Introduction
Esophageal squamous cell carcinoma (ESCC), one of
the most common and deadly malignancies, is the eighth most common
cancer and the sixth most frequent cause of cancer-associated
mortality worldwide (1). The
incidence of ESCC in China is higher when compared with western
populations, with 477,900 cases reported and the incidence rate
accounted for 11.1% of all malignant tumors. The estimated number
of mortalities was 375,000, accounting for 13.3% of
cancer-associated mortalities in China in 2015 (2,3). Because
early symptoms of ESGG are not typical, numerous patients are at an
advanced stage at the time of diagnosis, and the majority of
patients succumb to the disease due to recurrence or metastasis
(4,5). This is corroborated by the 5-year
survival rate, which was only 10–25% in China in 2015 (4,5). The
precise molecular mechanisms underlying the occurrence, development
and lymphatic metastasis of ESCC remain to be fully elucidated. In
addition, there are few effective diagnostic markers and
therapeutic targets for patients with ESCC, which account in part
for its poor prognosis. Therefore, there is an urgent need for the
identification of biomarkers or therapeutic targets to improve the
clinical outcome of ESCC.
Circular RNAs (circRNAs) are endogenous non-coding
RNAs that regulate transcriptional and post-transcriptional gene
expression, similar to long non-coding RNAs (lncRNAs) (6). However, unlike lncRNAs, the unique
circular structure of circRNAs, without 5′ to 3′ polarity and
without a polyadenylated tail, can prevent degradation by RNA
exo-enzymes, which ensures good stability (7). The circRNA loop contains multiple
microRNA (miRNA) binding sites. The miRNA binds specifically to
corresponding miRNA response elements (MREs) on the circRNA
according to the principles of Watson-Crick base pairing, acting as
an intracellular miRNA sponge (8).
This reduces the extent of miRNA binding to the original target
gene, thereby increasing gene expression (6,8). The
sponge function of circRNA has been identified in numerous types of
cancer, including, lung (9,10), liver (11,12),
bladder (13), pancreatic cancer
(14,15) and skin cancer (16). However, only a few studies have
demonstrated the differential expression of circRNAs between ESCC
and healthy tissues (17,18), and the specific molecular mechanisms
governing this process remain unclear.
In a clinical setting, we found that patients
present with varying levels of lymphatic metastasis; even those
with early tumor invasion may exhibit advanced lymphatic
metastases. The present study investigated the expression of
circRNAs and the associations with lymphatic metastasis in ESCC. A
circRNA microarray analysis was performed to identify variations in
the expression of circRNAs in a range of cancerous tissues with
different levels of lymphatic metastases. The present study also
investigated potential capabilities through predicting the
interactions of circRNA/miRNAs.
Materials and methods
Patients and samples
A total of six patients aged from 50 to 74 years (5
men and 1 woman) with esophageal cancer and three healthy
volunteers aged from 45 to 64 years (1 men and 2 women) who
attended Fujian Medical University Union Hospital (Fuzhou, China)
between May 2016 and January 2017 were included in the present
study. The clinical information of these individuals is presented
in Table I. The 6 patients underwent
minimally invasive esophagectomy and lymph node dissection, and
tumor pathological staging was determined according to the American
Joint Committee on tumor-node-metastasis staging criteria (7th
edition) (19). Healthy esophageal
epithelial tissue was obtained during an esophagoscopy from the 3
healthy volunteers with no recorded tumor complications. The
subjects were divided into three groups: Early tumor stage
associated with advanced nodal stage (T1N2-3M0; T1 group; n=3),
advanced tumor stage associated with early nodal stage (T3N0M0; T2
group; n=3), and the healthy volunteers as the control group (C
group; n=3).
 | Table I.Clinical characteristics of patients
with ESCC and healthy volunteers subjected to circRNA expression
profile chip assay. |
Table I.
Clinical characteristics of patients
with ESCC and healthy volunteers subjected to circRNA expression
profile chip assay.
| Sample no. | Group | Age, years | Sex | TNM stage |
|---|
| 1 | T1 | 71 | Male | T1N2M0 |
| 2 | T1 | 74 | Male | T1N2M0 |
| 3 | T1 | 50 | Male | T1N3M0 |
| 4 | T2 | 65 | Male | T3N0M0 |
| 5 | T2 | 65 | Male | T3N0M0 |
| 6 | T2 | 68 | Female | T3N0M0 |
| 7 | C | 64 | Female | N/A |
| 8 | C | 45 | Female | N/A |
| 9 | C | 52 | Male | N/A |
All tissues samples were snap-frozen in liquid
nitrogen immediately following resection, and were then transferred
to the Institute of Cardiothoracic Surgery of Fujian Medical
University Union Hospital (Fuzhou, China) and cryopreserved at
−80°C until use. Total RNA was extracted from the nine specimens
with TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Transformant 1.5% agarose gel electrophoresis was used to assess
RNA integrity and remove genomic DNA contamination. The purity and
concentration of total RNA were determined using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). All individuals provided written informed consent prior to
inclusion in the study. The study protocol was approved by the
Ethics Committee on Human Research of Fujian Medical University
(Fuzhou, China).
RNA labeling and hybridization
RNA marker and array hybridization was performed
using the Arraystar Super RNA Tag kit (Arraystar Inc.), according
to the manufacturer's protocol. Total RNA was treated with RNase R
and incubated at 37°C for 1 h (Epicentre; Illumina, Inc.) according
to the manufacturer's protocol to remove linear RNA and enrich
circRNA. Following the protocol of the Arraystar Super RNA Tag kit,
enriched circRNAs were amplified and transcribed into fluorescent
circRNA utilizing a random priming method. The labelled circRNAs
(pmol Cy3/µg cRNA) were purified using the TargetAmp 1-Round RNA
Amplification kit 103 (Epicentre; Illumina, Inc.) and the
concentration and radioactivity of the labeled circRNA were
determined using a NanoDrop ND-1000 spectrophotometer.
Subsequently, to cleave circRNA, 5 µl 10× blocker and 1 µl 25×
lysis buffer (Arraystar Super RNA Tag kit; Arraystar) were added
per 1 µg of sample, and the mixture was heated at 60°C for 30 min
until the labeled circRNA became fragmented. Next, 25 µl 2×
hybridization buffer (Arraystar Super RNA Tag kit; Arraystar) was
added to dilute the labeled circRNA samples. Finally, 50 ul labeled
circRNA hybridization solution was injected onto the microarray
slide and hybridized onto the Arraystar Human Circular RNA
Microarray (8_15k; Arraystar, Inc.). After incubation in a 65°C
Agilent hybridization oven (SureHyb Microarray Hybridization
Chamber; Agilent Technologies, Inc.) for 17 h, the hybrid arrays
were washed twice with wash buffer at room temperature for 5–10 sec
and fixed in the fixation buffer at 25°C for 10 min, then scanned
by a GenePix 4000B Microarray Scanner (Roche Diagnostics) to
generate expression differences in the circRNA expression profile
chip.
