Introduction
Multiple myeloma (MM) is a clonal plasma cell
malignancy that strongly depends on interactions with its
microenvironment (1). A major
complication of MM is the development of osteolytic lesions, which
are caused by an imbalance between osteoclastic bone resorption and
impaired osteoblastic bone formation and lead to severe bone pain,
fractures, osteoporosis and hypercalcemia (2). Mesenchymal stem cells (MSCs) are
involved in bone repair and regeneration as they can differentiate
into osteoblasts and osteocytes (3).
A previous study has indicated that the osteogenic differentiation
of MSCs obtained from patients with MM (MM-MSCs) is impaired
(4). Thus, an efficient strategy
that induces osteogenic differentiation of MM-MSCs is required to
improve the wellbeing of patients with MM.
MicroRNAs (miRNAs/miRs) are a large class of small,
noncoding RNA molecules of 17–25 nucleotides that are involved in
gene regulation at the post-transcriptional level by binding to the
3′-untranslated regions (UTRs) of target mRNAs and have been
demonstrated to serve critical roles in a number of biological
processes (5). Previous studies have
suggested that miRNAs contribute to bone development (6–8). Several
miRNAs have been identified as regulators of osteogenesis; for
instance, miR-133a-5p has been demonstrated to target runt-related
transcription factor 2 (RUNX2) and inhibit the expression of
osteoblast differentiation-associated markers (9). In addition, miR-20a promotes osteoblast
differentiation and bone formation of human MSCs by co-regulating
bone morphogenetic protein signaling (10). miR-203a is expressed in keratinocytes
and affects their growth, differentiation and function (11). Additionally, miR-203a has been
demonstrated to increase tumor growth in a number of types of
cancer (12,13). A recent study reported that
miR-203-3p inhibited osteogenesis in the jaws of diabetic rats and
in rat bone marrow mesenchymal stem cells cultured in high-glucose
medium by directly targeting mothers against decapentaplegic (Smad)
homolog 1 (Smad1) (14). However, to
the best of our knowledge, the role of miR-203a-3p.1 in the
osteogenic differentiation of MM-MSCs has not been identified.
Thus, the aim of the present study was to characterize the
expression of miR-203a-3p.1 in MM-MSCs and to investigate its
effects on osteoblast differentiation, as well as the potential
molecular mechanisms.
Materials and methods
Patients and subjects
A total of five patients with newly diagnosed stage
IIIA-IIIB of MM (age range, 40–63 years; 2 males, 3 females) and 5
normal healthy subjects (age range, 32–48 years; 3 males, 2
females) were recruited in the present study. The bone marrow
samples were obtained at The General Hospital of Western Theater
Command (Chengdu, China) from April 2017 to April 2018 according to
the institutional guidelines. The present study was approved by the
General Hospital of Western Theater Command (Chengdu, China). All
volunteers provided written informed consent.
Bone marrow (BM)-MSC isolation and
propagation
MSCs were isolated from BM samples. Briefly, the BM
fluid was mixed with an equal volume of Ficoll (Tianjin Haoyang
Biological Products Technology Co., Ltd.), and mononuclear cells
were obtained following centrifugation at 450 × g for 20 min at
room temperature. The cells were seeded in a T25 cell culture
bottle at 5,000 cells/cm2 with Dulbecco's modified
Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.). The medium was replaced twice weekly until the
cultures attained 80% confluence.
MSC identification by flow
cytometry
In the primary cells, CD44-PE, CD90-FITC and
CD105-PC5.5 were used to isolate MSCs. Briefly, MSCs were
resuspended in 4 ml PBS following digestion and centrifugation.
Subsequently, MSCs were incubated with 40 µl mouse anti-human
CD90-FITC (1:100), CD44-PE (1:100) and CD105-PerCP-Cy™5.5 (1:100)
for 30 min at 4°C. MSCs were isolated using a FACScalibur flow
cytometer (BD Biosciences). Following 4 culture passages, the MSCs
with very low fluorescence value (Blank) were sorted, and single
staining was used to identify MSCs. Following washing, trypsin
digestion and centrifugation, the sorted MSCs were resuspended in
100 µl PBS (1×106 cells) and stained with 5 µl mouse
anti-human CD90-FITC (1:20), CD44-PE (1:20), CD34-FITC (1:20),
CD105-PerCP-Cy™5.5 (1:20) and CD45-PE antibodies (1:20) for 30 min
at 4°C. After rinsing twice with PBS and resuspending in 500 µl of
PBS, the cells were analyzed using a FACScalibur flow cytometer (BD
Biosciences) and Cell Quest 3.3 software (BD Biosciences), based on
their characteristic immunophenotype of CD44+,
CD90+, CD105+, CD34− and
CD45−. The antibodies against CD45-PE (cat. no. 304008)
and CD34-FITC (cat. no. 343503) were purchased from BioLegend, Inc.
The BD Stemflow™ Human MSC Analysis kit (cat. no. 562245; BD
Biosciences) included CD90-FITC, CD105- PerCP-Cy™5.5 and CD44-PE
antibodies.
MSC differentiation
Following 3 passages, MSCs were seeded in 6-well
plates at 1×105 cells/well with DMEM supplemented with
10% FBS and cultured until cell confluence reached 60–80%. Fresh
DMEM containing 10% FBS, 1×10−8 mol/l dexamethasone, 10
mmol/l sodium β-glycerophosphate and 50 µg/ml vitamin C was used to
incubate the MSCs for 2 weeks (15)
and replaced twice a week to induce MSC differentiation into
osteoblasts (16).
Alizarin Red S staining
Following osteogenesis differentiation, MSCs were
washed with PBS and fixed in 4% paraformaldehyde for 20 min at room
temperature. MSCs were stained with Alizarin Red S (1%, pH 4.2;
Novon Scientific; Beijing Xinhua Luyuan Technology Co., Ltd.) for 5
min at room temperature. Images of the cells were captured using a
light microscope.
Cell transfection
MSCs were seeded into a 6-well plate at a density of
1×105 cells/well the day prior to transfection.
Transfection was performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol with 50 pmol/ml miR-203a-3p inhibitor or
mimic (Guangzhou RiboBio Co., Ltd.) at 37°C. Fresh medium was added
at 6 h. Cells were harvested for analysis following transfection
for 24 h.
