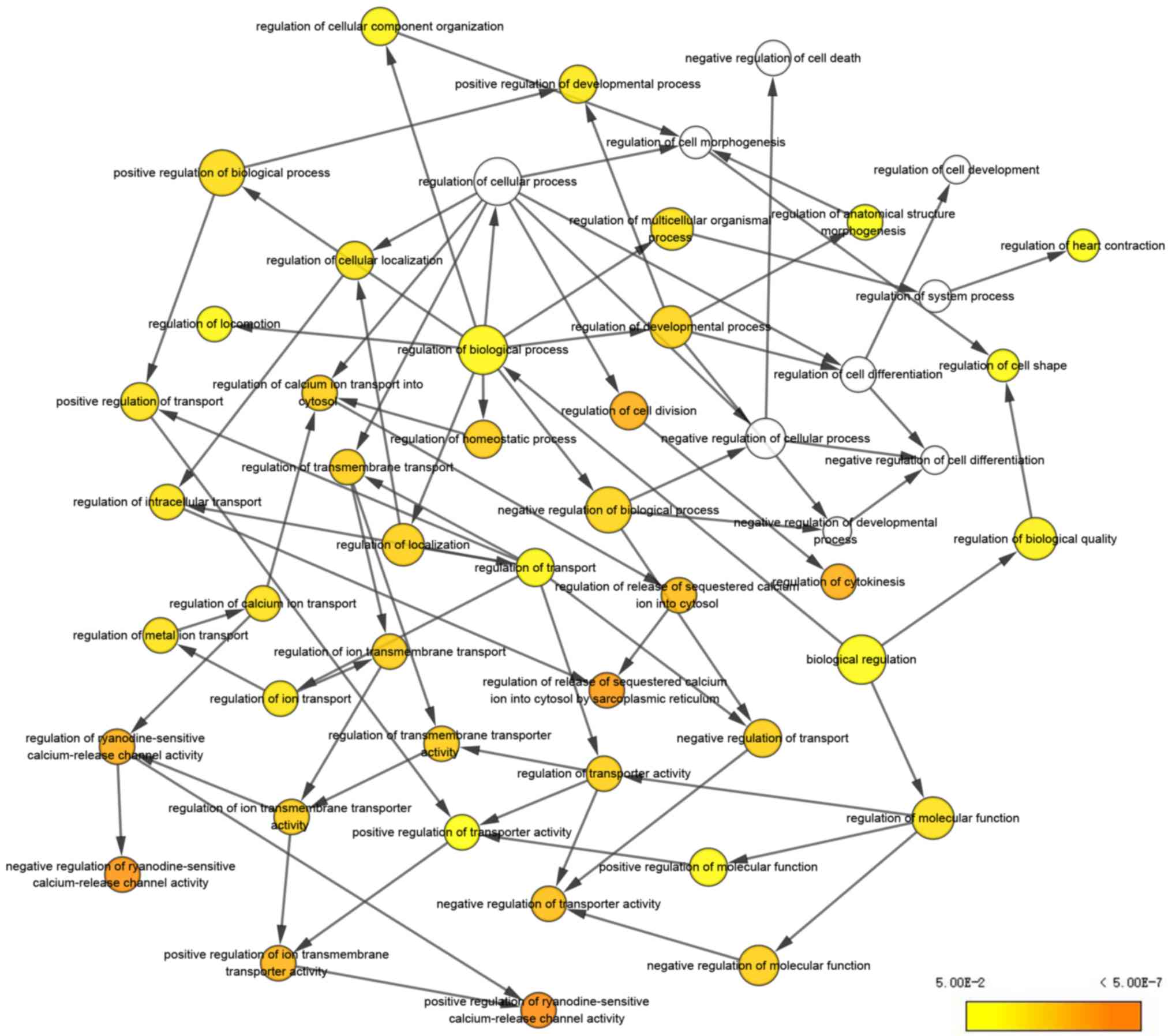
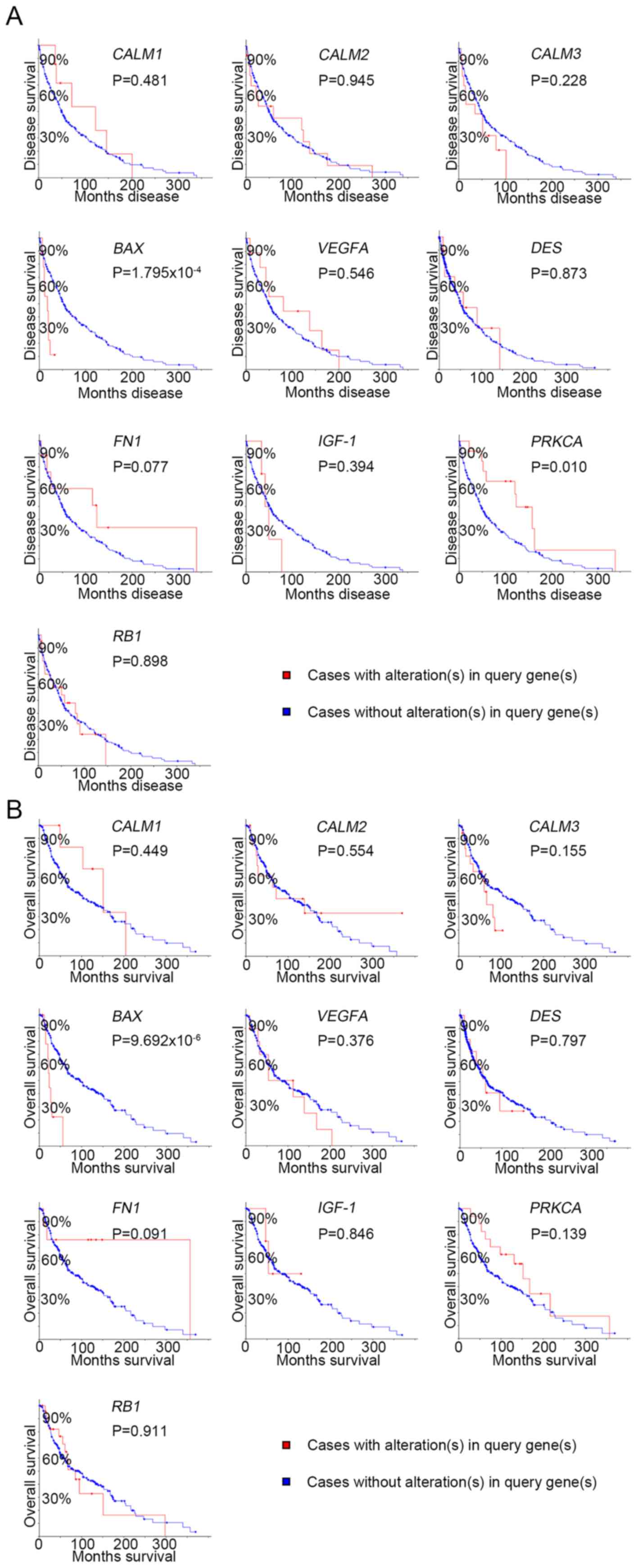
|
1
|
Qin J, Li S, Zhang C, Gao DW, Li Q, Zhang
H, Jin XD and Liu Y: Apoptosis and injuries of heavy ion beam and
x-ray radiation on malignant melanoma cell. Exp Biol Med (Maywood).
242:953–960. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ci C, Tang B, Lyu D, Liu W, Qiang D, Ji X,
Qiu X, Chen L and Ding W: Overexpression of CDCA8 promotes the
malignant progression of cutaneous melanoma and leads to poor
prognosis. Int J Mol Med. 43:404–412. 2019.PubMed/NCBI
|
|
3
|
Bai X, Kong Y, Chi Z, Sheng X, Cui C, Wang
X, Mao L, Tang B, Li S, Lian B, et al: MAPK pathway and TERT
promoter gene mutation pattern and its prognostic value in melanoma
patients: A retrospective study of 2,793 cases. Clin Cancer Res.
23:6120–6127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shalem O, Sanjana NE, Hartenian E, Shi X,
Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG and
Zhang F: Genome-scale CRISPR-Cas9 knockout screening in human
cells. Science. 343:84–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nissan MH, Pratilas CA, Jones AM, Ramirez
R, Won H, Liu C, Tiwari S, Kong L, Hanrahan AJ, Yao Z, et al: Loss
of NF1 in cutaneous melanoma is associated with RAS activation and
MEK dependence. Cancer Res. 74:2340–2350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Burd CE, Liu W, Huynh MV, Waqas MA,
Gillahan JE, Clark KS, Fu K, Martin BL, Jeck WR, Souroullas GP, et
al: Mutation-specific RAS oncogenicity explains NRAS codon 61
selection in melanoma. Cancer Discov. 4:1418–1429. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Agrawal P, Fontanals-Cirera B, Sokolova E,
Jacob S, Vaiana CA, Argibay D, Davalos V, McDermott M, Nayak S,
Darvishian F, et al: A systems biology approach identifies FUT8 as
a driver of melanoma metastasis. Cancer Cell. 31:804–819.e7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Talantov D, Mazumder A, Yu JX, Briggs T,
Jiang Y, Backus J, Atkins D and Wang Y: Novel genes associated with
malignant melanoma but not benign melanocytic lesions. Clin Cancer
Res. 11:7234–7242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hoek K, Rimm DL, Williams KR, Zhao H,
Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et
al: Expression profiling reveals novel pathways in the
transformation of melanocytes to melanomas. Cancer Res.
