Introduction
Glioma is an aggressive brain cancer with high
morbidity and mortality rates (1)
characterized by aggressive proliferation, invasion and dismal
prognosis (2). Although great
progress has been made in the therapeutic field, the prognosis of
patients with glioma remains poor (3). Recent studies have revealed a number of
aberrantly expressed genes, which may be involved in the
progression of glioma (4,5). These findings could provide a new
perspective for the development of novel therapeutic strategies for
glioma.
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNAs with a length of 20–24 nucleotides (6). miRNA dysregulation has been widely
reported in numerous human diseases, especially cancer (7). miRNAs regulate the expression of target
messenger RNAs through binding to their 3′-untranslated region
(8). Several miRNAs have been
identified to serve a crucial role in glioma progression and
multiple cellular processes, including cell proliferation,
differentiation, apoptosis, migration and invasion. For example,
Zhang et al (9) indicated
that miR-770 expression was markedly lower in human glioma tissues
and cell lines, and that miR-770 overexpression suppressed tumor
cell proliferation and induced apoptosis via targeting CDK8. Lu
et al (10) demonstrated that
the upregulation of miR-6807-3p facilitated carcinogenesis in human
glioma through promoting cancer cell proliferation and migration by
targeting dachshund homolog 1. Downregulation of miR-139-3p
expression was also detected in glioma tissues, which may be
involved in controlling behaviors associated with the malignant
progression of glioma, including cell proliferation, migration and
invasiveness (11). Thus, there is a
great interest in using aberrantly expressed miRNAs as prognosis
prediction markers for glioma. However, it may be challenging to
detect miRNAs involved in glioma occurrence and progression.
In the present study, a number of differentially
expressed miRNAs in glioma were detected via bioinformatics
analysis. OncomiR analysis suggested that three miRNAs were able to
accurately predict overall cancer survival in glioma. The
differential expression level of miR-455-3p was found to be the
most significant. Its clinical value has not been investigated.
Therefore, miR-455-3p levels were further validated in patients
with glioma, and its clinical value was explored.
Materials and methods
Microarray data
Firstly, datasets focusing on miRNA expression were
searched using the following search terms: ‘Non-coding RNA
profiling by array AND glioblastoma’ in the National Center for
Biotechnology Information Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/gds/),
and only miRNA gene expression profiles were included.
Additionally, only datasets based on the comparison of glioma
tissues and healthy samples were included. Datasets based on glioma
serum, glioma cell lines or animal models were excluded. The
GSE90603 and GSE103229 (12,13) datasets met the inclusion criteria and
were subsequently assessed using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) for
differential genetic analysis. The combination of the two datasets
has not been mentioned in previous studies. GSE90603 contained 16
patients with glioblastoma (GBM) and 7 healthy individuals, while
GSE103229 contained 5 GBM samples and 5 normal samples.
Identification of differentially
expressed miRNAs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/info/geo2r.html) is an
online tool used to compare two or more groups of samples in GEO
series for the identification of differentially expressed miRNAs
under different experimental conditions. In the present study,
GEO2R was used for the identification of differentially expressed
miRNAs in glioma, and the adjusted P-value <0.05 and |Log fold
change (FC)|>2 were chosen as cut-off levels.
miRNA expression-based survival signature analysis.
OncomiR is able to analyze miRNA-derived survival outcome
signatures dynamically for one or more types of cancer (14). In the present study, the glioma
patients were divided into a high-risk and low-risk cohort
according to the expression level of the selected miRNAs, overall
cancer survival rates were predicted using OncomiR, which was
presented with a Kaplan-Meier survival curve.
Patients and sample collection
A total of 108 patients with primary glioma who
underwent primary tumor resection between June 2010 and March 2011
at The 900 Hospital of the Joint Logistics Team were recruited in
the present study. The clinicopathological features of glioma
patients were collected, including age, sex, tumor size, World
Health Organization (WHO) grade and Karnofsky performance scale
(KPS). A total of 95 normal brain tissue samples were collected as
controls from patients who had undergone surgery for cerebral
injury and cerebral hemorrhage at the same hospital. All tissue
specimens were immediately snap-frozen in liquid nitrogen and
stored at −80°C until further processing. No participants received
radiotherapy or chemotherapy prior to surgery. After surgery, all
patients were followed up by special clinic reexamination, letters
or telephones to calculate the 5-year survival rate. The interval
follow-up time was ~half or one year. All procedures were reviewed
and approved by the Ethics Committee of The 900 Hospital of the
Joint Logistics Team. Written informed consent was obtained from
each participant.
Reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) of miR-455-3p. RT-qPCR was used to determine the
expression levels of miR-455-3p every two weeks or so since the
sample collection. Each experiment was repeated three times. Total
RNA was extracted from glioma and normal brain tissues using TRIzol
reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The miScript Reverse Transcription Kit
(Qiagen) was used for reverse transcription reactions. Next, qPCR
was performed using the SYBR Green I Master Mix Kit (Invitrogen;
Thermo Fisher Scientific, Inc.) and the 7300 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer
sequences were as follows: miR-455-3p forward,
5′-GGGGCAGTCCATGGGCAT-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; and
U6 forward, CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGAATTTGCGT.