For the transfection of Smad9, lentiviruses
overexpressing Smad9 (lv-Smad9; Smad9 mRNA sequence, NM_001127217;
vector name, GV492) and the corresponding control lentiviruses
(lv-green fluorescent protein) were purchased from Shanghai
GeneChem Co., Ltd. MSCs were seeded into a 6-well plate at a
density of 1×105 cells/well. Lentivirus
(1×108 TU/ml) infection was performed when the cells
reached 20–40% confluence. The lentiviruses were transfected into
MSCs with a multiplicity of infection of 30. After transfection for
8–12 h, fresh medium was added for further incubation. Cells were
collected for RT-qPCR or western blotting analysis 72 h after
transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the cells
(1×105 cells/well) using TRIzol® reagent
(Takara Biotechnology, Co., Ltd.). Following isolation, 4 µg RNA
was reverse-transcribed into cDNA using PrimeScript™ RT reagent kit
(Takara Biotechnology, Co., Ltd.) according to the manufacturer's
protocol at 37°C for 15 min and 85°C for 5 sec. qPCR was performed
on an Applied Biosystems 7500 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Premix Ex
Taq II (Takara Biotechnology, Co., Ltd.) in a 25 µl mixture
containing 2 µl cDNA templates, 12.5 µl 2X SYBR Premix Ex Taq II, 1
µl each primer and 8.5 µl DNase/RNase-free H2O. The
thermocycling conditions were as follows: 3 min at 95°C; 40 cycles
of 95°C for 5 sec and 60°C for 30 sec; followed by 72°C for 30 sec.
For the quantification of miRNA expression, RT was performed at
42°C for 60 min and 70°C for 10 min using Bulge-Loop™ miRNA RT-qPCR
Primer and Bulge-Loop™ miRNA RT-qPCR Starter kit (cat. no.
C10211-1; Guangzhou RiboBio Co., Ltd.). Gene expression levels were
quantified at 95°C for 10 min, followed by 40 cycles of 95°C for 2
sec, 60°C for 20 sec and 70°C for 10 sec. The 2−∆∆Cq
value was used for comparative quantitation (17). β-actin and U6 small nuclear RNA genes
were used as endogenous normalization controls. The primer
sequences (Sangon Biotech Co., Ltd.) are listed in Table I. The catalogue number of miR-203a-3p
primer was miRA0000264-1-200, and the catalogue number of U6 was
miRAN0002-1-200.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| ALP | F:
GACCTCCTCGGAAGACACTCTG |
|
| R:
CGCCTGGTAGTTGTTGTGAGC |
| OPN | F:
GCCGACCAAGGAAAACTCACT |
|
| R:
GGCACAGGTGATGCCTAGGA |
| OC | F:
CCAGGCGCTACCTGTATCAATG |
|
| R:
ATGTGGTCAGCCAACTCGTCA |
| Smad9 | F:
GCAGCCTCAAGGTCTTCAACAAC |
|
| R:
CATGAAGATGAATCTCAATCCAGGA |
| β-actin | F:
TGGCACCCAGCACAATGAA |
|
| R:
CTAAGTCATAGTCCGCCTAGAAGCA |
Western blot analysis
The MSCs from three normal healthy subjects were
randomly selected and seeded into 6-well plates at 1×105
cells/well after four culture passages. Total protein was extracted
using radioimmunoprecipitation assay lysis buffer (Wuhan Boster
Biological Technology, Ltd.). Protein concentration was quantified
using a BCA Protein Assay kit (Wuhan Boster Biological Technology,
Ltd.). The protein samples (20 µg) were separated by 10% SDS-PAGE
and transferred onto a polyvinylidene difluoride membrane (EMD
Millipore). Following blocking with 5% skimmed milk powder at 37°C
for 1 h, the membranes were incubated with primary antibodies
against Smad9 (cat. no. ab115900; 1:500; Abcam), Wnt3a (cat. no.
ab28472; 1:500; Abcam), β-catenin (cat. no. 8480; 1:1,000; Cell
Signaling Technology, Inc.), glycogen synthase kinase (GSK)-3β
(cat. no. 12456; 1:1,000; Cell Signaling Technology, Inc.) and
β-actin (cat. no. 4970; 1:1,000; Cell Signaling Technology, Inc.)
at 4°C overnight, followed by a HRP-conjugated goat anti-rabbit
immunoglobulin G secondary antibody (cat. no. BA1054; 1:5,000;
Wuhan Boster Biological Technology, Ltd.) at room temperature for 1
h. The protein bands were visualized using a ChemiDoc™ MP imaging
system (Bio-Rad Laboratories, Inc.). Protein levels were calculated
relative to β-actin.
Luciferase assay
miR-203a-3p.1 targets were predicted using
bioinformatics software, including TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org), DIANA TOOLS (http://diana.imis.athena-innovation.gr) and venny
2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
MSCs were plated in 24-well plates (4×104 cells/well).
When the cultures attained 50% confluence, cells were
co-transfected with the Renilla luciferase pRL-TK plasmid
(100 ng/ml; Shanghai GenePharma Co., Ltd.) plus the recombinant
Firefly luciferase pGL3 reporters containing the 3′-untranslated
region (3′-UTR) of human Smad9 (2 µg/ml; Shanghai GenePharma Co.,
Ltd.) in combination with miR-203a-3p.1 mimic and miR-203a-3p.1
inhibitor using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.). Luciferase activity was detected at 24 h using a
Dual-Luciferase Reporter Assay kit (cat. no. E1910; Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity for each tested well.
Statistical analysis
Statistical analysis was performed using SPSS 20.0
(IBM Corp.). The data are expressed as the mean ± standard
deviation. Comparisons between two groups were analyzed by unpaired
Student's t-test (for parametric data) or Mann-Whitney U test (for
non-parametric data). Differences among multiple groups were
compared by one-way analysis of variance (ANOVA) with Dunnett's
post hoc test or two-way ANOVA with Bonferroni's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference, and P<0.01 was considered to indicate a highly
significant difference.
Results
MSCs from patients with MM exhibit
decreased osteogenic differentiation
Following 7 days of primary culture, the adherent
cells exhibited colony growth and reached >40% confluence. The
cells were fusiform and pleomorphic. Following 14 days of primary
culture, the cells attained 60–70% confluence and had regular
morphology and a long spindle shape (Fig. 1A). After 4 culture passages, the cell
surface markers were detected by flow cytometry and the results
revealed that MSCs were negative for CD34 and CD45, but positive
for the CD44, CD90 and CD105 markers (Fig. 1B). These results suggested that the
cultured cells were MSCs.