64:5270–5282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smith AP, Hoek K and Becker D:
Whole-genome expression profiling of the melanoma progression
pathway reveals marked molecular differences between nevi/melanoma
in situ and advanced-stage melanomas. Cancer Biol Ther.
4:1018–1029. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Ser B (Methodological). 57:289–300.
1995.
|
|
13
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast delayed enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin X, Sun R, Zhao X, Zhu D, Zhao X, Gu Q,
Dong X, Zhang D, Zhang Y, Li Y and Sun B: C-myc overexpression
drives melanoma metastasis by promoting vasculogenic mimicry via
c-myc/snail/Bax signaling. J Mol Med (Berl). 95:53–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brecht IB, Garbe C, Gefeller O, Pfahlberg
A, Bauer J, Eigentler TK, Offenmueller S, Schneider DT and Leiter
U: 443 paediatric cases of malignant melanoma registered with the
German central malignant melanoma registry between 1983 and 2011.
Eur J Cancer. 51:861–868. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shimizu A, Kaira K, Yasuda M, Asao T and
Ishikawa O: Decreased expression of class III β-tubulin is
associated with unfavourable prognosis in patients with malignant
melanoma. Melanoma Res. 26:29–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song S, Jacobson KN, McDermott KM, Reddy
SP, Cress AE, Tang H, Dudek SM, Black SM, Garcia JG, Makino A and
Yuan JX: ATP promotes cell survival via regulation of cytosolic
[Ca2+] and Bcl-2/Bax ratio in lung cancer cells. Am J Physiol Cell
Physiol. 310:C99–S114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi L, Zhang G, Zheng Z, Lu B and Ji L:
Andrographolide reduced VEGFA expression in hepatoma cancer cells
by inactivating HIF-1α: The involvement of JNK and MTA1/HDCA. Chem
Biol Interact. 273:228–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Merino D, Lok SW, Visvader JE and Lindeman
GJ: Targeting BCL-2 to enhance vulnerability to therapy in estrogen
receptor-positive breast cancer. Oncogene. 35:1877–1887. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signaling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Delgado-Bellido D, Serrano-Saenz S,
Fernandez-Cortés M and Oliver FJ: Vasculogenic mimicry signaling
revisited: Focus on non-vascular VE-cadherin. Mol Cancer.
16:652017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin CY, Cho CF, Bai ST, Liu JP, Kuo TT,
Wang LJ, Lin YS, Lin CC, Lai LC, Lu TP, et al: ADAM9 promotes lung
cancer progression through vascular remodeling by VEGFA, ANGPT2,
and PLAT. Sci Rep. 7:151082017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim M, Jang K, Miller P, Picon-Ruiz M,
Yeasky TM, El-Ashry D and Slingerland JM: VEGFA links self-renewal
and metastasis by inducing Sox2 to repress miR-452, driving Slug.
Oncogene. 36:5199–5211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fahmy K, Gonzalez A, Arafa M, Peixoto P,
Bellahcène A, Turtoi A, Delvenne P, Thiry M, Castronovo V and
Peulen O: Myoferlin plays a key role in VEGFA secretion and impacts
tumor-associated angiogenesis in human pancreas cancer. Int J
Cancer. 138:652–663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cheng CY, Ho TY, Hsiang CY, Tang NY, Hsieh
CL, Kao ST and Lee YC: Angelica sinensis Exerts Angiogenic and
Anti-apoptotic effects against cerebral ischemia-reperfusion injury
by activating p38MAPK/HIF-1[Formula: See text]/VEGF-A signaling in
rats. Am J Chin Med. 45:1683–1708. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Lv Z, Xu J, Chen C, Ge Q, Li P,
Wei D, Wu Z and Sun X: MicroRNA-134 inhibits osteosarcoma
angiogenesis and proliferation by targeting the VEGFA/VEGFR1
pathway. FEBS J. 285:1359–1371. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Boczek NJ, Gomez-Hurtado N, Ye D, Calvert
ML, Tester DJ, Kryshtal D, Hwang HS, Johnson CN, Chazin WJ,
Loporcaro CG, et al: Spectrum and prevalence of CALM1-, CALM2- and
CALM3-encoded calmodulin variants in long QT syndrome and
functional characterization of a novel long QT syndrome-associated
calmodulin missense variant, E141G. Circ Cardiovasc Genet.
9:136–146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Limpitikul WB, Dick IE, Tester DJ, Boczek
NJ, Limphong P, Yang W, Choi MH, Babich J, DiSilvestre D, Kanter
RJ, et al: A precision medicine approach to the rescue of function
on malignant Calmodulinopathic long-QT syndrome. Circ Res.