The PCR conditions were as follows: 95°C for 5 min, followed by 40
cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 40 sec.
Relative miR-455-3p expression was normalized to the expression of
U6 and was quantified using the 2−ΔΔCq method (15).
Statistical analysis
SPSS software (version 18.0; SPSS, Inc.) and
GraphPad Prism software (version 5.0; GraphPad Software, Inc.) were
used for data analysis. Measurement data between groups were
evaluated using the Student's t test, whereas counting data were
evaluated using the χ2 test. The mean expression level
of miR-455-3p in glioma tissues was used as the cutoff, and the
patients were divided into low- and high-expression groups. The
differences between the two groups were compared using the
χ2 test. The overall survival of patients with glioma
was assessed using the Kaplan-Meier curve and log-rank test. Cox
regression analysis was performed to further assess the prognostic
value of miR-455-3p in glioma. P<0.05 was considered to indicate
a statistically significant difference.
Results
Identification of differentially
expressed miRNAs
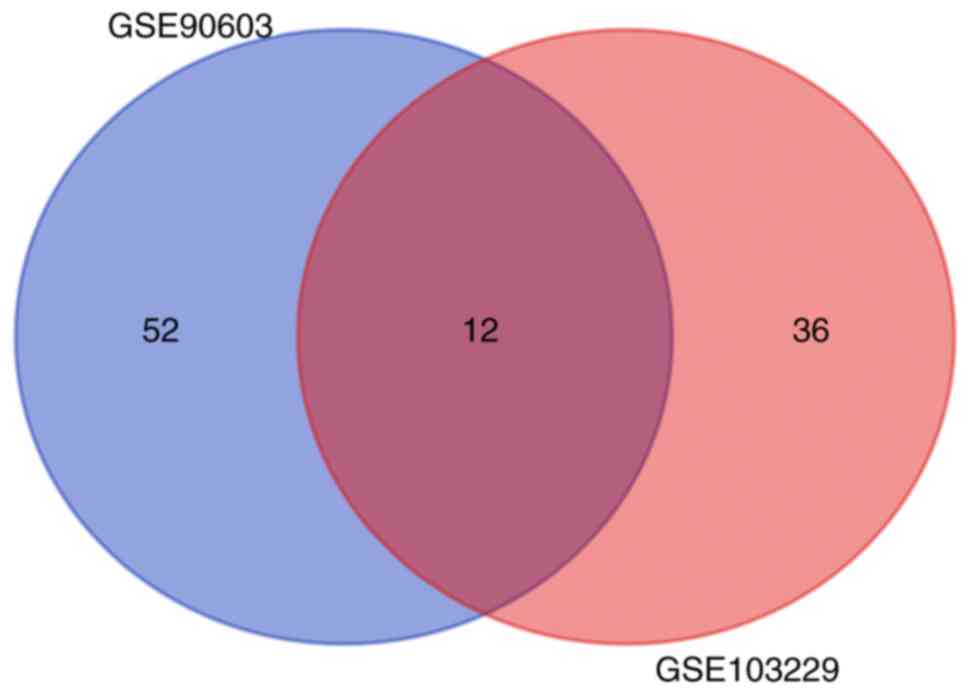
Two miRNA expression profiles (GSE90603 and
GSE103229) were obtained from the GEO database. GSE90603 contained
16 GBM and 7 normal tissue samples, and GSE103229 contained 5 GBM
and 5 normal tissue samples. A total of 64 differentially expressed
miRNAs were identified in the GSE90603 dataset, and a total of 48
were found in GSE103229. Twelve differentially expressed miRNAs
were observed to overlap between these two datasets (Fig. 1), and the analysis results are
presented in Table I.
 | Table I.Twelve differentially expressed miRNAs
in the overlap of GSE90603 and GSE103229 datasets. |
Table I.
Twelve differentially expressed miRNAs
in the overlap of GSE90603 and GSE103229 datasets.