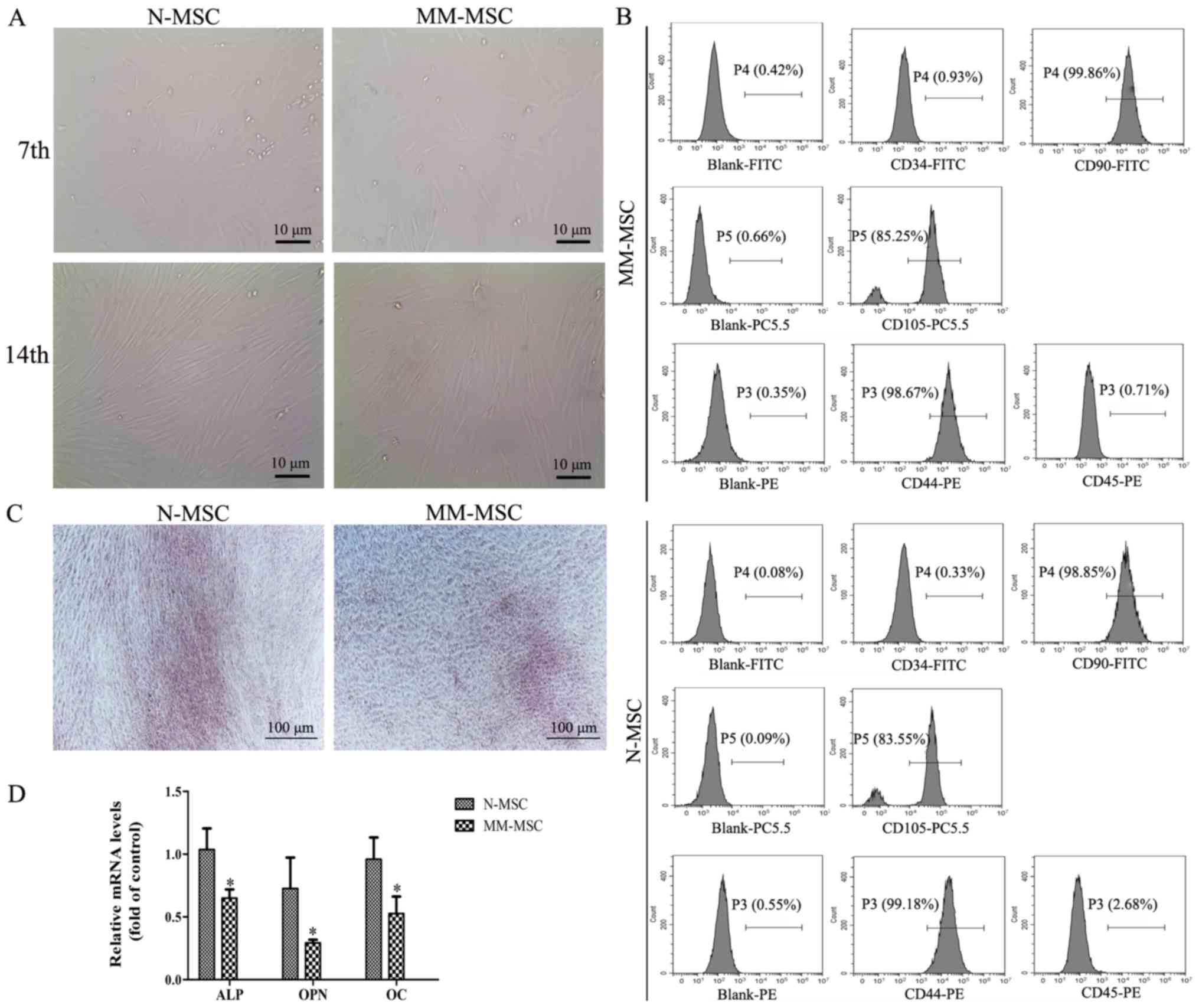 | Figure 1.MM-MSCs exhibit a reduced osteogenic
differentiation capacity. (A) Images of MSCs in primary culture on
the 7th and 14th day captured using an inverted microscope
(magnification, ×400). (B) The surface markers of the third
generation MM-MSCs and N-MSCs were identified by flow cytometry
Blank group, isolated MSCs from the primary cells at the fourth
generation with no fluorescence detected. (C) Following 21 days of
osteogenic induction, calcium deposition was evaluated using
Alizarin Red S staining (magnification, ×40). (D) Reverse
transcription-quantitative PCR was performed to detect the mRNA
levels of ALP, OPN and OC following osteogenic induction in N-MSCs
and MM-MSCs. *P<0.05 vs. N-MSC. MM, multiple myeloma; N, normal;
MSCs, mesenchymal stem cells; ALP, alkaline phosphatase; OPN,
osteopontin; OC, osteocalcin; CD, cluster of differentiation. |
The osteogenic differentiation capacity of MSCs from
patients with MM and normal subjects was investigated using
Alizarin Red S staining, which revealed that the calcium deposition
of MM-MSCs was lower compared with MSCs derived from normal healthy
subjects (N-MSCs) (Fig. 1C). RT-qPCR
results demonstrated an decrease in mRNA expression levels of
typical osteoblast differentiation markers in MM-MSCs compared with
N-MSCs, including alkaline phosphatase (ALP), osteopontin (OPN) and
osteocalcin (OC) (Fig. 1D). These
results indicated that the osteogenic differentiation capacity of
MM-MSCs may be reduced.
Effects of miR-203a-3p.1 on osteogenic
differentiation in MM-MSCs
To determine whether miR-203a-3p.1 is associated
with osteogenesis, the expression levels of miR-203a-3p.1 in MSCs
from patients with MM and normal subjects were analyzed by RT-qPCR.
The results revealed that the expression of miR-203a-3p.1 in
MM-MSCs was significantly lower compared with that in N-MSCs. In
addition, the expression of miR-203a-3p.1 in N-MSCs was
significantly decreased following osteoblast induction, whereas no
evident change was observed in MM-MSCs; therefore, it was
hypothesized that the decrease in the expression of miR-203a-3p.1
contributes to osteogenic differentiation in N-MSCs, whereas in
MM-MSCs, the decreased expression of miR-203a-3p.1 was inhibited,
which resulted in the reduced osteogenic differentiation capacity
of MM-MSCs. To study the role of miR-203a-3p.1 on osteogenic
differentiation, MM-MSCs cells were transfected with an
miR-203a-3p.1 mimic and inhibitor. RT-qPCR results revealed that
the miR-203a-3p.1 mRNA expression levels were significantly
increased in the mimic group and decreased in the inhibitor group
compared with the corresponding negative control groups (Fig. 2B). In N-MSCs transfected with the
miR-203a-3p.1 inhibitor, mRNA expression levels of ALP, OPN and OC
increased; however, the overexpression of miR-203a-3p.1 had no
significant effects on osteogenic differentiation marker expression
(Fig. 2C). These results indicate
that miR-203a-3p.1 inhibition may increase osteogenic
differentiation.