Microarray data analysis
Data generated from the circRNA microarray underwent
data summarization, quantile normalization and quality control
using GeneSpring software version 12.0 (Agilent Technologies,
Inc.). circRNA for which at least three out of six samples had
flags in ‘Present’ or ‘Marginal’ (‘All Targets Value’) were
selected for further data analysis. To compare differences in
circRNA expression profiles among different groups, the fold-change
was calculated between the groups for each circRNA. When comparing
the three groups for profile differences, the fold-change for each
circRNA was computed, and one-way ANOVA followed by Tukey's
post-hoc test were used to analyze statistical differences among
three groups (T1, T2 and C) using SPSS version 24 software (IBM
Corp.). P<0.05 and fold-change >1.5 indicated a statistically
significant difference. The acquired data were filtered and the
differentially expressed circRNAs were ranked based on the defined
P-value and fold-change parameters.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) validation of candidate
circRNAs
RT-qPCR was used to verify differentially expressed
circRNAs between the T1 and T2 groups. Total RNA was extracted from
tissues using the RNA Extraction Kit (Takara Bio, Inc.). cDNA was
generated from total RNA using 5X ALL-In-One RT MasterMix (Applied
Biosystems) according to the manufacturer's instructions. RT-qPCR
reactions were performed as follows: 25°C for 10 min, followed by
42°C for 50 min and 85°C for 5 min. The relative expression of