| miRNA_ID | adj.P.Val | Log FC |
|---|
| hsa-miR-455-3p |
2.03×10−6 | 2.30159 |
| hsa-miR-218-5p |
1.95×10−4 | −2.90172 |
| hsa-miR-7-5p |
3.69×10−4 | −3.25590 |
| hsa-miR-139-5p |
6.13×10−4 | −2.88092 |
|
hsa-miR-129-1-3p |
6.46×10−4 | −2.13994 |
|
hsa-miR-3200-3p |
9.75×10−4 | −2.00359 |
| hsa-miR-124-3p |
1.43×10−3 | −4.08201 |
| hsa-miR-129-5p |
2.31×10−3 | −2.68730 |
| hsa-miR-21-3p |
3.74×10−3 | 2.308139 |
| hsa-miR-338-5p |
4.85×10−3 | −2.08672 |
| hsa-miR-138-5p |
6.96×10−3 | −3.01343 |
| hsa-miR-491-5p |
9.93×10−3 | −2.03053 |
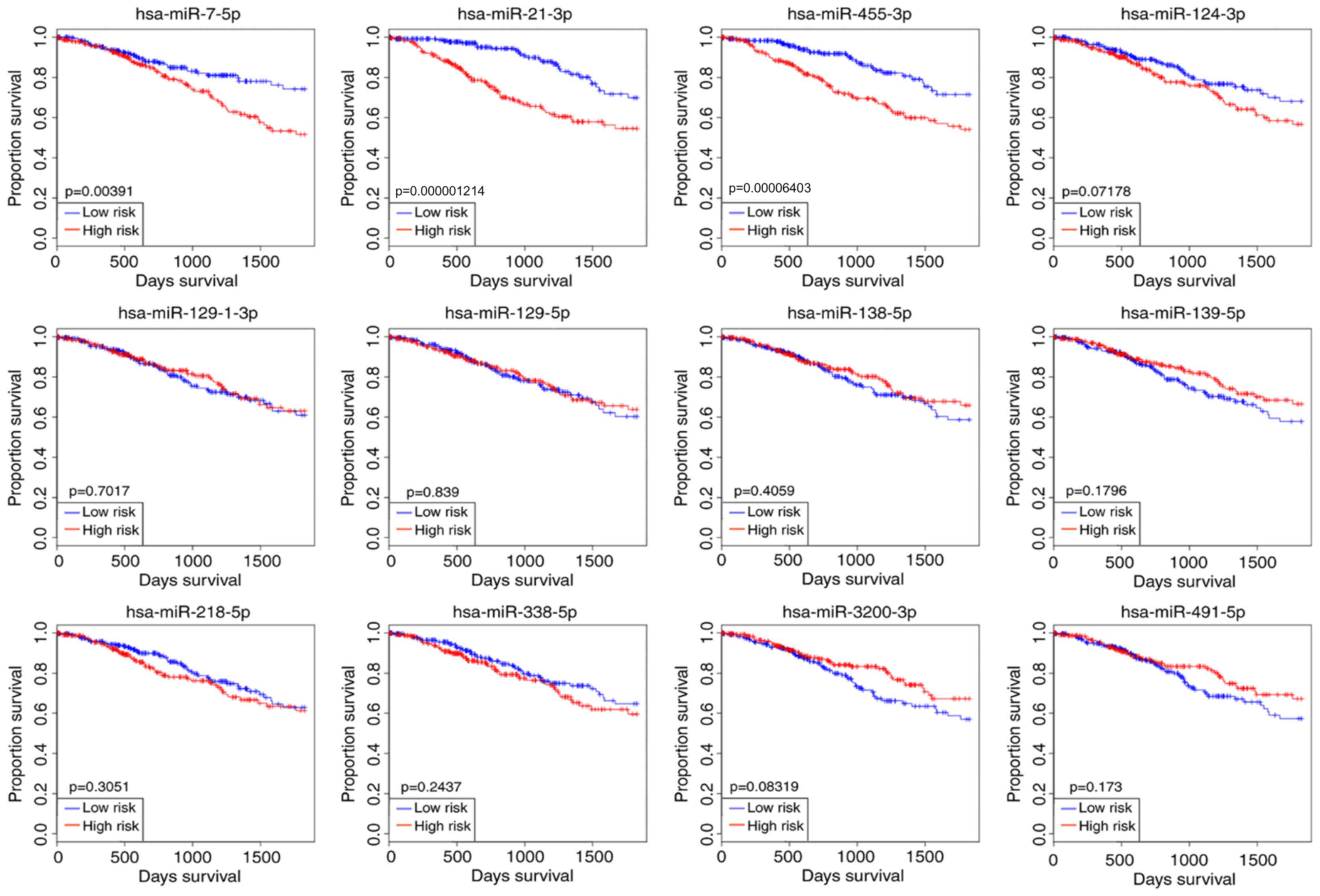
Kaplan-Meier survival curves of 12
differentially expressed miRNAs
The overall cancer survival was predicted using
OncomiR, and the results were presented with a Kaplan-Meier
survival curve. In the present study, the glioma patients were
divided into high-risk and low-risk cohorts according to the
expression levels of the selected miRNAs. The survival analysis
results suggested that only three miRNAs reached significant levels
among the 12 differentially expressed miRNAs when predicting
overall cancer survival, namely hsa-miR-7-5p, hsa-miR-21-3p and
hsa-miR-455-3p (Fig. 2). The
clinical value of miR-455-3p in glioma has not been reported in any
previous studies and its differential expression level was the most
significant (Table I).