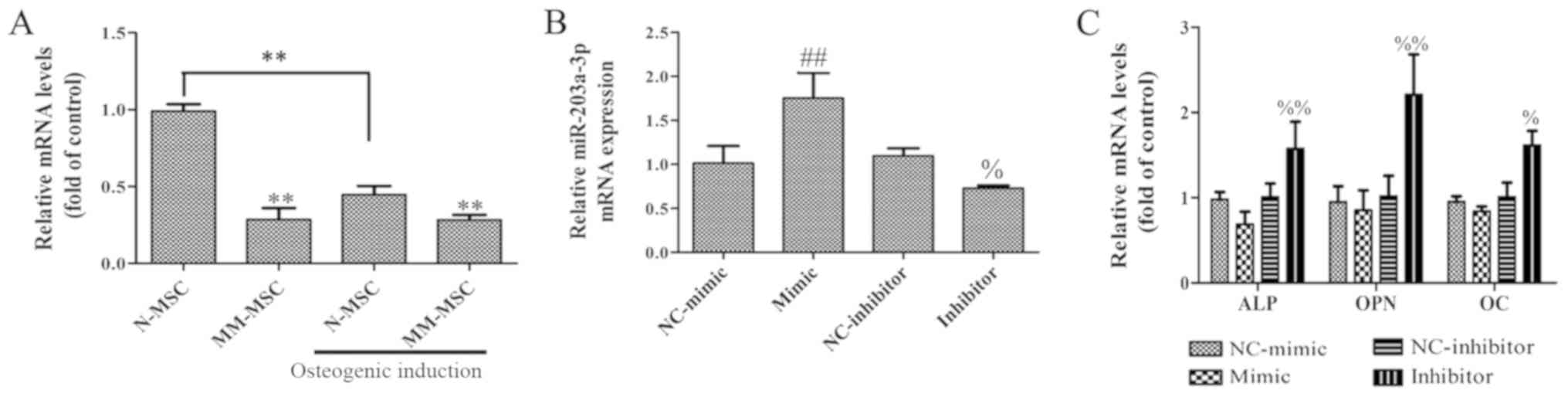 | Figure 2.Effects of miR-203a-3p.1 inhibition
on the osteogenic differentiation of N-MSCs. (A) The miR-203a-3p.1
mRNA expression levels in N-MSCs and MM-MSCs were detected by
RT-qPCR prior to and following osteoblast induction. (B) The
miR-203a-3p.1 mRNA expression levels in N-MSCs were detected by
RT-qPCR following transfection with miR-203a-3p.1 mimic or
inhibitor. (C) The mRNA expression levels of ALP, OPN and OC in
N-MSCs were detected by RT-qPCR following transfection with
miR-203a-3p.1 mimic or inhibitor. **P<0.01 vs. N-MSC;
##P<0.01 vs. NC-mimic; %P<0.05 and
%%P<0.01 vs. NC-inhibitor. miR, microRNA; MM,
multiple myeloma; N, normal; MSCs, mesenchymal stem cells; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; ALP,
alkaline phosphatase; OPN, osteopontin; OC, osteocalcin; NC,
negative control; mimic, miR-203a-3p.1-mimic; inhibitor,
miR-203a-3p.1-inhibitor. |
Smad9 is a target of
miR-203a-3p.1
The miRNA target prediction databases TargetScan,
miRDB, DIANA TOOLS and venny 2.1.0 were used to identify the target
genes of miR-203a-3p.1 in osteogenesis. A conserved putative target
site for miR-203a-3p.1 was identified in the 3′-UTR of the Smad9
gene (Fig. 3A). To assess whether
Smad9 may be regulated by miR-203a-3p.1, the N-MSCs were
transfected with miR-203a-3p.1 mimic and inhibitor, respectively.
The luciferase assay revealed that the miR-203a-3p.1 mimic
significantly repressed the luciferase activity and the
miR-203a-3p.1 inhibitor increased luciferase activity (Fig. 3B). RT-qPCR and western blotting
results demonstrated that the mRNA and protein levels of Smad9 were
reduced when miR-203a-3p.1 was overexpressed; by contrast, the
expression of Smad9 increased following treatment with
miR-203a-3p.1 inhibitor (Fig. 3C and
D). These results indicated that miR-203a-3p.1 may target Smad9
and negatively regulate Smad9 expression.
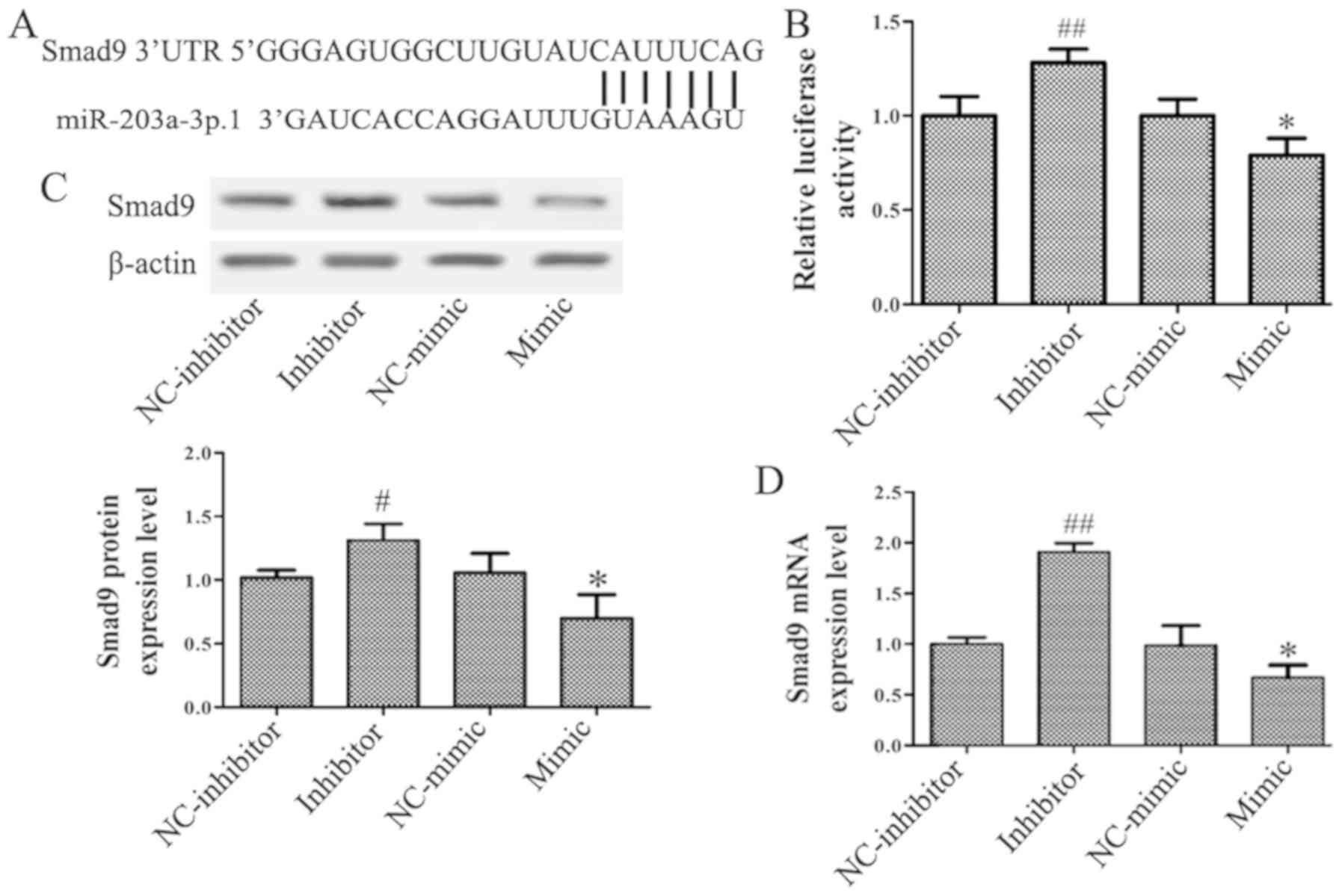 | Figure 3.miR-203a-3p.1 targets Smad9 in
N-MSCs. (A) Putative miR-203a-3p.1-binding sequence of Smad9. (B)
N-MSCs were co-transfected with miR-203a-3p.1 mimic or inhibitor
and the reporter plasmid containing the 3′-UTR of Smad9.