circRNA was then determined using a sequence-specific
oligonucleotide primer designed to generate a 200 bp sequence with
the ViiA 7 real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using Hieff™qPCR SYBR Green Master Mix (Shanghai
Yeasen Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The cycling parameters of the PCR reaction were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec
and 60°C for 60 sec. circRNA levels were normalized using β-actin
as an internal control. Data were calculated and run three times
using the 2−ΔΔCq method (20) and quantitative PCR. Primer sequences
are presented in Table II. The
differences between three groups (n=3) were analyzed by one-way
ANOVA followed by Tukey's post-hoc test.
 | Table II.Primers used for RT-qPCR analysis of
circRNA levels. |
Table II.
Primers used for RT-qPCR analysis of
circRNA levels.
|
| Primer sequence
(5′-3′) |
|
|
|---|
|
|
|
|
|
|---|
| Gene name | Forward | Reverse | Annealing
temperature, °C | Length, bp |
|---|
| β-actin |
GTGGCCGAGGACTTTGATTG |
CCTGTAACAACGCATCTCATATT | 60 | 73 |
|
hsa_circRNA_402458 |
GCACAGTCAGCCAGCCTAATC |
TTTTCTCGCACATCCGTTTG | 60 | 125 |
|
hsa_circRNA_067567 |
GGACTAGGCCCCAATTTAGTG |
TCGTGTTTTTACAACTTCCAGTG | 60 | 96 |
|
hsa_circRNA_100873 |
TGGCCATCCAGGAGATCAT |
GGGGAGGTTTCACACTTTATG | 60 | 122 |
|
hsa_circRNA_002554 |
TGCCAGTTAACAAATAAAATGGA |
CAGACTCCTGATGGACCACAAT | 60 | 96 |
Prediction of circRNA/miRNA
interactions
The TargetScan (http://www.targetscan.org/vert_72/) and miRanda
(http://www.microrna.org/microrna/home.do) databases