120:39–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cai R, Zhang C, Zhao Y, Zhu K, Wang Y,
Jiang H, Xiang Y and Cheng B: Genome-wide analysis of the IQD gene
family in maize. Mol Genet Genomics. 291:543–558. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bürstenbinder K, Möller B, Plötner R,
Stamm G, Hause G, Mitra D and Abel S: The IQD family of
Calmodulin-binding proteins links calcium signaling to
microtubules, membrane subdomains and the nucleus. Plant Physiol.
173:1692–1708. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shoshan-Barmatz V, Krelin Y and
Shteinfer-Kuzmine A: VDAC1 functions in Ca2+ homeostasis
and cell life and death in health and disease. Cell Calcium.
69:81–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu Z, Ding Y, Ye N, Wild C, Chen H and
Zhou J: Direct activation of bax protein for cancer therapy. Med
Res Rev. 36:313–341. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gil J, Ramsey D, Szmida E, Leszczynski P,
Pawlowski P, Bebenek M and Sasiadek MM: The BAX gene as a candidate
for negative autophagy-related genes regulator on mRNA levels in
colorectal cancer. Med Oncol. 34:162017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Del Principe MI, Dal Bo M, Bittolo T,
Buccisano F, Rossi FM, Zucchetto A, Rossi D, Bomben R, Maurillo L,
Cefalo M, et al: Clinical significance of bax/bcl-2 ratio in
chronic lymphocytic leukemia. Haematologica. 101:77–85. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kowalczyk AE, Krazinski BE, Godlewski J,
Kiewisz J, Kwiatkowski P, Sliwinska-Jewsiewicka A, Kiezun J, Sulik
M and Kmiec Z: Expression of the EP300, TP53 and BAX genes in
colorectal cancer: Correlations with clinicopathological parameters
and survival. Oncol Rep. 38:201–210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rust R, Visser L, van der Leij J, Harms G,
Blokzijl T, Deloulme JC, van der Vlies P, Kamps W, Kok K, Lim M, et
al: High expression of calcium-binding proteins, S100A10, S100A11
and CALM2 in anaplastic large cell lymphoma. Br J Haematol.
131:596–608. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Haddad SA, Lunetta KL, Ruiz-Narvaez EA,
Bensen JT, Hong CC, Sucheston-Campbell LE, Yao S, Bandera EV,
Rosenberg L, Haiman CA, et al: Hormone-related pathways and risk of
breast cancer subtypes in African American women. Breast Cancer Res
Treat. 154:145–154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cai H, Xu J, Han Y, Lu Z, Han T, Ding Y
and Ma L: Integrated miRNA-risk gene-pathway pair network analysis
provides prognostic biomarkers for gastric cancer. Onco Targets
Ther. 9:2975–2986. 2016.PubMed/NCBI
|
|
49
|
Li CF, Yan ZK, Chen LB, Jin JP and Li DD:
Desmin detection by facile prepared carbon quantum dots for early
screening of colorectal cancer. Medicine (Baltimore). 96:e55212017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Y, Li Y, Chen Z, Wang T, Gu J, Wu X,
Yin Y, Wang M and Pan Z: The evaluation of colorectal cancer risk
in serum by anti-DESMIN-conjugated CdTe/CdS quantum dots. Clin Lab.
63:579–586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ekinci O, Ogut B, Celik B and Dursun A:
Compared with elastin Stains, h-Caldesmon and desmin offer superior
detection of vessel invasion in gastric, pancreatic and colorectal
adenocarcinomas. Int J Surg Pathol. 26:318–326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Trevisan F, Tregnago AC, Lopes Pinto CA,
Urvanegia ACM, Morbeck DL, Bertolli E, Riva Neto FR, Duprat Neto JP
and de Macedo MP: Osteogenic melanoma with desmin expression. Am J
Dermatopathol. 39:528–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ku SY, Rosario S, Wang Y, Mu P, Seshadri
M, Goodrich ZW, Goodrich MM, Labbé DP, Gomez EC, Wang J, et al: Rb1
and Trp53 cooperate to suppress prostate cancer lineage plasticity,
metastasis and antiandrogen resistance. Science. 355:78–83. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang X, Zhu Q, Lin Y, Wu L, Wu X, Wang K,
He Q, Xu C, Wan X and Wang X: Crosstalk between TEMs and
endothelial cells modulates angiogenesis and metastasis via
IGF1-IGF1R signalling in epithelial ovarian cancer. Br J Cancer.
117:1371–1382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ohshima Y, Takata N, Suzuki-Karasaki M,
Yoshida Y, Tokuhashi Y and Suzuki-Karasaki Y: Disrupting
mitochondrial Ca2+ homeostasis causes tumor-selective TRAIL
sensitization through mitochondrial network abnormalities. Int J
Oncol. 51:1146–1158. 2017. View Article : Google Scholar : PubMed/NCBI
|