miR-203a-3p.1 inhibitor enhanced luciferase activity, while the
miR-203a-3p.1 mimic reduced luciferase activity. (C and D) Smad9
protein and mRNA expression levels were detected by western blot
and RT-qPCR following transfection with miR-203a-3p.1 inhibitor or
mimic, respectively. miR-203a-3p.1 inhibitor increased the
expression levels of Smad9 protein and mRNA, while miR-203a-3p.1
mimic decreased the expression levels of Smad9 protein and mRNA.
*P<0.05 vs. NC-mimic; #P<0.05 and
##P<0.01 vs. NC-inhibitor. miR, microRNA; MM,
multiple myeloma; MSCs, mesenchymal stem cells; UTR, untranslated
region; Smad9, mothers against decapentaplegic homolog 9; NC,
negative control; mimic, miR-203a-3p.1-mimic; inhibitor,
miR-203a-3p.1-inhibitor. |
Inhibition of miR-203a-3p.1 mediates
osteogenic differentiation through Smad9
To investigate the association between miR-203a-3p.1
and Smad9 during osteogenic differentiation, a lentiviral vector
overexpressing Smad9 was used. Western blotting results
demonstrated that Smad9 protein expression levels were
significantly increased in N-MSCs following lentiviral vector
transfection (Fig. 4A). Furthermore,
co-transfection of miR-203a-3p.1 inhibitor with lv-Smad9 increased
calcium deposition and mRNA expression levels of ALP, OPN and OC in
MM-MSCs (Fig. 4B and C).
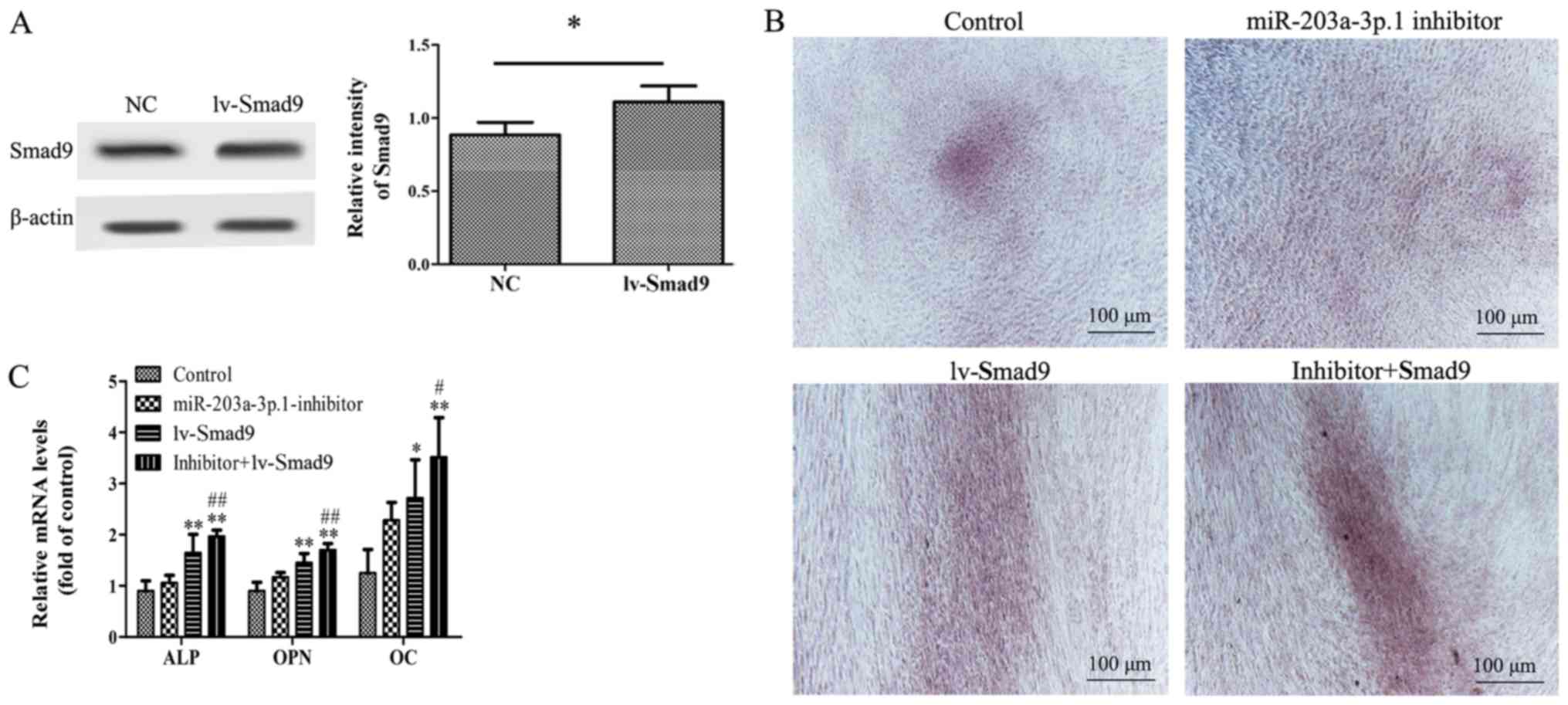 | Figure 4.Effects of Smad9 overexpression on
miR-203a-3p.1 inhibitor-mediated osteogenic differentiation in
N-MSCs. (A) Smad9 protein expression levels were detected by
western blot analysis following transfection with lentivirus. (B)
N-MSCs transfected with miR-203a-3p.1 inhibitors were infected with
lv-Smad9. Images of Alizarin Red S staining were captured using an
inverted microscope (magnification, ×40). Co-transfection of
miR-203a-3p.1 inhibitor with lv-Smad9 increased the calcium
deposition. (C) The ALP, OPN and OC mRNA expression levels were
detected by reverse transcription-quantitative polymerase chain
reaction following transfection with miR-203a-3p.1 inhibitor or
lentiviral vector. *P<0.05 and **P<0.01 vs. control group;
#P<0.05 and ##P<0.01 vs. miR-203a-3p.1
inhibitor group. miR, microRNA; MM, multiple myeloma; MSCs,
mesenchymal stem cells; ALP, alkaline phosphatase; OPN,
osteopontin; OC, osteocalcin; Smad9, mothers against
decapentaplegic homolog 9; lv-, lentivirus; inhibitor:
miR-203a-3p.1-inhibitor. |
Thus, overexpression of Smad9 appeared to enhance
the miR-203a-3p.1-knockdown-mediated promotion of osteogenic
differentiation. These results indicated that the inhibition of
miR-203a-3p.1 increased osteogenic differentiation, in part via the
upregulation of Smad9 expression.