were used to facilitate the prediction of circRNA/microRNA
interactions, and differentially expressed circRNAs were annotated
using circRNA/miRNA interaction information.
Results
Differentially expressed circRNAs
based on microarray analysis
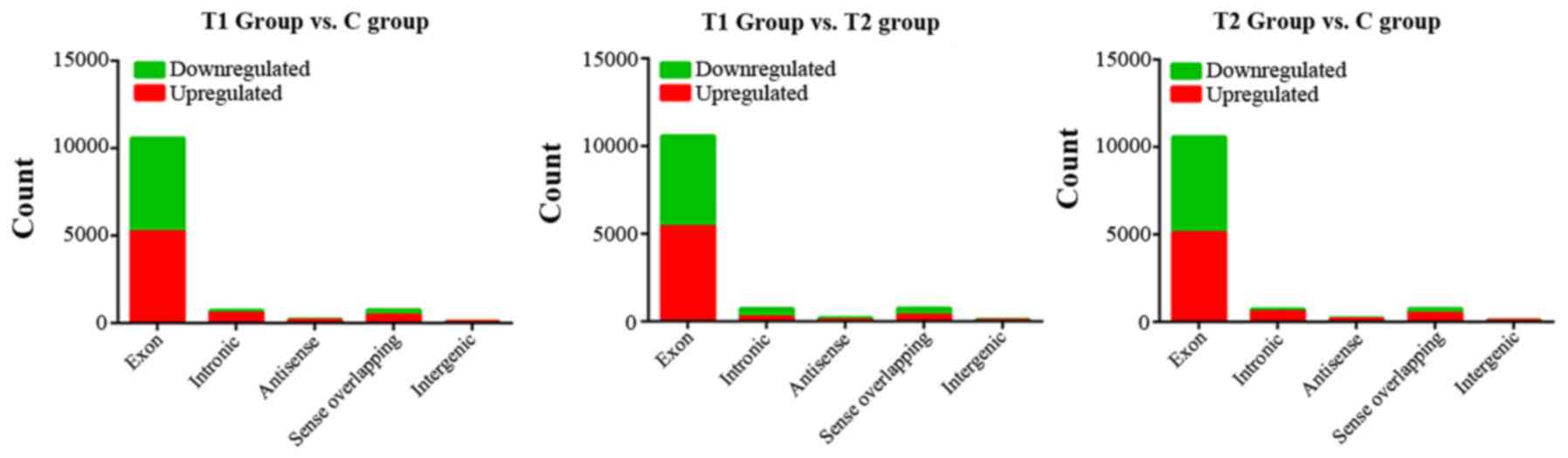
The present study detected a total of 12,275
circRNAs, including 861 circRNAs that were significantly
differentially expressed between groups. The distribution of sample
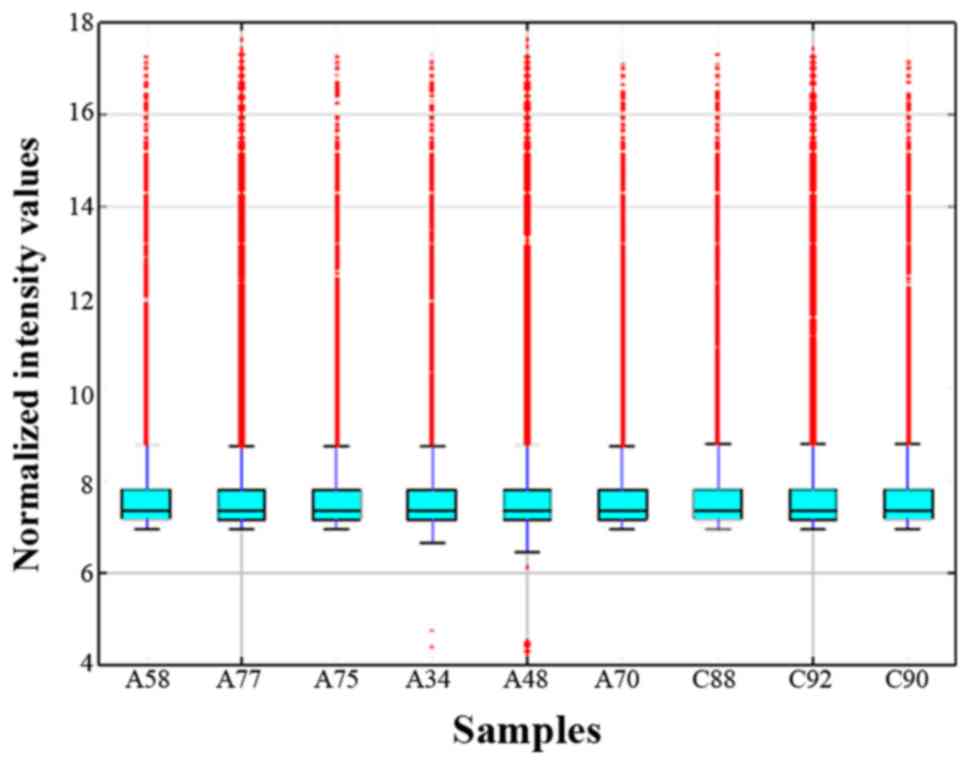
intensities was compared using a box plot, and following
standardization, the distribution of log2 ratios for circRNAs was
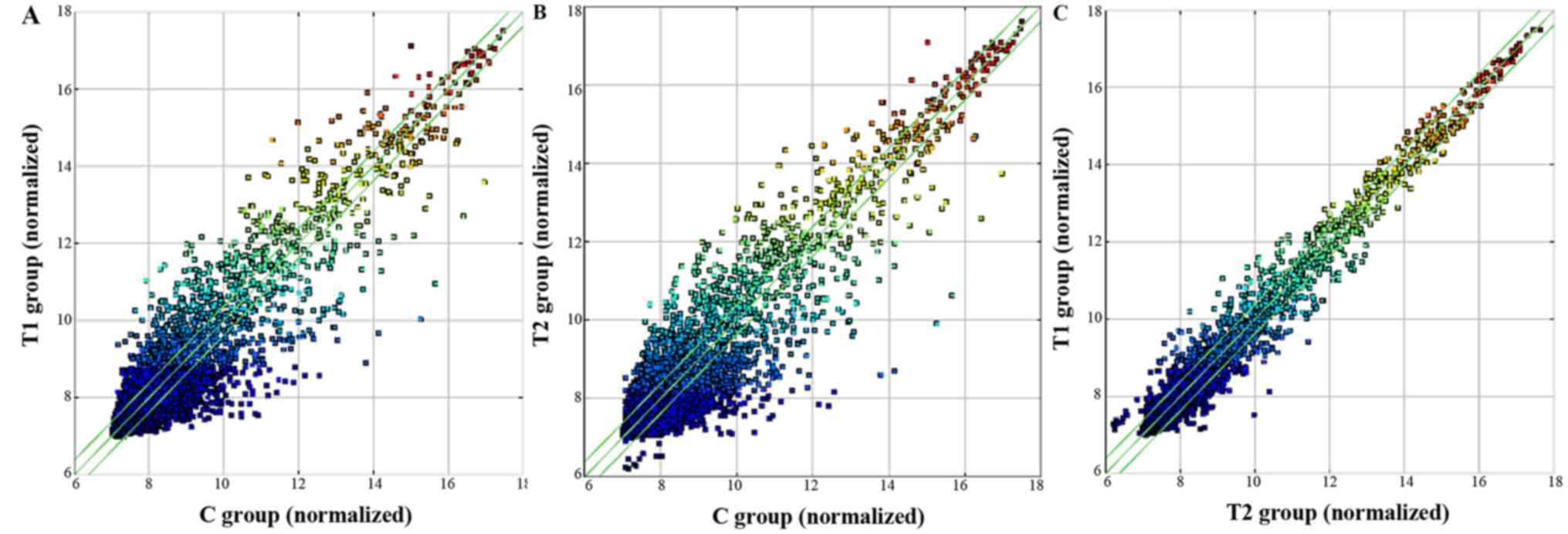
almost identical in all tested samples (Fig. 1). Scatter plots were used to present
differences in the expression of circRNAs among the T1, T2 and C
groups. The circRNAs above the top and below the bottom green lines
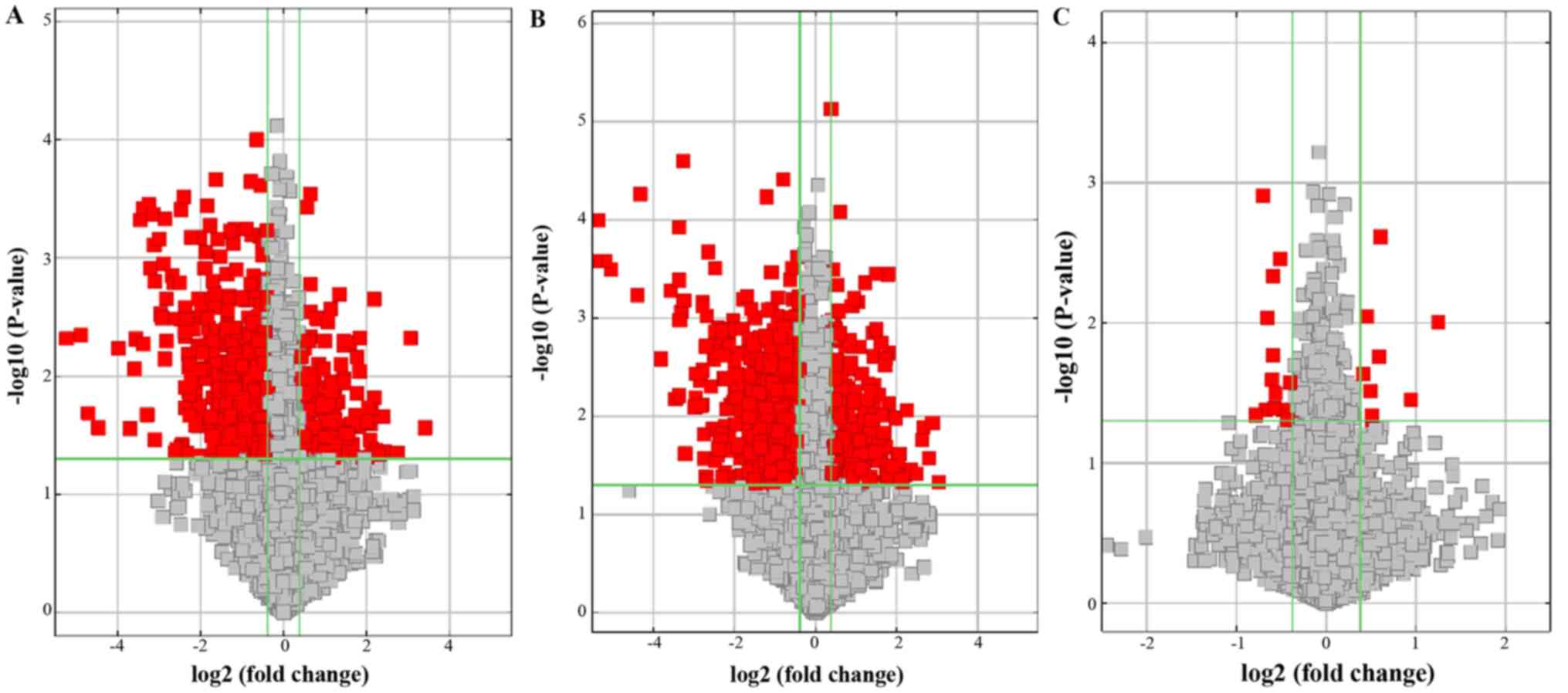
exhibited a fold-change >1.5 between the two groups (Fig. 2). Volcano plots were used to
visualize the differential expression of circRNAs, with red points
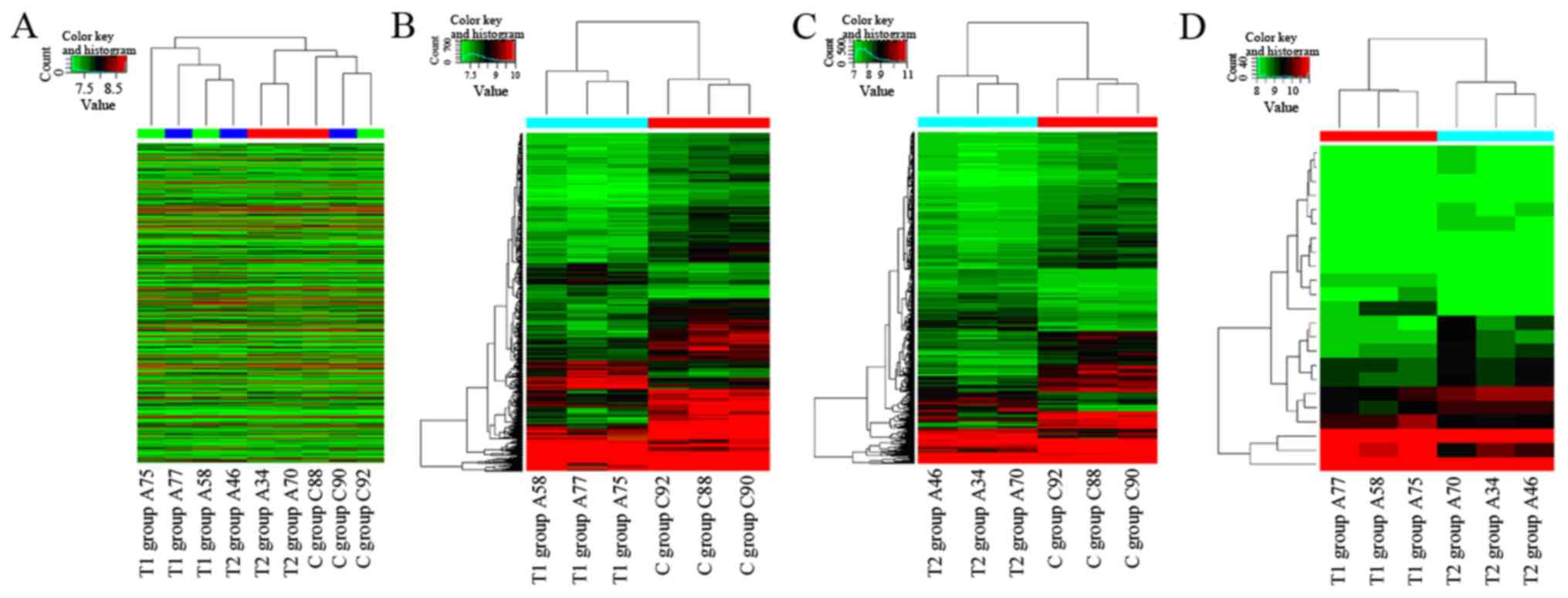
representing statistical significance (Fig. 3). The analysis of circRNA expression
by hierarchical clustering of heat maps aids hypotheses regarding
the association between samples. Hierarchical clustering
demonstrated that the circRNA expression profiles were
distinguishable in the samples (Fig.
4), and following integration with microarray data differences
were detected in the circRNA profiles among the three groups
(Figs. 5 and 6).
