Introduction
Lung cancer is the most commonly diagnosed
malignancy and the main cause of cancer mortality worldwide,
accounting for 1.6 million deaths per year (1). In particular, non-small cell lung
cancer (NSCLC) accounts for ~80% of lung cancer cases (2). Smoking and exposure to pollutants are
key factors in NSCLC tumorigenesis (3). It has been suggested that cigarette and
air pollutants promote the progression of cancer through oxidative
stress in cells and tissues (4).
Similarly, endogenous oxidants are formed continuously (3). However, in the case of excess oxidants,
reactive oxidative species (ROS) are generated and induce a redox
imbalance, modifying numerous biomolecular markers (5,6).
Following this imbalance, ROS readily react with proteins, lipids
and DNA resulting in a number of pathological consequences,
including induced cell death, apoptosis and senescence, and an
increased risk for developing cancer (7).
ROS have been reported to be involved in the
pathogenesis of cancer (8,9); the mechanism by which this occurs has
been extensively investigated (10).
Smoking and pollutants, as well as internal metabolism, may induce
ROS production, while hypoxia may also induce ROS to promote tumor
growth (11). Furthermore,
alterations in the signaling pathways in cancer, such as integrin
activation, are also caused by an increase in ROS production
(12). A previous study reported
that ROS prevent cancer progression by regulating cancer cell death
(13). However, the molecular
mechanism underlying oxidants in NSCLC remain unknown.
Serine protease inhibitor Kazal-type 1 (SPINK1),
also known as pancreatic secretory trypsin inhibitor or pancreatic
secretory trypsin inhibitor, is a trypsin kinase inhibitor that
encodes a 56 amino acid secreted peptide (14). The normal function of SPINK1 is
thought to be the inhibition of serine proteases, such as trypsin,
involved in inflammation and cell proliferation (15). The altered expression of SPINK1 has
been demonstrated to be associated with disease prognosis and has
also been identified in different types of cancer, including lung,
bladder, kidney, prostate, testis, ovary, cervix and breast
cancers, and has been used in targeted therapy (16,17). In
the present study, it was revealed that SPINK1 may be a prognostic
factor for NSCLC and that SPINK1 regulates redox homeostasis
through the nuclear factor erythroid 2-related factor 2 (NRF2)
pathways.
Materials and methods
Human tissue collection
Paraffin-embedded tissue samples, including tumor
and adjacent tissues, were obtained from January 2010 to December
2018 at The Department of Pathology, The Affiliated Hospital of
Jining Medical University (Jining, China). Written informed consent
was obtained from the patients and the study was approved by The
Affiliated Hospital of Jining Medical University. All patients were
diagnosed and confirmed to have NSCLC by histology. Patients who
had received radiotherapy or chemotherapy were excluded. In total,
100 patients were included, 49 male and 51 female, with a median
age of 61 years (range, 35–81 years). Staging was based on the
pathological findings according to the 7th edition of The American
Joint Committee on Cancer guidelines (18). Overall survival time ranged from 6 to
98 months, with a median of 28 months. All patients were followed
up by telephone. The follow-up period ceased in December 2018. In
addition, liquid nitrogen frozen samples of NSCLC tissue samples
and adjacent normal tissues (2 cm from the lesion) were obtained by
surgical resection from 20 patients. These 20 patients were in
addition to the 100 patients aforementioned, and included 15 males
and 5 females, with a median age of 60 years (range, 37–74
years).
Immunohistochemical staining
Briefly, tissue section (4 µm) were deparaffinized
in xylene and rehydrated in a graded alcohol series (100, 95, 85
and 75% ethanol) and blocked with 3% H2O2 for
15 min at room temperature. Antigen retrieval was performed after
heating in citrate buffer at 98°C for 10 min. The sections were
incubated with an antibody against SPINK1 (1:100; catalog no.
ab183034; Abcam) at 4°C overnight and with horseradish peroxidase
universal immunoglobulin G secondary antibody (cat. no. sc69786;
1:1,000; Santa Cruz Biotechnology, Inc.) for 30 min at 37°C. The
signal was detected with 3,3′-diaminobenzidine solution using a
light microscope (magnification, ×200; IX71; Olympus Corporation).
For SPINK1 protein, all visual fields were analyzed; the entire
tissue specimen irrespective of its size was assessed, taking into
account the percentage of positively stained cells and the
intensity of staining.