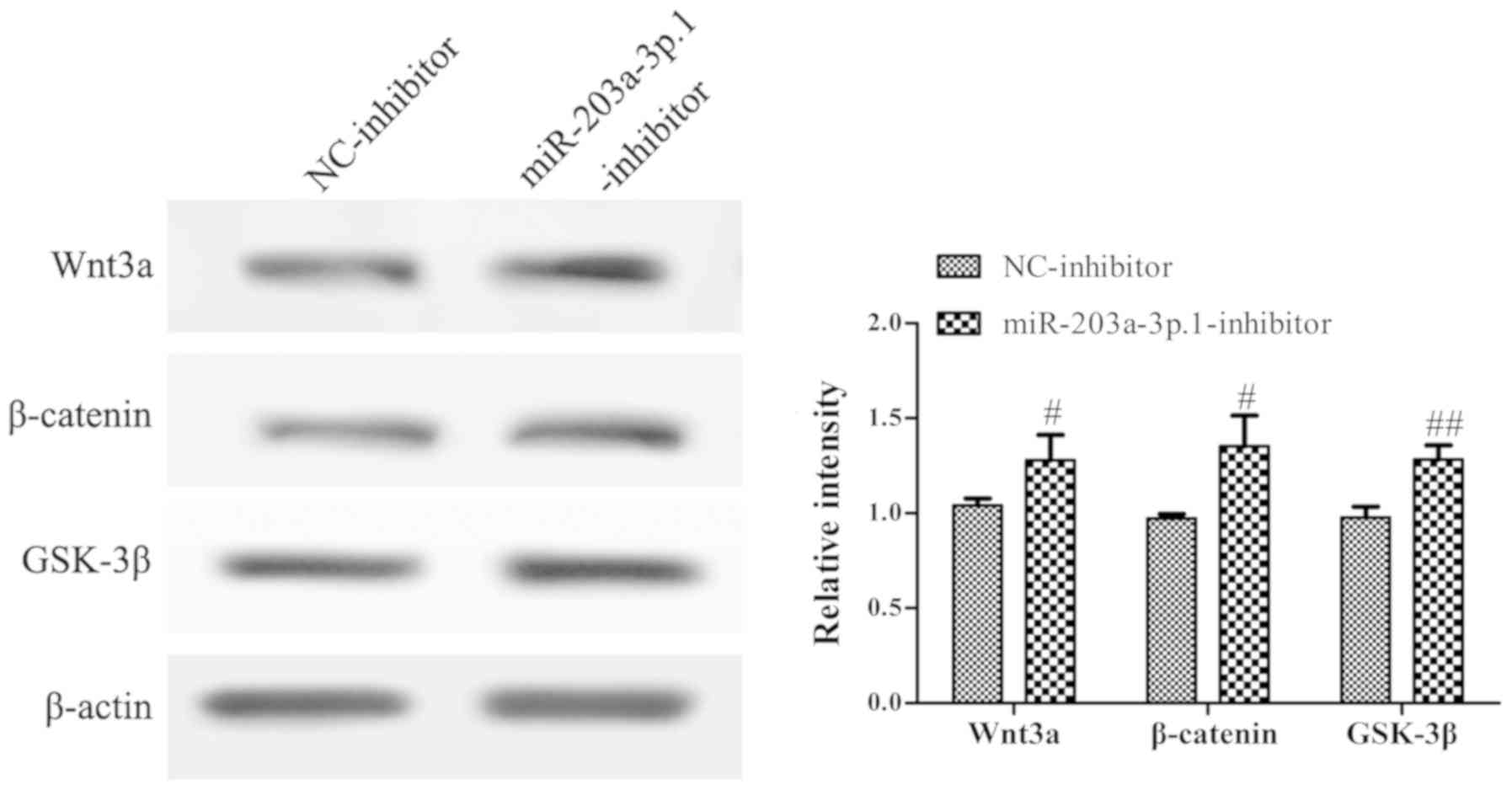
miR-203a-3p.1 may inhibit the
osteogenic differentiation of N-MSCs by inhibiting the
Wnt3a/β-catenin signaling pathway
To assess whether the impairment of osteogenic
differentiation in N-MSCs was due to abnormalities in the
Wnt3a/β-catenin signaling pathway, the protein expression levels of
genes involved in the Wnt3a/β-catenin signaling pathway were
analyzed by western blotting. The results revealed that the
miR-203a-3p.1 inhibitor significantly increased the expression of
Wnt3a, β-catenin and GSK-3β (Fig.
5).
Discussion
MM is a common malignancy characterized by the
abnormal proliferation of clonal plasma cells in BM (18). One of the characteristics of MM is
bone lesions due to a severe imbalance in bone remodeling (19). MSCs serve as a basic cellular unit of
embryonic bone formation, which has a key role in fracture repair
and bone regeneration (20).
However, the osteogenic differentiation of MSCs obtained from
patients with MM is impaired, which leads to decreased
osteogenesis, increased adipogenesis and osteonecrosis (4,21,22).