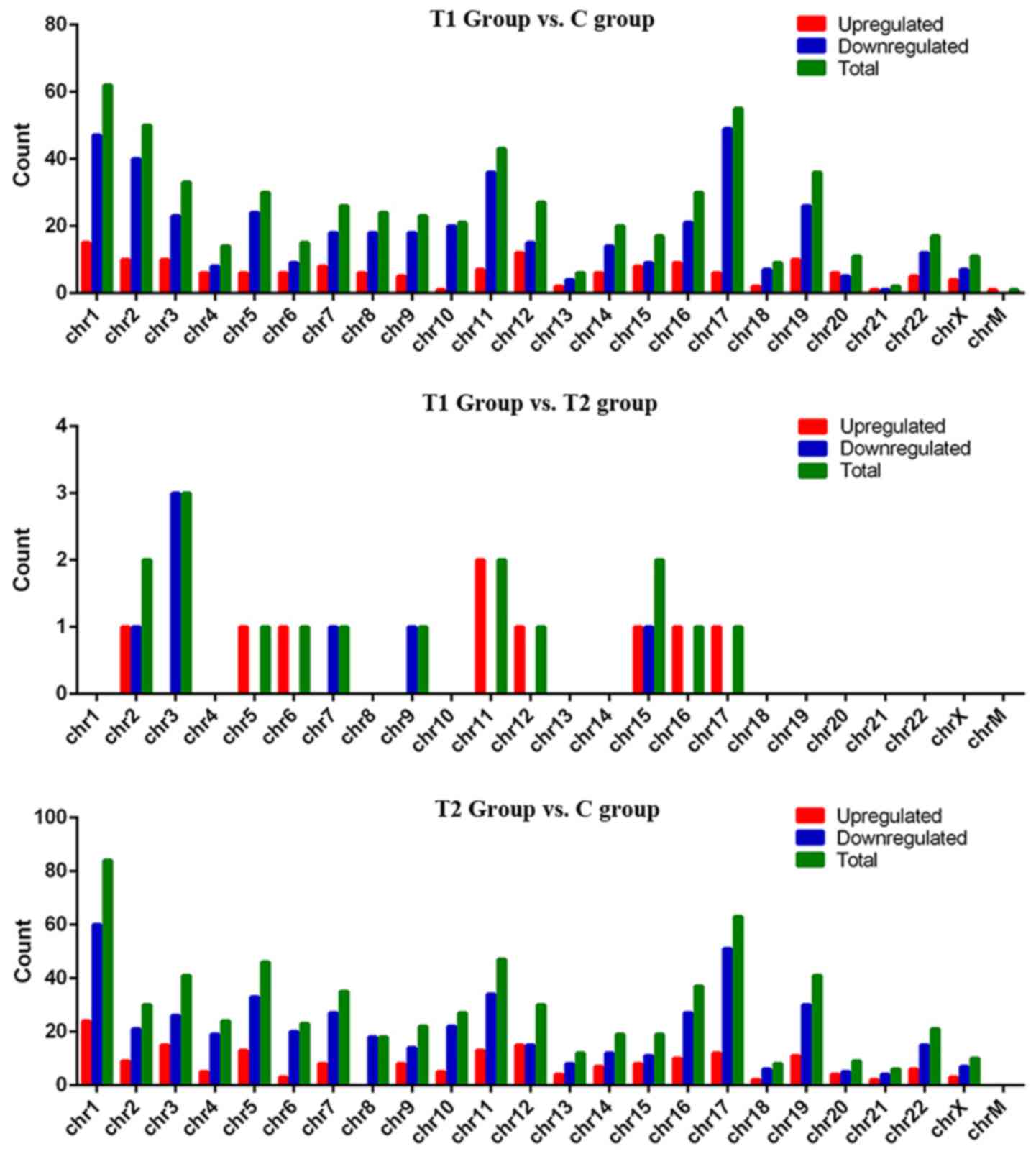
From the aforementioned analyses, circRNAs were
identified that were differentially expressed among the three
groups. Of these, there were 152 upregulated circRNAs and 431
downregulated circRNAs in the T1 group compared with the C group,
and 187 upregulated circRNAs and 481 downregulated circRNAs in the
T2 group compared with the C group (Fig.
4). Additionally, there were four upregulated circRNAs in the
T1 group compared with the T2 group. Two of these (circRNA_402458
and circRNA_100873) were also differentially expressed in the T1
group or T2 group compared with the C group. Seven circRNAs in the
T1 group were downregulated compared with the T2 group, of which
two (circRNA_002554 and circRNA_067567) were also differentially
expressed in the T1 group or T2 group compared with the C
group.
Validation of selected circRNAs using
RT-qPCR
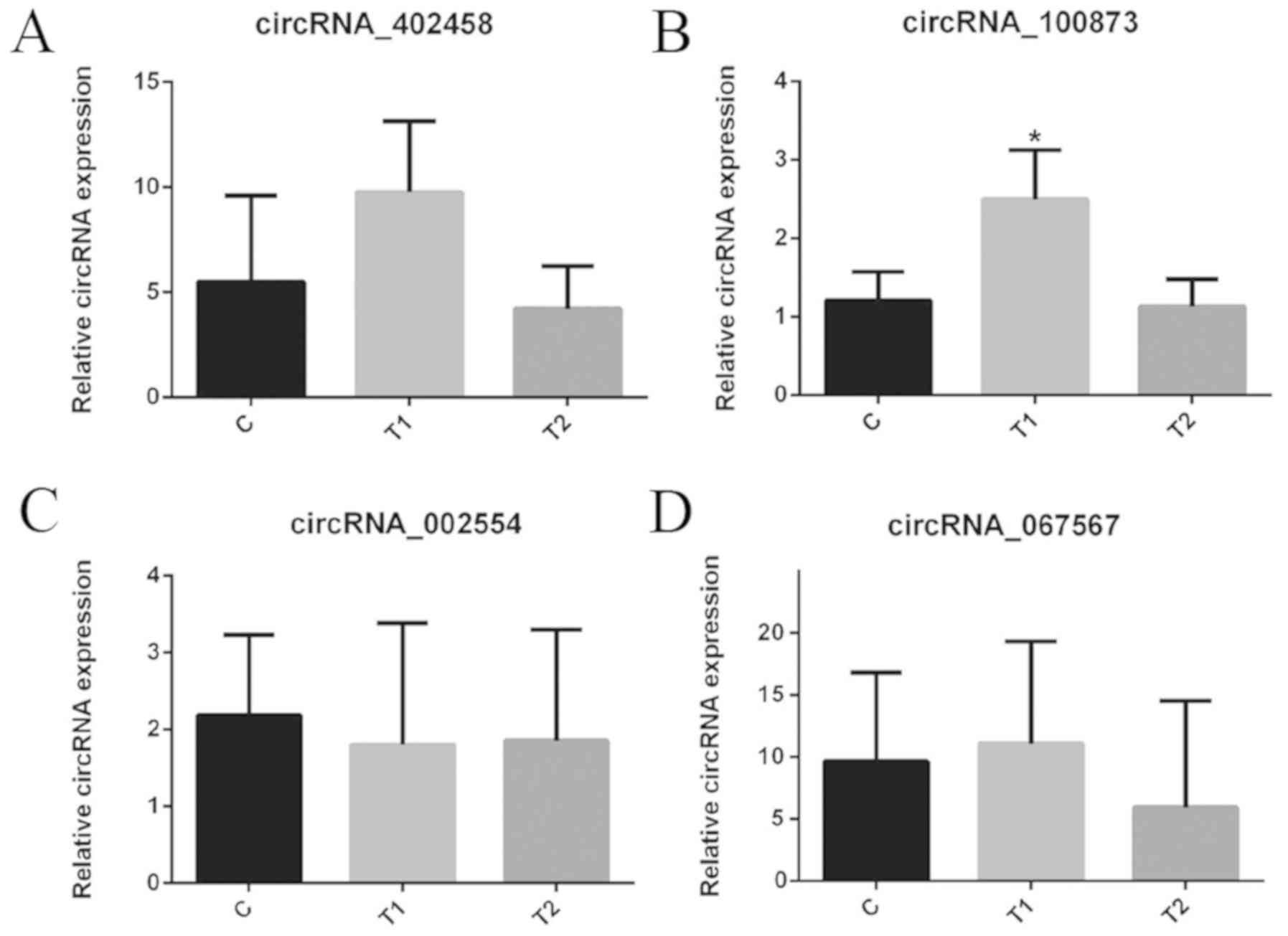
The present study selected the following circRNAs:
circRNA_402,458, circRNA_100873, circRNA_002554 and circRNA_067567,
which were differentially expressed in the T1 group compared with
the T2 group and in the T1/T2 group compared with the C group, to
verify the three groups of samples by RT-qPCR (Fig. 7). hsa_circRNA_100873 (Table III) demonstrated an expression
pattern consistent with microarray data analysis for the four
circRNAs. There was a significant difference in hsa_circRNA_100873
expression between tissues in the T1 group and T2/C groups
(Table IV).