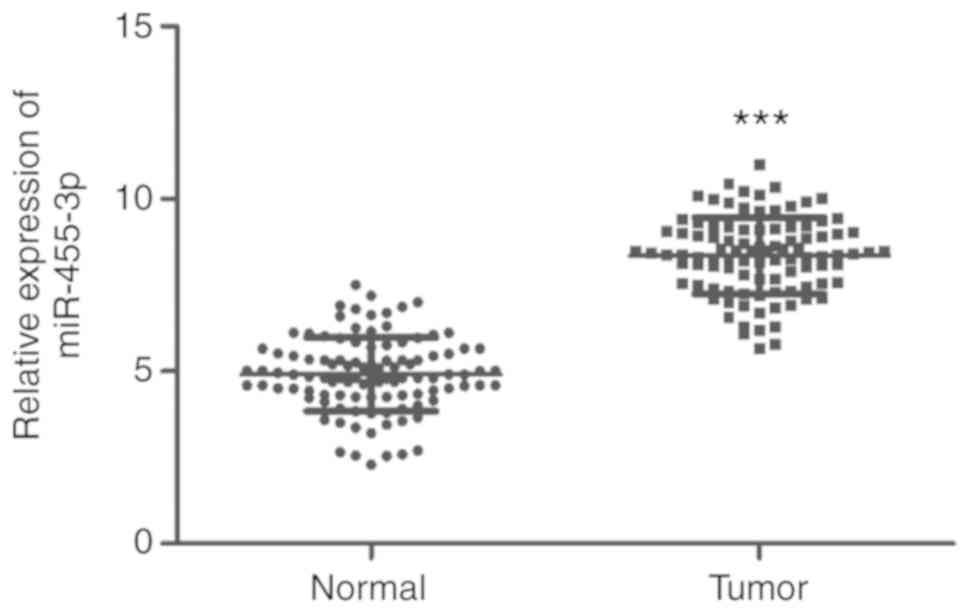
Upregulation of miR-455-3p in glioma
tissues
The expression levels of miR-455-3p were examined in
108 glioma and 95 normal brain tissue samples. The RT-qPCR results
suggested that the expression of miR-455-3p was significantly
upregulated in glioma tissues compared with normal tissues
(P<0.001; Fig. 3).
Association of miR-455-3p expression
with clinicopathological features of patients with glioma
The clinical data of 108 patients with glioma are
presented in Table II. The mean
expression level of miR-455-3p in glioma tissues was identified as
a cut-off level.
 | Table II.Association of miR-455-3p with the
clinicopathological features of patients with glioma. |
Table II.
Association of miR-455-3p with the
clinicopathological features of patients with glioma.
|
|
| miR-455-3p
expression |
|
|---|
|
|
|
|
|
|---|
| Feature | n=108 | Low (n=44) | High (n=64) | P-value |
|---|
| Age (years) |
|
|
| 0.383 |
|
≤60 | 51 | 23 | 28 |
|
|
>60 | 57 | 21 | 36 |
|
| Sex |
|
|
| 0.942 |
|
Male | 56 | 23 | 33 |
|
|
Female | 52 | 21 | 31 |
|
| Tumor size
(cm) |
|
|
| 0.354 |
|
≥5.0 | 67 | 25 | 42 |
|
|
<5.0 | 41 | 19 | 22 |
|
| WHO grade |
|
|
| 0.006a |
|
I–II | 64 | 33 | 31 |
|
|
III–IV | 44 | 11 | 33 |
|
| KPS |
|
|
| 0.037b |
|
<80 | 62 | 20 | 42 |
|
|
≥80 | 46 | 24 | 22 |
|
The patients were divided into low (n=44) and high
miR-455-3p expression (n=64) groups. Differences between groups
were compared using the χ2 test. It was observed that
the upregulation of miR-455-3p expression demonstrated significant
association with high WHO grade (P=0.006) and low KPS, P=0.037) in
glioma patients. However, miR-455-3p expression was not
significantly associated with other clinicopathological features
including age, sex and tumor size (all P>0.05; Table II).