Maintaining a balance between the levels of osteoclasts and
osteoblasts regulates bone homeostasis (23). Osteoblasts promote bone formation by
secreting ALP and the bone matrix protein that induces bone matrix
mineralization (24). In the present
study, the osteogenic differentiation capacity of MM-MSCs was
significantly lower compared with that of N-MSCs, as indicated by
the decrease in calcium deposition and the mRNA expression levels
of the typical osteoblast differentiation markers, including ALP,
OPN and OC. These results demonstrated that the osteogenic
differentiation of MSCs was inhibited in patients with MM. Thus,
understanding the mechanisms leading to the decreased osteogenic
differentiation capability of MM-MSCs may explain the osteogenesis
defects in patients with MM. In further studies, more bone marrow
samples will be used to verify this result and further study the
mechanism underlying decreased osteogenic differentiation of MSCs
in patients with MM.
miRNAs have emerged as essential regulatory
molecules of gene expression that participate in the regulation of
bone homeostasis through transcriptional inhibition or mRNA
cleavage (25). The majority of
miRNAs have been described as signaling network nodes that serve a
vital role in osteoblastic differentiation processes; for instance,
miR-99a serves as a novel regulator of lysine demethylase 6B to
regulate the osteogenic differentiation of BM stromal cells
(26). In addition, miR-590-5p
promotes osteoblast differentiation by indirectly protecting and
stabilizing the Runx2 protein by targeting Smad7 gene expression in
MSCs (27). Tang et al
(14) demonstrated that miR-203-3p
participates in the suppression of diabetes-associated osteogenesis
in the jaw bone through targeting smad1. The results of the present
study revealed decreased expression levels of miR-203a-3p.1 in
MM-MSCs. Following osteoblast induction, the miR-203a-3p.1 mRNA
expression level in N-MSCs was significantly decreased, whereas no
change was observed in MM-MSCs. These results indicated that the
downregulation of miR-203a-3p.1 may contribute to the osteogenic
differentiation of normal MSCs. In agreement with these results,
the typical osteoblast differentiation markers ALP, OPN and OC were
upregulated in MM-MSCs following treatment with the miR-203a-3p.1
inhibitor. No changes were observed in ALP, OPN and OC mRNA levels
in MM-MSCs overexpressing miR-203a-3p.1. These results suggested
that the inhibition of miR-203a-3p.1 may increase osteoblast
differentiation of MM-MSCs.
Transforming growth factor (TGF)-β signaling is an
important pathway in osteoblastic differentiation. Through putative
target prediction, the present study identified that miR-203a-3p.1
may target Smad9, which is an important component of the TGF-β
signaling pathway. Previous studies have reported that several
SMADs, including Smad3, Smad7 and Smad5, are involved in bone
formation, remodeling and maintenance (27–29).
Smad9 is upregulated during chondrocyte differentiation (30), whereas its expression and role in
osteoclast differentiation has not been studied. In the present
study, a conserved putative target site for miR-203a-3p.1 was
identified in the 3′-UTR of Smad9. Furthermore, the RT-qPCR and
western blotting assays revealed a negative association between
Smad9 and miR-203a-3p.1. Luciferase reporter analysis indicated
that Smad9 may be a direct target of miR-203a-3p.1 in MM-MSCs.
Rescue experiments demonstrated that overexpression of Smad9
significantly enhanced the effect of the miR-203a-3p.1 inhibitor on
osteoblast marker expression, which indicated that inhibition of
miR-203a-3p.1 mediated promotion of osteogenic differentiation
partially by upregulating Smad9. In further studies, Smad9 gene
silencing will be used to validate the effects of Smad9
overexpression on osteoblast marker expression.
Wnt proteins are a large family of highly conserved
secreted signaling molecules that mediate essential biological
processes such as embryogenesis, organogenesis and tumorigenesis
(31). In addition, Wnt/β-catenin is
an important signaling pathway that regulates osteoblast
differentiation and bone formation. Chen et al (32) reported that the knockdown of
Sirtuin-7 increased the osteogenic differentiation of human BM-MSCs
by activating the Wnt/β-catenin signaling pathway. In addition,
baicalein promoted the osteogenic differentiation of human
periodontal ligament cells by activating the Wnt/β-catenin
signaling pathway (33). In the
present study, the partial inhibition of miR-203a-3p.1 increased
the protein expression levels of Wnt3a, β-catenin and GSK-3β in
N-MSCs. These results indicated that the miR-203a-3p.1 inhibitor
enhanced the osteoblast differentiation of MM-MSCs potentially by
activating the Wnt/β-catenin signaling pathway.
In conclusion, the present study demonstrated that
miR-203a-3p.1 was downregulated in MM-MSCs and may participate in
osteogenic differentiation. Furthermore, the inhibition of
miR-203a-3p.1 increased osteoblast differentiation by directly
targeting Smad9. The potential mechanism may be associated with the
activation of the Wnt3a/β-catenin signaling pathway. The present
study revealed a potential function of miR-203a-3p.1 in the
osteogenic differentiation of MM-MSCs, which may be targeted to
develop a promising therapeutic against myeloma bone disease in the
future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key
Research and Development Program of China (grant no.
2017YFA0105502), the Surface Project of National Natural Science
Foundation of China (grant no. 81570097), the Basic and Frontier
Research Project of Chongqing (grant no. cstc2015jcyjBX0077) and
the Science and Technology Innovation Special Project of Social
Undertakings and People's Livelihood Security of Chongqing (grant
no. cstc2016shms-ztzx10003).
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FYF, RD, YS and XZ conceived and designed the
experiments. FYF, RD, LQ, QW, YZ and LG performed the experiments.
CZ, PK, JZ, NZ and ZL analyzed the data and assisted with the
experiments. FYF, RD, YS and XZ wrote the manuscript. YS and XZ
revised the manuscript and supervised the study. All authors have
read and approved the final version of this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of General Hospital of Western Theater Command (Chengdu,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
López-Corral L, Gutiérrez NC, Vidriales
MB, Mateos MV, Rasillo A, García-Sanz R, Paiva B and San Miguel JF:
The progression from MGUS to smoldering myeloma and eventually to
multiple myeloma involves a clonal expansion of genetically
abnormal plasma cells. Clin Cancer Res. 17:1692–1700. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson DC, Weinhold N, Mitchell J, Chen
B, Stephens OW, Försti A, Nickel J, Kaiser M, Gregory WA, Cairns D,
et al: Genetic factors influencing the risk of multiple myeloma
bone disease. Leukemia. 30:883–888. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Robey PG, Kuznetsov SA, Ren J, Klein HG,
Sabatino M and Stroncek DF: Generation of clinical grade human bone
marrow stromal cells for use in bone regeneration. Bone. 70:87–92.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raje N and Roodman GD: Advances in the
biology and treatment of bone disease in multiple myeloma. Clin
Cancer Res. 17:1278–1286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen J, Qiu M, Dou C, Cao Z and Dong S:
MicroRNAs in bone balance and osteoporosis. Drug Dev Res.