 | Table III.Expression of has_circRNA_10087. |
Table III.
Expression of has_circRNA_10087.
| circRNA | P-value | FDR | Fold change | Regulation | Chrom | circRNA type | Strand | Best
transcript | Gene symbol |
|---|
|
hsa_circRNA_100873 | 0.035 | 0.999915 | 1.93 | Up | chr11 | Exonic | + | NM_003626 | PPFIA1 |
 | Table IV.Four circRNAs demonstrate
differential expression in the three groups. |
Table IV.
Four circRNAs demonstrate
differential expression in the three groups.
| Comparison |
hsa_circRNA_402458 |
hsa_circRNA_100873 |
hsa_circRNA_002554 |
hsa_circRNA_067567 |
|---|
| T1 vs. C | 1.790 | 2.120 | 0.930 | 1.030 |
| P-value | 0.382 | 0.037 | 0.989 | 0.996 |
| T2 vs. C | 0.760 | 0.940 | 0.960 | 0.880 |
| P-value | 0.903 | 0.981 | 0.996 | 0.919 |
| T1 vs. T2 | 2.350 | 2.270 | 0.970 | 1.170 |
| P-value | 0.227 | 0.030 | 0.998 | 0.882 |
Prediction of circRNA/miRNA
interactions
According to the competing endogenous RNA hypothesis
and previous reports, circRNAs can interact with specific miRNAs
through base complementation via MREs, in a function known as the
miRNA sponge (6,21–23).
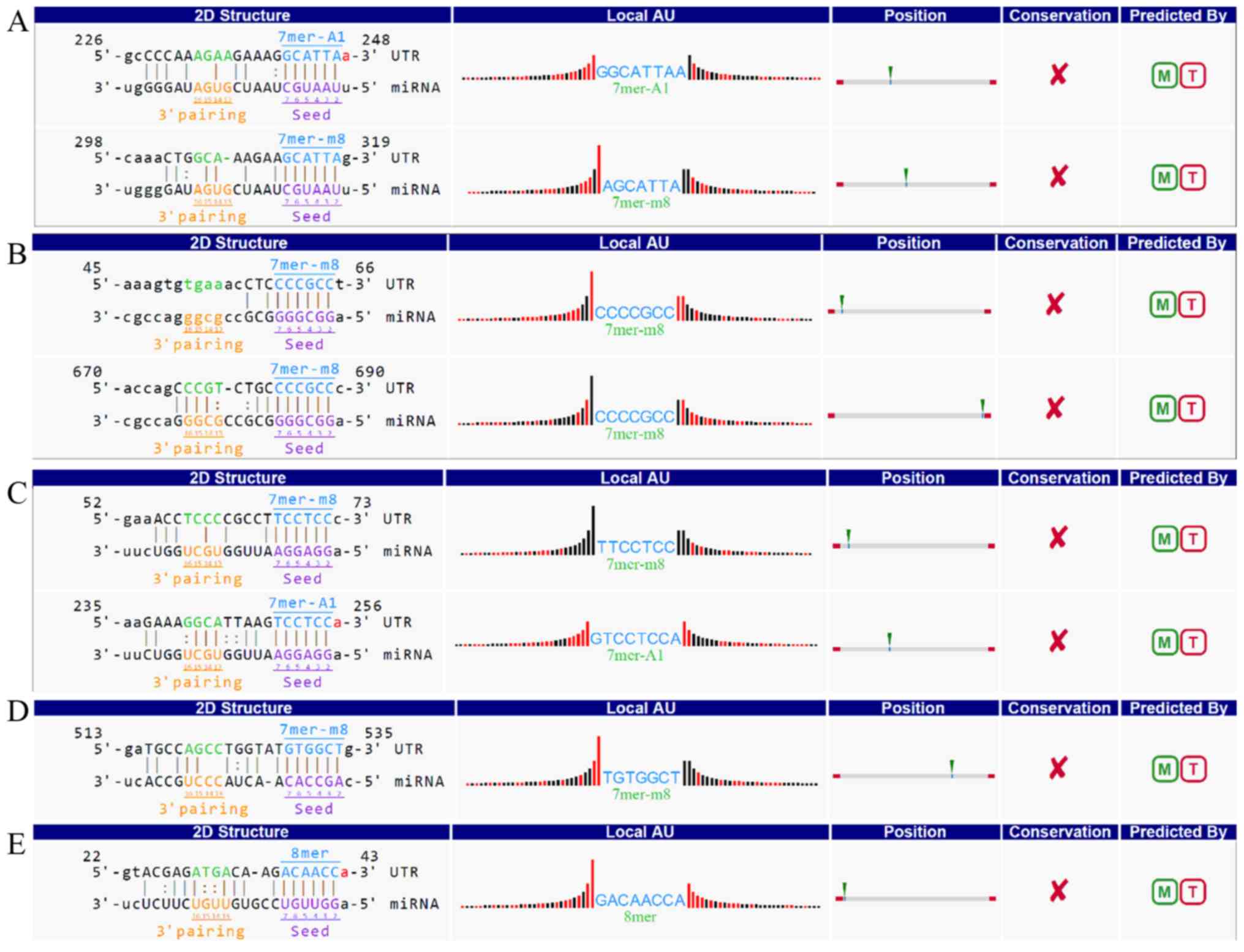
Therefore, the present study used an in-house miRNA target
prediction software from Arraystar to predict miRNAs that may bind
to the selected circRNAs. The top five miRNAs predicted to pair
with circRNA_100873 were identified as hsa-miR-1236-3p,
hsa-miR-3064-5p, hsa-miR-6504-5p, hsa-miR-943 and hsa-miR-522-3p
(Fig. 8).
