|
1
|
Zhou C: Lung cancer molecular epidemiology
in China: Recent trends. Transl Lung Cancer Res. 3:270–279.
2014.PubMed/NCBI
|
|
2
|
Stewart BW and Wild CP: World Cancer
Report 2014IARC; Lyon: 2014
|
|
3
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goodgame B, Viswanathan A, Miller CR, Gao
F, Meyers B, Battafarano RJ, Patterson A, Cooper J, Guthrie TJ,
Bradley J, et al: A clinical model to estimate recurrence risk in
resected stage I non-small cell lung cancer. Am J Clin Oncol.
31:22–28. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kerr KM, Galler JS, Hagen JA, Laird PW and
Laird-Offringa IA: The role of DNA methylation in the development
and progression of lung adenocarcinoma. Dis Markers. 23:5–30. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu XF, Zhu BS, Wu FM and Hu HB: DNA
methylation biomarkers for the occurrence of lung adenocarcinoma
from TCGA data mining. J Cell Physiol. 233:6777–6784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han L, Xu G, Xu C, Liu B and Liu D:
Potential prognostic biomarkers identified by DNA methylation
profiling analysis for patients with lung adenocarcinoma. Oncol
Lett. 15:3552–3557. 2018.PubMed/NCBI
|
|
10
|
Sandoval J, Mendez-Gonzalez J, Nadal E,
Chen G, Carmona FJ, Sayols S, Moran S, Heyn H, Vizoso M, Gomez A,
et al: A prognostic DNA methylation signature for stage I
non-small-cell lung cancer. J Clin Oncol. 31:4140–4147. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuo IY, Jen J, Hsu LH, Hsu HS, Lai WW and
Wang YC: A prognostic predictor panel with DNA methylation
biomarkers for early-stage lung adenocarcinoma in Asian and
Caucasian populations. J Biomed Sci. 23:582016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao J and Zhang S: A bayesian extension of