Scoring of immunohistochemistry
Double-blind scoring was performed independently by
two pathologists from the Department of Pathology, Affiliated
Hospital of Jining Medical University. If different scores were
assigned for the same sample, the sample was revaluated and, if
needed, further discussed to determine a final score. SPINK1
expression was scored according to a previously described staining
intensity and percentage of positive cells method (19). The percentage of positive cells was
scored as follows: 0, no positive cells; 1, 1–10% positive cells;
2, 11–50% positive cells; 3, ≥51% positive cells. Staining
intensity was scored as follows: 0, no staining; 1, weak staining;
2, moderate staining; 3, dark staining. Comprehensive score =
staining percentage × intensity. SPINK1 expression: <2 low
expression, ≥2 high expression.
Cells and RNA interference (RNAi)
The HBE, H460, H1299 and A549 cell lines were
obtained from The American Type Culture Collection. A549 cells were
cultured in F-12K medium with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere
with 5% CO2. The other cell lines were cultured in
RPMI-1640 medium with 10% FBS in a humidified atmosphere with 5%
CO2 at 37°C, until they reached 100% confluence. Small
interfering (si)RNA targeting SPINK1 (SPINK1-si1, sense
5′-AAGAAAGAUGCCUGUUACCUU-3′, antisense 5′-GGUAACAGGCAUCUUUCUUCU-3′
and SPINK1-si2, sense 5′-AGUGUUACCAGAUAGACUCAA-3′, antisense
5′-GAGUCUAUCUGGUAACACUGG-3′; Shanghai GenePharma Co, Ltd.) was used
to knockdown the expression of SPINK1. Negative control siRNA (4
µg; non-targeting) or SPINK1 siRNA (Shanghai GenePharma Co, Ltd.)
were transiently transfected using 8 µl Lipofectamine®
RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.). The
transfection efficiency was determined by western blotting 24 h
following transfection.
Cell proliferation and apoptosis
Proliferation assays were performed by seeding 4,500
cells into 96-well plates and using the Cell Counting Kit-8 (CCK-8;
Dojindo Laboratories), according to the manufacturer's protocol.
The Annexin V-FITC/propidium iodide (PI) apoptosis detection kit I
(BD Biosciences) was used to determine the level of apoptosis.
Briefly, the collected cells were centrifuged at ×500 g for 5 min
at room temperature and washed three times with PBS, and
resuspended with 100 µl binding buffer with 5 µl Annexin V-FITC and
5 µl PI solution. Cells were incubated in the dark for 15 min at
4°C. The cells were added to 400 µl binding buffer and analyzed
using a FACS LSR II (BD Biosciences) flow cytometer, and analyzed
using CellQuest software (version 5.1; BD Biosciences).
Western blot analysis
The protein levels of SPINK1 from cell lines were
determined using western blot analysis. Briefly, proteins were
lysed in RIPA (Beyotime Institute of Biotechnology). Protein
concentration was determined using a bicinchoninic acid assay kit
(Beyotime institute of Biotechnology). Protein (20 µg) were
separated by 10% SDS-PAGE and transferred onto a nitrocellulose
membrane. After the transfer, 5% bovine serum albumin (Thermo
Fisher Scientific, Inc.) was used to block the membranes for 1 h at
room temperature. The following primary antibodies were incubated
overnight at 4°C: SPINK1 (1:1,000; catalog no. ab183034; Abcam),
β-actin (1:5,000; cat. no. ab8226; Abcam). Membranes were washed
with TBST three times. The secondary antibody anti-mouse (1:1,000;
cat. no. 7076; Cell Signaling Technology, Inc.) and anti-rabbit
(1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) was
incubated for 1 h at room temperature. Protein bands were
visualized using ECL reagent (Thermo Fisher Scientific, Inc.),
images were captured using a Tanon detection system, and
densitometry analysis was performed using ImageQuant TL software
version 1.1 (GE Healthcare Life Sciences).