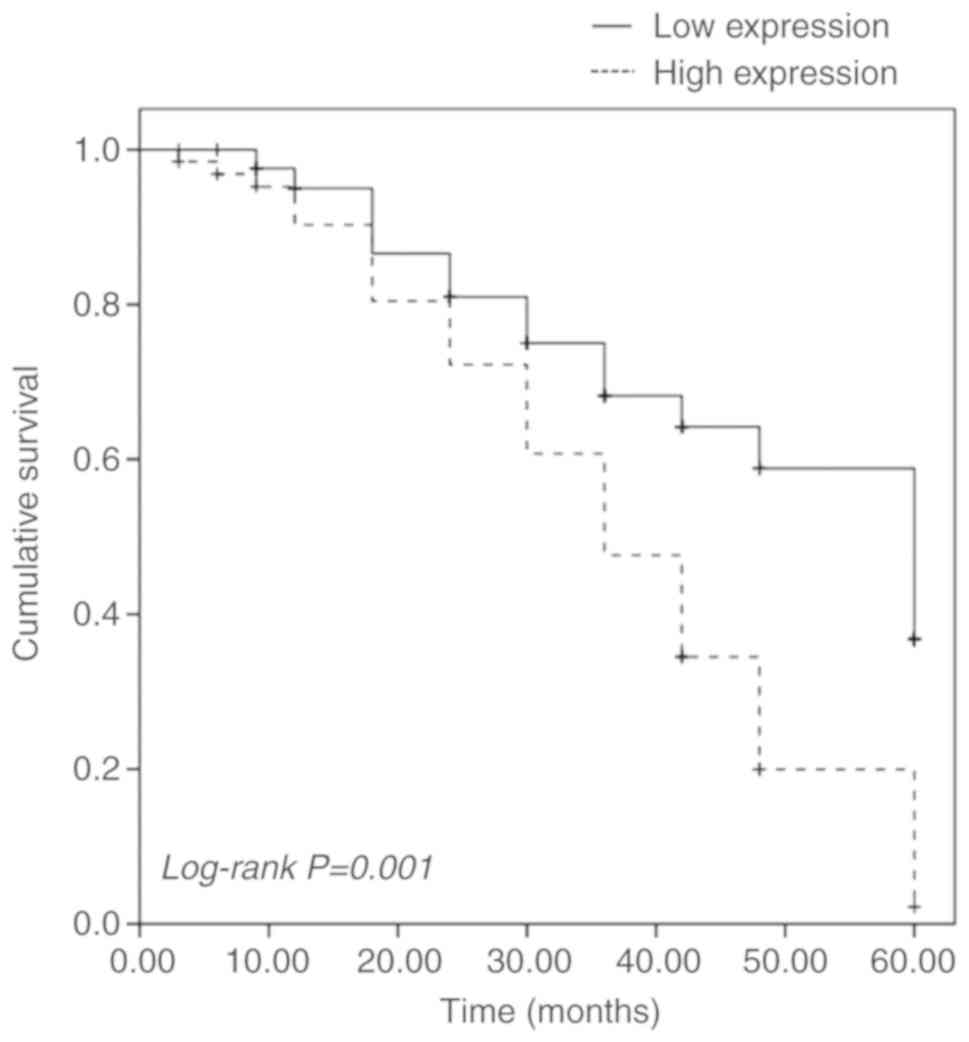
Prognostic value of miR-455-3p
expression in patients with glioma
The Kaplan-Meier survival curve and the log-rank
test were performed to evaluate the association between miR-455-3p
expression and patient survival. It was identified that patients
with high miR-455-3p expression had significantly lower 5-year
overall survival than those with low miR-455-3p expression
(log-rank P=0.001; Fig. 4). The
multivariate Cox regression analysis further suggested that
miR-455-3p (hazard ratio=2.136; 95% CI=1.177–3.877; P=0.013) was an
independent prognostic indicator for overall survival in patients
with glioma (Table III).
 | Table III.Multivariate Cox regression analysis
for miR-455-3p in patients with glioma. |
Table III.
Multivariate Cox regression analysis
for miR-455-3p in patients with glioma.
|
| Multivariate
analysis |
|
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value |
|---|
| miR-455-3p | 2.136 | 1.177–3.877 | 0.013a |
| Age | 0.899 | 0.551–1.468 | 0.671 |
| Sex | 1.115 | 0.693–1.793 | 0.654 |
| Tumor size | 0.808 | 0.490–1.332 | 0.403 |
| WHO grade | 1.607 | 0.973–2.654 | 0.064 |
| KPS | 0.789 | 0.472–1.319 | 0.367 |
Discussion
Glioma is an aggressive central nervous system
malignancy, which greatly affects human health (16). Owing to rapid proliferation,
metastasis and angiogenesis, this disease is relatively hard to
treat (17,18). The prognosis of patients with glioma
is relatively poor due to metastasis and recurrence (3,19). The
incidence and mortality rates of glioma are continuously increasing
(1). Although numerous epigenetic
changes have been identified to be associated with glioma, the
pathogenesis is not fully understood. A recent study reported that
a number of abnormal miRNAs were found to be involved in the
transcriptional regulatory network associated with tumor initiation
and progression (20). Therefore,
systematic and integrative analysis is required to explore key
miRNAs that may contribute to tumor progression.