Discussion
circRNAs exist in the cytoplasm of eukaryotic cells
and their contents are very similar to classical linear RNAs
(7). However, their circular
structure is more stable than linear structures; therefore, the
circular configuration is often adopted in the cell (7,8,24). Several studies have demonstrated that
circRNAs exhibit tissue specificity during normal tissue
differentiation and development, and have multiple regulatory
functions during the generation and progression of disease,
including the regulation of Wnt signaling pathways and
epithelial-mesenchymal transition (6,8,17,25). One
of the best known is the sponge function of circRNA discovered by
Hansen et al (6). This
involves circRNAs binding to specific miRNAs, which reduces the
miRNA content and weakens inhibition of the original target mRNA,
thus changing the expression of the corresponding protein (6).
Numerous examples of circRNA sponges have been
identified, including ciRS-7 (6),
sex determination region Y (SRY) (6,21),
hsacirc001569 (22), circPVT1
(23), circTCF25 (26) and cir-E3 ubiquitin ligase (ITCH; 17).
ciRS-7 has >70 valid MREs, including miR-7 (6,8,27,28),
while SRY was demonstrated to be the sponge of miR-138 (6,21). These
circRNAs participate in the development of specific tumorigenesis
by binding to specific miRNAs. Thus, circRNAs are promising targets
for the diagnosis and treatment of disease.
Few circRNAs have been reported in the field of
ESCC. Li et al (17) reported
that cir-ITCH is expressed at low levels in ESCC and promotes the
ubiquitination and degradation of phosphorylated Dv12 by increasing
ITCH expression through the sponge action of miR-7, miR-17 and
miR-214. In turn, cir-ITCH inhibits the Wnt/β-catenin pathway. Xia
et al (29) reported a
significant increase in the novel circRNA hsa_circ_0067934 in the
cytoplasm of esophageal cancer, and revealed that small interfering
RNA-mediated silencing of hsa_circ_0067934 inhibits the invasion
and migration of ESCC cells in vitro and blocks cell cycle
progression, indicating its potential as a novel biomarker and
therapeutic target. Su et al (18) performed a circRNA microarray and
bioinformatics analysis on the expression of circRNA in
radioresistant and non-radioresistant ESCC cells. They identified
RNAs with significantly different expression and proposed that
>400 target genes were enriched in the Wnt signaling pathway. Of
these, circRNA_001059 and circRNA_000167 were the two largest nodes
in the circRNA/miRNA co-expression network. Sang et al
(30) reported significant
upregulation of ciRS-7 in ESCC, which was associated with
significant increases in the proliferation, migration and invasion
of ESCC cells. High-throughput experiments identified 19 miR-876-5p
binding sites in ciRS-7, suggesting it is a sponge for miR-876-5p.
In addition, upregulating ciRS-7 enhanced the growth and metastasis
of ESCC tumors by targeting the miR-876-5p/MAGE-A family axis. This
was further confirmed in animal studies (30). Thus, circRNAs serve an important role
in the development of ESCC.
We have previously detected differences, albeit
insignificant, in circRNA expression between different N stage
tumor tissues with the same T stage. Therefore, the present study
examined factors that influence metastasis and tumor development.
T1 was defined as a group of early tumor stage associated with
advanced nodal stage with extremely high metastatic ability, and a
T2 group of advanced tumor stage associated with early nodal stage
with relatively lower metastatic ability. The present study
investigated circRNA expression profiles in ESCC by comparing
low-invasive high-lymphatic metastasis, high-invasive low-lymphatic
metastasis and healthy tissue to identify differential
hsa_circRNA_100873 expression in the T1/C and T1/T2 groups, as
verified by RT-qPCR. The circular RNA differential expression was
not only between tumor tissue and healthy tissue, but also between
high-lymphatic metastasis tissue and low-lymphatic metastasis
tissue, which may suggest a key function in tumor metastasis.
Further prediction of circRNA/miRNA interactions identified five
potential binding sites in has_circRNA_100873: hsa-miR-155-5p,
hsa-miR-663a, hsa-miR-766-5p, hsa-miR-449b-3p and
hsa-miR-494-5p.
Shi et al (31,32)
observed that hsa-miR-663a directly targets
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ
in glioblastoma and downregulated three critical downstream
effectors of phosphorylated AKT and PIK3D, including cyclin D1,
matrix metallopeptidase (MMP)2 and MMP7, which inhibit tumors. They
also identified that hsa-miR-663a functioned as a tumor suppressor
by downregulating C-X-C chemokine receptor type 4. Zang et
al (33) demonstrated that
hsa-miR-663a and eEF1A2 were negatively correlated with each other
in pancreatic cancer, and that hsa-miR-663a inhibited the invasion
and metastasis of pancreatic cancer cells in vitro and in
vivo by directly targeting eEF1A2. Ma et al (34) reported that epithelial membrane
protein 3 (EMP3) is a target gene of hsa-miR-663a in gallbladder
carcinoma, and that hsa-miR-663a overexpression downregulates EMP3
and activates mitogen-activated protein kinase/extracellular
signal-regulated kinase signaling, which directly affects cell
invasion and metastasis. Additionally, in liver cancer and
non-small cell lung cancer (35,36),
hsa-miR-663a inhibits tumor cell proliferation and metastasis by
targeting high mobility group AT-hook 2 and JunD, respectively. The
role of hsa-miR-663a in inhibiting tumor growth has also been
reported in gastric cancer (37),
papillary thyroid carcinoma (38)
and chronic myeloid leukemia (39).
Therefore, the present study hypothesizes that hsa-miR-663a is
likely to serve an important role in the invasion and metastasis of
esophageal cancer as a sponge of hsa_circRNA_100873.
hsa_circRNA_100873 is an exon-derived RNA that may downregulate
miRNA hsa-miR-663a through direct binding, thereby regulating
metastasis and invasion in ESCC cells.
In summary, the present study examined the
expression patterns of circRNAs between a low-invasive
high-lymphatic metastasis group, a high-invasive low-lymphatic
metastasis group and a healthy esophageal epithelial tissue group.
It was observed that, compared with healthy tissues,
hsa_circRNA_100873 was upregulated in cancer tissues and
differentially expressed in low-invasive high-lymphatic metastasis
and high-invasive low-lymphatic metastasis ESCC, confirming its
regulatory role in the invasion and metastasis of ESCC cells.
However, the present study has certain limitations. Future
experiments should verify the circRNA expression levels in a higher
number of pathological samples and conduct a series of associated
phenotypic and mechanism experiments. This circRNA,
hsa_circRNA_100873, may be a potential novel target for the
diagnosis and treatment of ESCC, although its specific molecular
biological mechanism requires further research.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Fujian Provincial Key Project (grant no. 2014Y0024), the Fujian
Provincial Joint Research Project of Health Care and Education
(grant no. WKJ2016-2-09) and the Science and Technology Innovation
Joint Fund Project (grant no. 2018Y9058).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CC, BZ and WZ undertook study conception and design,
and provided administrative support. BZ, ZW and SX provided the
study materials. ZW, SX and WW undertook the collection and
assembly of data. HC, SZ, TZ and GX performed data analysis and
interpretation. All authors undertook manuscript writing and gave
final approval of manuscript to be published.