the hypergeometric test for functional enrichment analysis.
Biometrics. 70:84–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Eberly LE: Correlation and simple linear
regression. Methods Mol Biol. 404:143–164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
19
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goeman JJ: L1 penalized estimation in the
Cox proportional hazards model. Biom J. 52:70–84. 2010.PubMed/NCBI
|
|
21
|
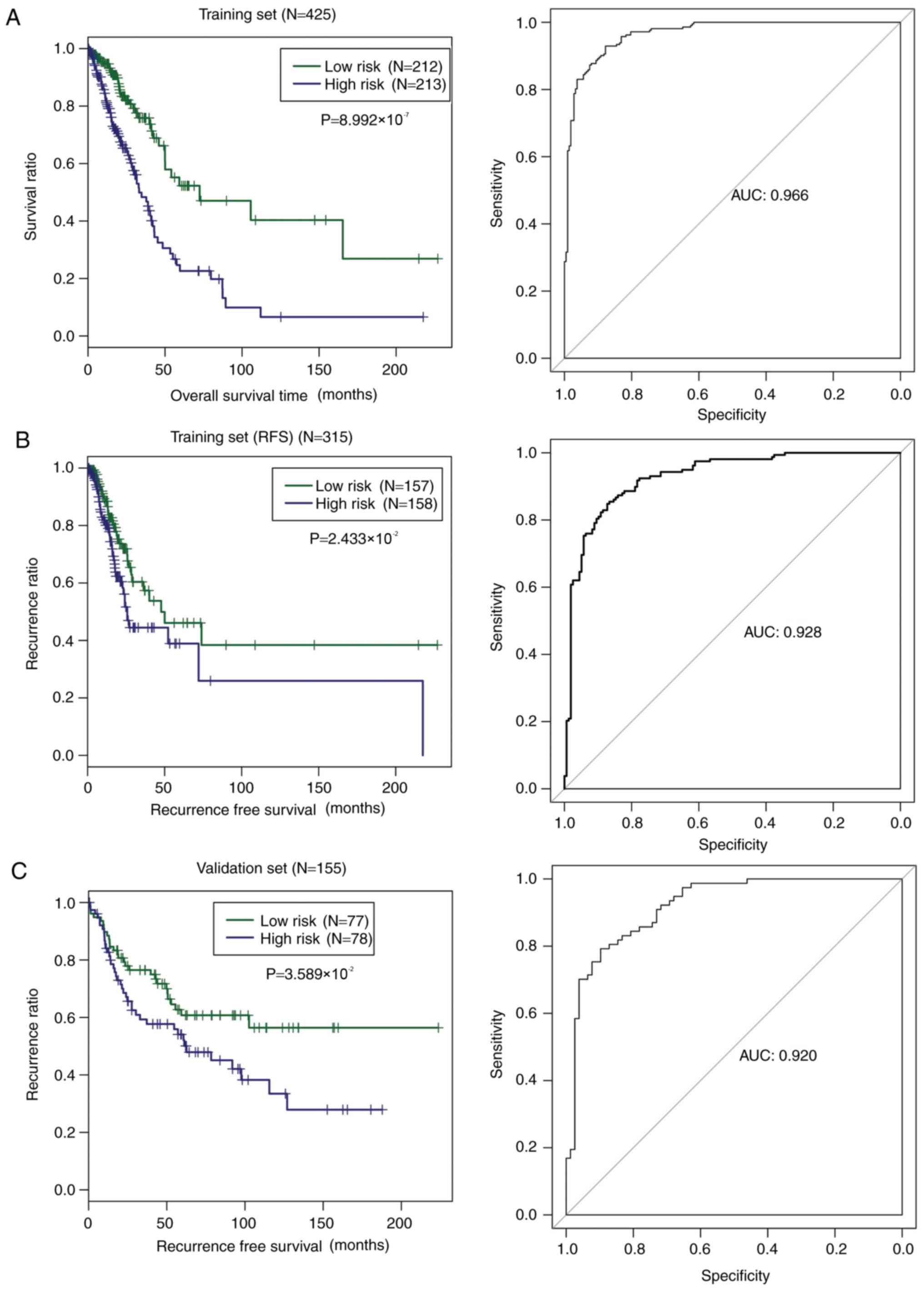
Goel MK, Khanna P and Kishore J:
Understanding survival analysis: Kaplan-Meier estimate. Int J
Ayurveda Res. 1:274–278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Greulich H: The genomics of lung
adenocarcinoma: Opportunities for targeted therapies. Genes Cancer.
1:1200–1210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Momparler RL and Bovenzi V: DNA
methylation and cancer. J Cell Physiol. 183:145–154. 2015.
View Article : Google Scholar
|
|
25
|
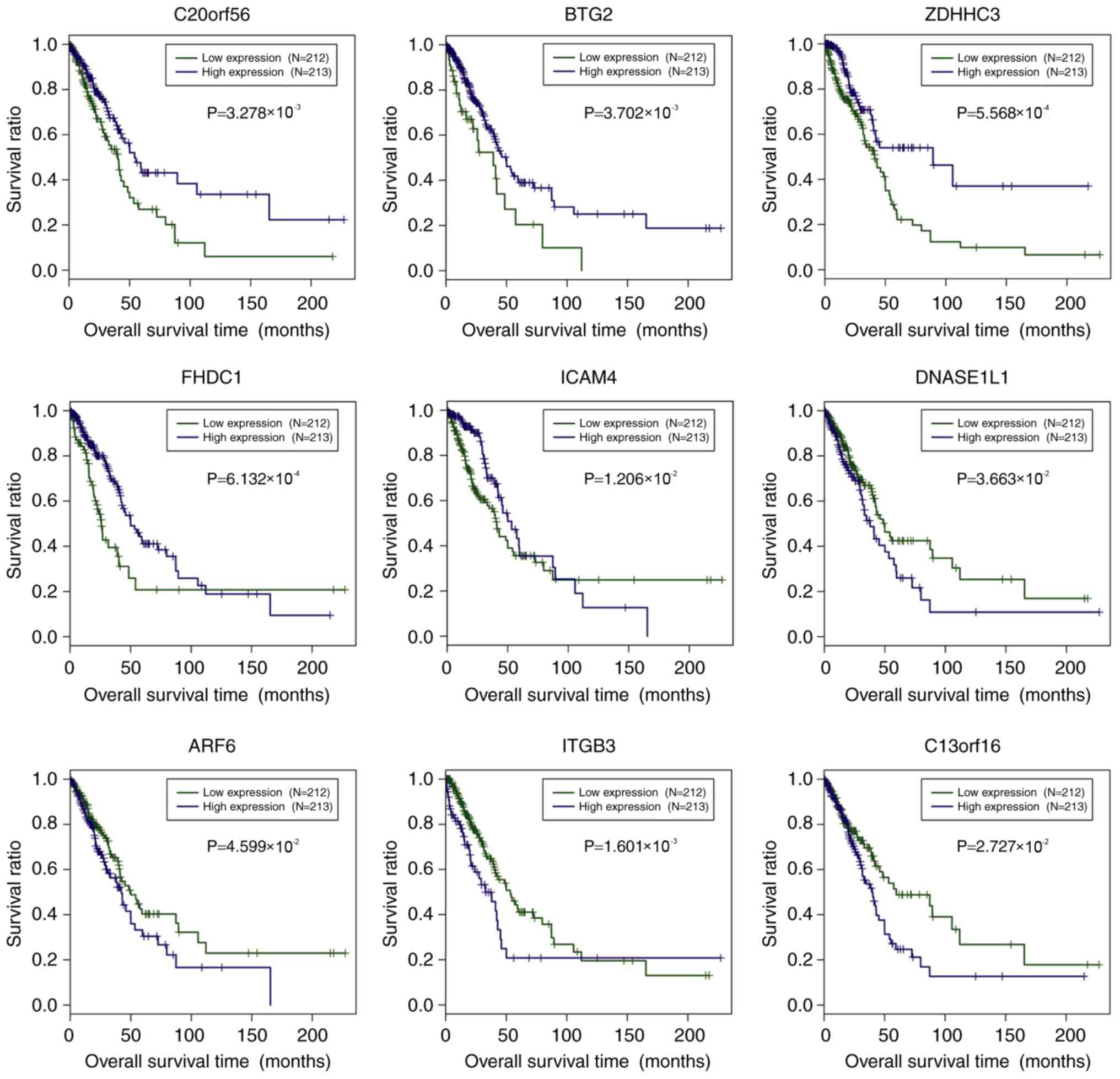
Mao B, Zhang Z and Wang G: BTG2: A rising
star of tumor suppressors (review). Int J Oncol. 46:459–464. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu M, Wu H, Liu T, Li Y, Wang F, Wan H,
Li X and Tang H: Regulation of the cell cycle gene, BTG2, by miR-21
in human laryngeal carcinoma. Cell Res. 19:828–837. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Struckmann K, Schraml P, Simon R,
Elmenhorst K, Mirlacher M, Kononen J and Moch H: Impaired
expression of the cell cycle regulator BTG2 is common in clear cell
renal cell carcinoma. Cancer Res. 64:1632–1638. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takahashi F, Chiba N, Tajima K, Hayashida