Currently, microarray and bioinformatics analyses
are widely used to investigate potential targets for the diagnosis
and therapy of different types of human cancer (21,22). In
the present study, a total of 64 differentially expressed miRNAs
were identified in GSE90603, and 48 were found in GSE103229. Among
them, 12 differentially expressed miRNAs were observed to overlap
between these two datasets. OncomiR is an online resource for
identifying aberrantly expressed miRNAs in cancer. In the present
study, OncomiR was used to examine the predictive value of the
differentially expressed miRNAs in the overall survival rate of
patients with glioma. Kaplan-Meier survival curves were generated
and the survival analysis suggested that only three miRNAs were
significantly upregulated among the 12 differentially expressed
miRNAs, namely hsa-miR-7-5p, hsa-miR-21-3p and hsa-miR-455-3p. All
results indicated the potential involvement of miR-7-5p, miR-21-3p
and miR-455-3p in glioma. Regarding miR-7-5p, Liu et al
(23) have reported its
downregulation in GBM microvessels compared with normal brain
capillaries. In the same study, it was demonstrated that miR-7-5p
may function as a tumor suppressor by inhibiting vascular
endothelial cell proliferation through Raf1 targeting. Furthermore,
different expression of miR-7-5p has been detected in other types
of human cancer, including bladder cancer, breast cancer and
melanoma (24–26). These findings indicate the crucial
role of miR-7-5p in cancer development. miR-21-3p, the passenger
strand of pre-miR-21, has been widely reported to be aberrantly
expressed in various types of human cancer. Tseng et al
(27) suggested that miR-21-3p
expression is significantly upregulated in oral squamous cell
carcinoma tissues compared with corresponding adjacent normal
tissues. In glioma, miR-21-3p was proven to regulate glioma cell
proliferation and apoptosis by downregulating the expression of
PTEN protein (28).
Regarding miR-455-3p, it has been reported to serve
a role in acquired temozolomide resistance in patients with glioma
(29–31). A recent study suggested that
miR-455-3p was significantly increased in glioma cell lines and may
regulate tumor cell progression (32). However, its expression levels and
clinical significance have not been assessed in patients with
glioma. In the present study, the bioinformatics analysis results
suggested that the differential expression level of miR-455-3p in
glioma was the most significant. Subsequently, its expression in
patients with glioma was examined. Consistent with the
bioinformatics analysis results, the expression levels of
miR-455-3p were significantly upregulated in glioma tissues
compared with normal tissues. Based on malignancy degree, WHO has
divided gliomas into four grades: i) Pilocytic astrocytoma (WHO
grade I), ii) diffuse astrocytoma (WHO grade II), iii) anaplastic
astrocytoma (WHO grade III); and iv) GBM multiform (WHO grade IV)
(33). In the present study, the
histopathological WHO grades and KPS score, which are currently
used to predict prognosis in patients with glioma, were recorded
(34). It was observed that the
upregulation of miR-455-3p was closely associated with high WHO
grade and low KPS score, indicating the potential prognostic value
of miR-455-3p in glioma. As suggested by the bioinformatics
analysis, miR-455-3p may be used as a potential prognostic marker
in glioma. Moreover, the Kaplan-Meier survival curves and log-rank
test were also performed, and it was confirmed that miR-455-3p
served as an independent prognostic indicator for overall survival
and that high miR-455-3p expression levels were associated with
poor prognosis. The abnormal expression of miR-455-3p and its
prognostic value have been widely reported in numerous types of
human cancer (35–37). For example, Gao et al
(37) reported that miR-455-3p was
significantly downregulated in non-small cell lung cancer, while
low miR-455-3p expression was an unfavorable prognostic factor for
overall survival. Another study suggested that miR-455-3p
functioned as a tumor suppressor in esophageal squamous cell
carcinoma (ESCC) and inhibited cell proliferation and invasion by
targeting FAM83F. In addition, low miR-455-3p expression was
identified as an unfavorable prognostic factor for the overall
survival of patients with ESCC (38). Nevertheless, the functional role and
underlying mechanism of miR-455-3p in glioma was not examined in
the present study, thus further studies are required. Additionally,
certain limitations are included in the present study. Firstly, it
is generally accepted that higher tumor grade is associated with
poorer prognosis, however KPS revealed no significant association
with the prognosis of patients with glioma, although results
indicated that the difference was close to statistical
significance. This may be due to the sample size being relatively
small; a larger study would be required to confirm the present
results of the present study. Secondly, according to the
bioinformatics analysis, several differentially expressed miRNAs
were identified to be associated with glioma prognosis, though only
one major miRNA was investigated. Additional miRNAs may be
investigated in future studies. Lastly, the clinical and
pathological information included in the present study were
limited. For example, although the mutation status of isocitrate
dehydrogenase 1 and 1p19q, which may serve an important role in the
prognosis of patients with glioma, have attracted broad attention
during the last decades (32,39);
these were not examined in the present study. In future studies,
these clinical and pathological parameters should be included.
In conclusion, 12 differentially expressed miRNAs
were identified via integrative analysis of GEO data and OncomiR
analysis suggested that among them, three miRNAs were able to
accurately predict overall cancer survival in glioma. Furthermore,
miR-455-3p upregulation was validated in patients with glioma, and
it was indicated as a potential prognostic biomarker for glioma.