76:235–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang S, Deng Y, Gu P and Fan X: MicroRNAs
regulate bone development and regeneration. Int J Mol Sci.
16:8227–8253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lian JB, Stein GS, van Wijnen AJ, Stein
JL, Hassan MQ, Gaur T and Zhang Y: MicroRNA control of bone
formation and homeostasis. Nat Rev Endocrinol. 8:212–227. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Wu Y, Shiozaki Y, Sugimoto Y,
Takigawa T, Tanaka M, Matsukawa A and Ozaki T: miRNA-133a-5p
inhibits the expression of osteoblast differentiation-associated
markers by targeting the 3′ UTR of RUNX2. DNA Cell Biol.
37:199–209. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan
G, Li G, Wang H, Lu G, Hu X, et al: MiRNA-20a promotes osteogenic
differentiation of human mesenchymal stem cells by co-regulating
BMP signaling. RNA Biol. 8:829–838. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma X, Li L, Jia T, Chen M, Liu G, Li C, Li
N and Yang D: miR-203a controls keratinocyte proliferation and
differentiation via targeting the stemness-associated factor ΔNp63
and establishing a regulatory circuit with SNAI2. Biochem Biophys
Res Commun. 491:241–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu G, Lai P, Liu M, Xu L, Guo Z, Liu H, Li
W, Wang G, Yao X, Zheng J and Xu Y: miR-203a regulates
proliferation, migration, and apoptosis by targeting glycogen
synthase kinase-3β in human renal cell carcinoma. Tumor Biol.
35:11443–11453. 2014. View Article : Google Scholar
|
|
13
|
Huo W, Du M, Pan X, Zhu X, Gao Y and Li Z:
miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma
cell growth and metastasis. FEBS Open Bio. 7:1085–1091. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Y, Zheng L, Zhou J, Chen Y, Yang L,
Deng F and Hu Y: miR-203-3p participates in the suppression of
diabetes-associated osteogenesis in the jaw bone through targeting
Smad1. Int J Mol Med. 41:1595–1607. 2018.PubMed/NCBI
|
|
15
|
Shi YY, Wang GL, Yang HL, Lu SZ, Zhang Y
and Cai X: Repairing rabbit femur bone defects by porous silk
fibroin/hydroxyapatite combined with adipose-derived stromal cells.
J Clin Rehabil Tissue Eng Res. 14:1341–1344. 2010.
|
|
16
|
Langenbach F and Handschel J: Effects of
dexamethasone, ascorbic acid and β-glycerophosphate on the
osteogenic differentiation of stem cells in vitro. Stem Cell Res
Ther. 4:1172013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo J, Fei C, Zhao Y, Zhao S, Zheng Q, Su
J, Wu D, Li X and Chang C: Lenalidomide restores the osteogenic
differentiation of bone marrow mesenchymal stem cells from multiple
myeloma patients via deactivating Notch signaling pathway.
Oncotarget. 121:55405–55421. 2017.
|
|
19
|
Yaccoby S: Advances in the understanding
of myeloma bone disease and tumour growth. Br J Haematol.
149:311–321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He X, Wang H, Jin T, Xu Y, Mei L and Yang
J: TLR4 activation promotes bone marrow MSC proliferation and
osteogenic differentiation via Wnt3a and Wnt5a signaling. PLoS One.
11:e01498762016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Giuliani N, Rizzoli V and Roodman GD:
Multiple myeloma bone disease: Pathophysiology of osteoblast
inhibition. Blood. 108:3992–3996. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhuang W, Ge X, Yang S, Huang M, Zhuang W,
Chen P, Zhang X, Fu J, Qu J and Li B: Upregulation of lncRNA MEG3
promotes osteogenic differentiation of mesenchymal stem cells from
multiple myeloma patients by targeting BMP4 transcription. Stem
Cells. 33:1985–1997. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tanaka Y, Nakayamada S and Okada Y:
Osteoblasts and osteoclasts in bone remodeling and inflammation.
Curr Drug Targets Inflamm Allergy. 4:325–328. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rodan GA and Martin TJ: Therapeutic
approaches to bone diseases. Science. 289:1508–1514. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pi C, Li YP, Zhou X and Gao B: The
expression and function of microRNAs in bone homeostasis. Front
Biosci (Landmark Ed). 20:119–138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Y, Zhang L, Tu T, Li Y, Murray D, Tu
Q and Chen JJ: MicroRNA-99a is a novel regulator of KDM6B-mediated
osteogenic differentiation of BMSCs. J Cell Mol Med. 22:2162–2176.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vishal M, Vimalraj S, Ajeetha R, Gokulnath
M, Keerthana R, He Z, Partridge NC and Selvamurugan N:
MicroRNA-590-5p stabilizes Runx2 by targeting Smad7 during
osteoblast differentiation. J Cell Physiol. 232:371–380. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao C, Yang S, Xu W, Shen JK, Ye S, Liu X,
Dong Z, Xiao B and Feng Y: MiR-708 promotes steroid-induced
osteonecrosis of femoral head, suppresses osteogenic
differentiation by targeting SMAD3. Sci Rep. 6:225992016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishibashi O, Ikegame M, Takizawa F,
Yoshizawa T, Moksed MA, Iizawa F, Mera H, Matsuda A and Kawashima
H: Endoglin is involved in BMP-2-induced osteogenic differentiation
of periodontal ligament cells through a pathway independent of
Smad-1/5/8 phosphorylation. J Cell Physiol. 222:465–473. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dexheimer V, Gabler J, Bomans K, Sims T,
Omlor G and Richter W: Differential expression of TGF-β superfamily
members and role of Smad1/5/9-signalling in chondral versus
endochondral chondrocyte differentiation. Sci Rep. 6:366552016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang H, Sun W, Ma J, Pan Y, Wang L and
Zhang WB: Biglycan mediates suture expansion osteogenesis via
potentiation of Wnt/β-catenin signaling. J Biomech. 48:432–440.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen EEM, Zhang W, Ye CCY, Gao X, Jiang
LLJ, Zhao TTF, Pan ZZJ and Xue DDT: Knockdown of SIRT7 enhances the
osteogenic differentiation of human bone marrow mesenchymal stem
cells partly via activation of the Wnt/b-catenin signaling pathway.
Cell Death Dis. 8:e30422017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen LJ, Hu BB, Shi XL, Ren MM, Yu WB, Cen
SD, Hu RD and Deng H: Baicalein enhances the osteogenic
differentiation of human periodontal ligament cells by activating
the Wnt/β-catenin signaling pathway. Arch Oral Biol. 78:100–108.
2017. View Article : Google Scholar : PubMed/NCBI
|