T, Shimada T, Takahashi M, Moriyama H, Brachtel E, Edelman EJ,
Ramaswamy S and Maheswaran S: Breast tumor progression induced by
loss of BTG2 expression is inhibited by targeted therapy with the
ErbB/HER inhibitor lapatinib. Oncogene. 30:3084–3095. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Huang H, Wu K, Wang M and Wu B:
Impact of BTG2 expression on proliferation and invasion of gastric
cancer cells in vitro. Mol Biol Rep. 37:2579–2586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei S, Hao C, Xin L, Zhao H, Chen J and
Zhou Q: Effects of BTG2 on proliferation inhibition and
anti-invasion in human lung cancer cells. Tumour Biol.
33:1223–1230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tague SE, Muralidharan V and
D'Souza-Schorey C: ADP-ribosylation factor 6 regulates tumor cell
invasion through the activation of the MEK/ERK signaling pathway.
Proc Natl Acad Sci USA. 101:9671–9676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Muralidharan-Chari V, Hoover H, Clancy J,
Schweitzer J, Suckow MA, Schroeder V, Castellino FJ, Schorey JS and
D'Souza-Schorey C: ADP-ribosylation factor 6 regulates tumorigenic
and invasive properties in vivo. Cancer Res. 69:2201–2209. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seguin L, Desgrosellier JS, Weis SM and
Cheresh DA: Integrins and cancer: Regulators of cancer stemness,
metastasis, and drug resistance. Trends Cell Biol. 25:234–240.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gahmberg CG, Tolvanen M and Kotovuori P:
Leukocyte adhesion-structure and function of human leukocyte
beta2-integrins and their cellular ligands. Eur J Biochem.
245:215–232. 1997. View Article : Google Scholar : PubMed/NCBI
|