The present study identified a series of miRNAs with potential
functional roles in the pathogenesis of glioma, and provides
evidence for the use of miR-455-3p as a promising biomarker for
glioma prognosis.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science
Foundation of Fujian Province (grant nos. 2018Y0067 and 2017J0105),
the Startup Fund for scientific research of Fujian Medical
University (grant no. 2017XQ2049), 900th Hospital of the Joint
Logistics Team Innovation Team Fund (grant nos. 2014CXTD07 and
2018Z03) and the Fujian Science and Technology Plan Key Project
(grant no. 2014Y0036).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WW and SM conceived and designed the study, acquired
the data and drafted the manuscript. QZ was involved in drafting
the manuscript and data analysis. LX and SW were responsible for
data interpretation and critically revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures of the present study were reviewed
and approved by the Ethics Committee of The 900th Hospital of the
Joint Logistics Team. Written informed consent was obtained from
each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang D, Yuan Y, Zhang S, Zhao K, Li F, Ren
H, Zhang Z and Yu Y: Association between IL-13 Gene rs20541
polymorphism and glioma susceptibility: A meta-analysis. Oncol Res
Treat. 41:14–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bas Ayata H, Ceylan C, Kilic A, Guden M
and Engin K: Comparison of multiple treatment planning techniques
for high-grade glioma tumors near to critical organs. Oncol Res
Treat. 41:514–519. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hua S, Li H, Liu Y, Zhang J, Cheng Y and
Dai C: High expression of GALNT7 promotes invasion and
proliferation of glioma cells. Oncol Lett. 16:6307–6314.
2018.PubMed/NCBI
|
|
4
|
Su C, Li H and Gao W: TRIM28 is
overexpressed in glioma and associated with tumor progression. Onco
Targets Ther. 11:6447–6458. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai X, Deng Z, Guo H, Zhu X and Tu W: HP1α
is highly expressed in glioma cells and facilitates cell
proliferation and survival. Cancer Biomark. 20:453–460. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paul S: Integration of miRNA and mRNA
expression data for understanding etiology of gynecologic cancers.
Methods Mol Biol. 1912:323–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang S, Chen Y, Yu X, Lu Y, Wang H, Wu F
and Teng L: miR-129-5p attenuates cell proliferation and epithelial
mesenchymal transition via HMGB1 in gastric cancer. Pathol Res
Pract. 215:676–682. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang X, Liang M, Dittmar R and Wang L:
Extracellular microRNAs in urologic malignancies: Chances and
challenges. Int J Mol Sci. 14:14785–14799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang JF, Zhang JS, Zhao ZH, Yang PB, Ji
SF, Li N, Shi QD, Tan J, Xu X, Xu CB and Zhao LY: MicroRNA-770
affects proliferation and cell cycle transition by directly
targeting CDK8 in glioma. Cancer Cell Int. 18:1952018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu GF, Geng F, Xiao Z, Chen YS, Han Y, You
CY, Gong NL, Xie ZM and Pan M: MicroRNA-6807-3p promotes the
tumorigenesis of glioma by targeting downstream DACH1. Brain Res.
1708:47–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian W, Wu W, Li X, Rui X and Wu Y:
MiRNA-139-3p inhibits the proliferation, invasion, and migration of
human glioma cells by targeting MDA-9/syntenin. Biochem Biophys Res
Commun. 508:295–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gulluoglu S, Tuysuz EC, Sahin M, Kuskucu
A, Kaan Yaltirik C, Ture U, Kucukkaraduman B, Akbar MW, Gure AO,
Bayrak OF and Dalan AB: Simultaneous miRNA and mRNA transcriptome
profiling of glioblastoma samples reveals a novel set of OncomiR
candidates and their target genes. Brain Res. 1700:199–210. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang
H, Wan Z, Sheng F, Ma Y, Li W, et al: Differential expression of
circular RNAs in glioblastoma multiforme and its correlation with
prognosis. Transl Oncol. 10:271–279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wong NW, Chen Y, Chen S and Wang X:
OncomiR: An online resource for exploring pan-cancer microRNA
dysregulation. Bioinformatics. 34:713–715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Q, Wang C, Hou Z, Wang G, Lv J, Wang
H, Yang J, Zhang Z and Zhang H: Serum microRNA-376 family as
diagnostic and prognostic markers in human gliomas. Cancer Biomark.
19:137–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Chen X, Sun L, Bi X, He H, Chen L
and Pang J: The function of MMP-28/TGF-β induced cell apoptosis in
human glioma cells. Exp Ther Med. 16:2867–2874. 2018.PubMed/NCBI
|
|
18
|
Xu L, Yu QW, Fang SQ, Zheng YK and Qi JC:
MiR-650 inhibits the progression of glioma by targeting FAM83F. Eur
Rev Med Pharmacol Sci. 22:8391–8398. 2018.PubMed/NCBI
|
|
19
|
Hu S, Xu L, Li L, Luo D, Zhao H, Li D and
Peng B: Overexpression of lncRNA PTENP1 suppresses glioma cell
proliferation and metastasis in vitro. Onco Targets Ther.