Ethics approval and consent to
participate
All patients provided written informed consent for
participation, and the study protocol was approved by the Ethics
Committee on Human Research of Fujian Medical University (Fuzhou,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Jemal A, Grey N, Ferlay J and
Forman D: Global cancer transitions according to the human
development index (2008–2030): A population-based study. Lancet
Oncol. 13:790–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in china,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang W, Wei C, Li P, Wang L, Li W, Chen K,
Zhang J, Zhang W and Jiang G: Integrative analysis of mRNA and
lncRNA profiles identified pathogenetic lncRNAs in esophageal
squamous cell carcinoma. Gene. 661:169–175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mariette C, Balon JM, Piessen G, Fabre S,
Van Seuningen I and Triboulet JP: Pattern of recurrence following
complete resection of esophageal carcinoma and factors predictive
of recurrent disease. Cancer. 97:1616–1623. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu J, Shu Y, Xu T, Zhu W, Qiu T, Li J,
Zhang M, Xu J, Guo R, Lu K, et al: Microarray expression profiling
and bioinformatics analysis of circular RNA expression in lung
squamous cell carcinoma. Am J Transl Res. 10:771–783.
2018.PubMed/NCBI
|
|
10
|
Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX,
Yan PW and Wen J: Emerging roles of circular RNA hsa_circ_0000064
in the proliferation and metastasis of lung cancer. Biomed
Pharmacother. 96:892–898. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin Q, Ling YB, Chen JW, Zhou CR, Chen J,
Li X and Huang MS: Circular RNA circCDK13 suppresses cell
proliferation, migration and invasion by modulating the JAK/STAT
and PI3K/AKT pathways in liver cancer. Int J Oncol. 53:246–256.
2018.PubMed/NCBI
|
|
12
|
Jiang W, Wen D, Gong L, Wang Y, Liu Z and
Yin F: Circular RNA hsa_circ_0000673 promotes hepatocellular
carcinoma malignance by decreasing mir-767-3p targeting set.
Biochem Biophys Res Commun. 500:211–216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu C, Yu Y and Ding F: Microarray analysis
of circular RNA expression profiles associated with gemcitabine
resistance in pancreatic cancer cells. Oncol Rep. 40:395–404.
2018.PubMed/NCBI
|
|
15
|
Huang WJ, Wang Y, Liu S, Yang J, Guo SX,
Wang L, Wang H and Fan YF: Silencing circular RNA hsa_circ_0000977
suppresses pancreatic ductal adenocarcinoma progression by
stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett.
422:70–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sand M, Bechara FG, Gambichler T, Sand D,
Bromba M, Hahn SA, Stockfleth E and Hessam S: Circular RNA
expression in cutaneous squamous cell carcinoma. J Dermatol Sci.
83:210–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA itch has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
18
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. 7th. New York,
NY: Springer; 2010
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Granados-Riveron JT and Aquino-Jarquin G:
Does the linear Sry transcript function as a ceRNA for miR-138? The
sense of antisense. Version 2. F1000Res. 3:902014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circrna_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
23
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Alexandrov PN, Jaber V and Lukiw
WJ: Deficiency in the ubiquitin conjugating enzyme UBE2A in
Alzheimer's disease (AD) is linked to deficits in a natural
circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel). 7(pii):
E1162016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang
J, Xu Y, Hu J, Dong G, Xu PL and Yin R: Circular RNA
has_circ_0067934 is upregulated in esophageal squamous cell
carcinoma and promoted proliferation. Sci Rep. 6:355762016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sang M, Meng L, Sang Y, Liu S, Ding P, Ju
Y, Liu F, Gu L, Lian Y, Li J, et al: Circular RNA cirs-7
accelerates ESCC progression through acting as a miR-876-5p sponge
to enhance MAGE-a family expression. Cancer Lett. 426:37–46. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi Y, Chen C, Zhang X, Liu Q, Xu JL,
Zhang HR, Yao XH, Jiang T, He ZC, Ren Y, et al: Primate-specific
miR-663 functions as a tumor suppressor by targeting PIK3CD and
predicts the prognosis of human glioblastoma. Clin Cancer Rse.
20:1803–1813. 2014. View Article : Google Scholar
|
|
32
|
Shi Y, Chen C, Yu SZ, Liu Q, Rao J, Zhang
HR, Xiao HL, Fu TW, Long H, He ZC, et al: Mir-663 suppresses
oncogenic function of CXCR4 in glioblastoma. Clin Cancer Res.
21:4004–4013. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zang W, Wang Y, Wang T, Du Y, Chen X, Li M
and Zhao G: miR-663 attenuates tumor growth and invasiveness by
targeting eEF1A2 in pancreatic cancer. Mol Cancer. 14:372015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma Q, Zhang Y, Liang H, Zhang F, Liu F,
Chen S, Hu Y, Jiang L, Hao Y, Li M and Liu Y: EMP3, which is
regulated by miR-663a, suppresses gallbladder cancer progression
via interference with the MAPK/ERK pathway. Cancer Lett.
420:97–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang W, Li J, Guo X, Zhao Y and Yuan X:
miR-663a inhibits hepatocellular carcinoma cell proliferation and
invasion by targeting HMGA2. Biomed Pharmacother. 81:431–438. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Y, Zhou X, Xu X, Zhang M, Wang X,
Bai X, Li H, Kan L, Zhou Y, Niu H and He P: Waltonitone induces
apoptosis through mir-663-induced Bcl-2 downregulation in non-small
cell lung cancer. Tumour Biol. 36:871–876. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L,
Sun L, Liu J, Yang Z and Ran Y: Tumor-suppressive mir-663 gene
induces mitotic catastrophe growth arrest in human gastric cancer
cells. Oncol Rep. 24:105–112. 2010.PubMed/NCBI
|
|
38
|
Wang Z, Zhang H, Zhang P, Dong W and He L:
MicroRNA-663 suppresses cell invasion and migration by targeting
transforming growth factor beta 1 in papillary thyroid carcinoma.
Tumour Biol. 37:7633–7644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang Y, Wang LL, Wang HX, Guo ZK, Gao XF,
Cen J, Li YH, Dou LP and Yu L: The epigenetically-regulated miR-663
targets H-Ras in K-562 cells. FEBS J. 280:5109–5117. 2013.
View Article : Google Scholar : PubMed/NCBI
|