Reverse transcription-quantitative
(RT-qPCR)
Total RNA from cells or patients' tissues was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). The PrimeScript RT reagent kit (Takara
Bio, Inc.) was used to synthesize complementary DNA, which was used
in the qPCR (SYBR Green PCR kit; Takara Bio, Inc.) with a 7500 Fast
Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The RT reactions were performed as follows: 37°C for 15 min
and 85°C for 5 sec. PCR amplification was performed in a thermal
cycler for 40 cycles at the following cycle conditions: 95°C for 5
sec, 60°C for 30 sec, and 72°C for 30 sec. The following primers
were used: SPINK1, forward 5′-TGTCTGTGGGACTGATGGAA-3′, reverse
5′-AGGCCCAGATTTTTGAATGA-3′. NRF2, forward
5′-AGGTTGCCCACATTCCCAAA-3′, reverse 5′-AGTGACTGAAACGTAGCCGA-3′;
ME1, forward 5′-GAACCCTCACCTCAACAAGGTA-3′, reverse
5′-AGCCAAGAATACGCTCTCCA-3′; TXNRD1, forward
5′-GAAAGGCAAGAACGGCGATG-3′, reverse 5′-CGTGTGCATGTGGACCTACT-3′;
GCLC, forward 5′-ACTTCATTTCCCAGTACCTTAACA-3′, reverse
5′-CCGGCTTAGAAGCCCTTGAA-3′; GCLM, forward
5′-AAGCACTTTCTCGGCTACGA-3′, reverse 5′-GCGGGAGAGCTGATTCCAAA-3′; and
GAPDH, forward 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse
5′-AGGGGCCATCCACAGTCTTC-3′. GAPDH was used as the internal control.
Each experiment was performed in triplicate. The relative mRNA
expression of each gene was calculated with the comparative
quantitation cycle method 2−ΔΔcq (20,21).
ROS evaluation
Dichlorofluorescin diacetate (DCFH-DA; Beyotime
Institute of Biotechnology) staining was used to detect the
cytoplasmic ROS level. Briefly, cells were washed twice with PBS
and incubated with 10 µM DCFH-DA at 37°C for 30 min. The cells were
resuspended in ice-cold PBS and the fluorescence intensity of DCF
was determined at an excitation wavelength of 488 nm and an
emission wavelength at 530 nm.
The intracellular ATP level, NADPH/NADP+
ratio and NADH/NAD+ ratio was evaluated using the ATP
Assay kit (Beyotime Institute of Biotechnology), the NADP/NADPH
Quantitation Colorimetric kit (BioVision) and the NAD/NADH
Quantitation Colorimetric kit (BioVision), according to the
manufacturers' protocols. The GSH and GSSG Assay kit was purchased
from Beyotime Institute of Biotechnology and was performed
according to the manufacturer's protocol.
Dual-luciferase reporter assay
In total, 2×104 cells were seeded into
96-well plates. The plasmids (1 µg) REPO-ARE [a DNA sequence
containing the GST A1 ARE (−833 to −533 from the start codon)
inserted into a luciferase reporter vector as described previously
(22)] and 0.5 µg/1 µg
pCMV-SPINK1-flag (both from Shanghai GenePharma Co, Ltd.) were
transfected into each well using Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). The Renilla luciferase
plasmid (Promega Corporation) was transfected into each well to
normalize transfection efficiency. Luciferase activity
(Dual-Luciferase Assay System; Promega Corporation) was detected 48
h after transfection. Renilla luciferase gene was used as the
internal reference to verify the transfection efficiency and
calculate the relative luciferase activity as follows: Relative
luciferase activity=firefly luciferase activity/Renilla luciferase
activity. The experiments were performed twice.
Statistical analysis
The data are presented as the mean ± standard
deviation. For comparisons between two groups, two-tailed Student's
t-tests were used. Multiple group comparisons were calculated using
the one-way ANOVA analysis followed by the Dunnett's post hoc test.
Associations between the expression of SPINK1 and clinical
characteristics were analyzed using the χ2 test. The
Kaplan-Meier method and log-rank test was used to estimate overall
survival (OS). Univariate and multivariate COX regression analysis
was performed to estimate the relative risk of death associated
with SPINK1 expression and other prognostic variables for OS. Those
parameters with P<0.05 in the univariate analyses were included
in a Cox multivariate proportional hazards regression model.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SPINK1 is a prognostic marker of
NSCLC
In total, 100 paraffin-embedded tissue samples used
for detecting protein expression by IHC were collected in order to
investigate the expression and clinical features of SPINK1 in
patients with NSCLC in the present study. SPINK1 was expressed at a
higher level in tumor samples compared with adjacent normal tissue
(Table I). No differences in SPINK1
expression were observed for age, sex, smoking habits, histological
type or the Tumor (T) and Node (N) stage (Table II). However, SPINK1 expression was
associated with Tumor-Node-Metastasis (TNM) stage (P<0.001;
Table II). The univariate and
multivariate analysis indicated that, in addition to TNM stage,
SPINK1 may be a prognostic factor for patients with NSCLC
(P<0.001; Table III).
 | Table I.Comparisons with SPINK1 expression
between NSCLC and paired adjacent normal tissues. |
Table I.