12:147–156. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie C, Xu M, Lu D, Zhang W, Wang L, Wang
H, Li J, Ren F and Wang C: Candidate genes and microRNAs for glioma
pathogenesis and prognosis based on gene expression profiles. Mol
Med Rep. 18:2715–2723. 2018.PubMed/NCBI
|
|
21
|
Long T, Liu Z, Zhou X, Yu S, Tian H and
Bao Y: Identification of differentially expressed genes and
enriched pathways in lung cancer using bioinformatics analysis. Mol
Med Rep. 19:2029–2040. 2019.PubMed/NCBI
|
|
22
|
Rahman MR, Islam T, Gov E, Turanli B,
Gulfidan G, Shahjaman M, Banu NA, Mollah MNH, Arga KY and Moni MA:
Identification of prognostic biomarker signatures and candidate
drugs in colorectal cancer: Insights from systems biology Analysis.
Medicina (Kaunas). 55(pii): E202019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Z, Liu Y, Li L, Xu Z, Bi B, Wang Y and
Li JY: MiR-7-5p is frequently downregulated in glioblastoma
microvasculature and inhibits vascular endothelial cell
proliferation by targeting RAF1. Tumour Biol. 35:10177–10184. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Qiu M, An Y, Huang J and Gong C:
miR-7-5p acts as a tumor suppressor in bladder cancer by regulating
the hedgehog pathway factor Gli3. Biochem Biophys Res Commun.
503:2101–2107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Block I, Burton M, Sorensen KP, Andersen
L, Larsen MJ, Bak M, Cold S, Thomassen M, Tan Q and Kruse TA:
Association of miR-548c-5p, miR-7-5p, miR-210-3p, miR-128-3p with
recurrence in systemically untreated breast cancer. Oncotarget.
9:9030–9042. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Giles KM, Brown RA, Epis MR, Kalinowski FC
and Leedman PJ: miRNA-7-5p inhibits melanoma cell migration and
invasion. Biochem Biophys Res Commun. 430:706–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tseng HH, Tseng YK, You JJ, Kang BH, Wang
TH, Yang CM, Chen HC, Liou HH, Liu PF, Ger LP and Tsai KW:
Next-generation sequencing for microRNA profiling: MicroRNA-21-3p
promotes oral cancer metastasis. Anticancer Res. 37:1059–1066.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li SJ, Zhou J, Zhang L, Xiang W, Hu Q, He
YY and Chen LG: The effect of miR-21 on SWOZ2 glioma cells and its
biological mechanism. J BUON. 22:468–473. 2017.PubMed/NCBI
|
|
29
|
Ujifuku K, Mitsutake N, Takakura S,
Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K,
Nagata I and Yamashita S: miR-195, miR-455-3p and miR-10a(*) are
implicated in acquired temozolomide resistance in glioblastoma
multiforme cells. Cancer Lett. 296:241–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mizoguchi M, Guan Y, Yoshimoto K, Hata N,
Amano T, Nakamizo A and Sasaki T: Clinical implications of
microRNAs in human glioblastoma. Front Oncol. 3:192013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Dutta A and Abounader R: The role
of microRNAs in glioma initiation and progression. Front Biosci
(Landmark Ed). 17:700–712. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han J, Xiong Y, Deng H, Zhou J, Peng L,
Xiang W, Ming Y and Chen L: MiR-455-3p regulates glioma cell
proliferation by targeting PAX6. Tropical J Pharmaceutical Res.
18:689–695. 2019.
|
|
33
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang W, Zhao W, Ge C, Li X, Yang X, Xiang
Y and Sun Z: Decreased let-7b is associated with poor prognosis in
glioma. Medicine (Baltimore). 98:e157842019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo J, Liu C, Wang W, Liu Y, He H, Chen C,
Xiang R and Luo Y: Identification of serum miR-1915-3p and
miR-455-3p as biomarkers for breast cancer. PLoS One.
13:e02007162018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR,
Ding Y, Liang JQ, Li TT and Zhao J: MiR-497-5p, miR-195-5p and
miR-455-3p function as tumor suppressors by targeting hTERT in
melanoma A375 cells. Cancer Manag Res. 10:989–1003. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao X, Zhao H, Diao C, Wang X, Xie Y, Liu
Y, Han J and Zhang M: miR-455-3p serves as prognostic factor and
regulates the proliferation and migration of non-small cell lung
cancer through targeting HOXB5. Biochem Biophys Res Commun.
495:1074–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang H, Wei YN, Zhou J, Hao TT and Liu XL:
MiR-455-3p acts as a prognostic marker and inhibits the
proliferation and invasion of esophageal squamous cell carcinoma by
targeting FAM83F. Eur Rev Med Pharmacol Sci. 21:3200–3206.
2017.PubMed/NCBI
|
|
39
|
Chen H, Judkins J, Thomas C, Wu M, Khoury
L, Benjamin CG, Pacione D, Golfinos JG, Kumthekar P, Ghamsari F, et
al: Mutant IDH1 and seizures in patients with glioma. Neurology.
88:1805–1813. 2017. View Article : Google Scholar : PubMed/NCBI
|