Comparisons with SPINK1 expression
between NSCLC and paired adjacent normal tissues.
|
|
| SPINK1
expression |
|
|---|
|
|
|
|
|
|---|
| Tissues | Cases, n | Low | High | P-value |
|---|
| NSCLC tissues | 100 | 49 | 51 | 0.006 |
| Adjacent
tissue | 100 | 68 | 32 |
|
 | Table II.Association between the
clinicopathological characteristics and SPINK1 expression in
non-small cell lung cancer. |
Table II.
Association between the
clinicopathological characteristics and SPINK1 expression in
non-small cell lung cancer.
|
|
| SPINK1
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | N | Low (n=49) | High (n=51) | P-value |
|---|
| Age, years |
|
|
|
|
|
≤60 | 50 | 22 | 28 | 0.317 |
|
>60 | 50 | 27 | 23 |
|
| Sex |
|
|
|
|
|
Male | 55 | 26 | 29 | 0.702 |
|
Female | 45 | 23 | 22 |
|
| Smoking habit |
|
|
|
|
|
Never | 64 | 31 | 33 | 0.881 |
|
Ever | 36 | 18 | 18 |
|
| Histological
type |
|
|
|
|
|
Adenocarcinoma | 46 | 25 | 21 | 0.319 |
|
Squamous cell carcinoma | 36 | 14 | 22 |
|
|
Others | 18 | 10 | 8 |
|
| Grade |
|
|
|
|
|
Well-differentiated | 70 | 36 | 34 | 0.458 |
|
Moderate-poorly
differentiated | 30 | 13 | 17 |
|
| T stage |
|
|
|
|
|
T1+T2 | 30 | 14 | 16 | 0.711 |
| T3 | 69 | 35 | 34 |
|
| N stage |
|
|
|
|
| N0 | 39 | 22 | 17 | 0.236 |
|
N1+N2 | 61 | 27 | 34 |
|
| TNM stage |
|
|
|
|
|
I+II | 66 | 42 | 24 | <0.001 |
|
IIIA | 34 | 7 | 27 |
|
 | Table III.Summary of univariate and
multivariate Cox regression analysis of overall survival. |
Table III.
Summary of univariate and
multivariate Cox regression analysis of overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
Characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| SPINK1
expression |
|
|
|
|
|
|
|
Low | 1 |
|
| 1 |
|
|
|
High | 4.003 | 2.450–6.541 | <0.001 | 3.491 | 2.071–5.884 | <0.001 |
| Age, years |
|
|
|
|
|
|
|
≤60 | 1 |
|
|
|
|
|
|
>60 | 0.821 | 0.536–1.256 | 0.362 |
|
|
|
| Sex |
|
|
|
|
|
|
|
Male | 1 |
|
|
|
|
|
|
Female | 0.868 | 0.566–1.331 | 0.517 |
|
|
|
| Smoking habit |
|
|
|
|
|
|
|
Never | 1 |
|
|
|
|
|
|
Ever | 0.947 | 0.610–1.471 | 0.947 |
|
|
|
| Histological
type |
|
|
|
|
|
|
|
Adenocarcinoma | 1 |
|
|
|
|
|
|
Squamous cell carcinoma | 0.970 | 0.536–1.758 | 0.921 |
|
|
|
|
Other | 1.362 | 0.729–2.542 | 0.332 |
|
|
|
| Grade |
|
|
|
|
|
|
| Well
differentiated | 1 |
|
|
|
|
|
|
Moderate-poorly
differentiated | 1.500 | 0.951–2.368 | 0.081 |
|
|
|
| T stage |
|
|
|
|
|
|
|
T1+T2 | 1 |
|
|
|
|
|
| T3 | 0.731 | 0.416–1.284 | 0.275 |
|
|
|
| N stage |
|
|
|
|
|
|
| N0 | 1 |
|
|
|
|
|
|
N1+N2 | 0.858 | 0.684–1.076 | 0.186 |
|
|
|
| TNM stage |
|
|
|
|
|
|
|
I+II | 1 |
|
| 1 |
|
|
|
IIIA | 19.263 | 8.932–41.542 | <0.001 | 19.117 | 8.243–44.333 | <0.001 |
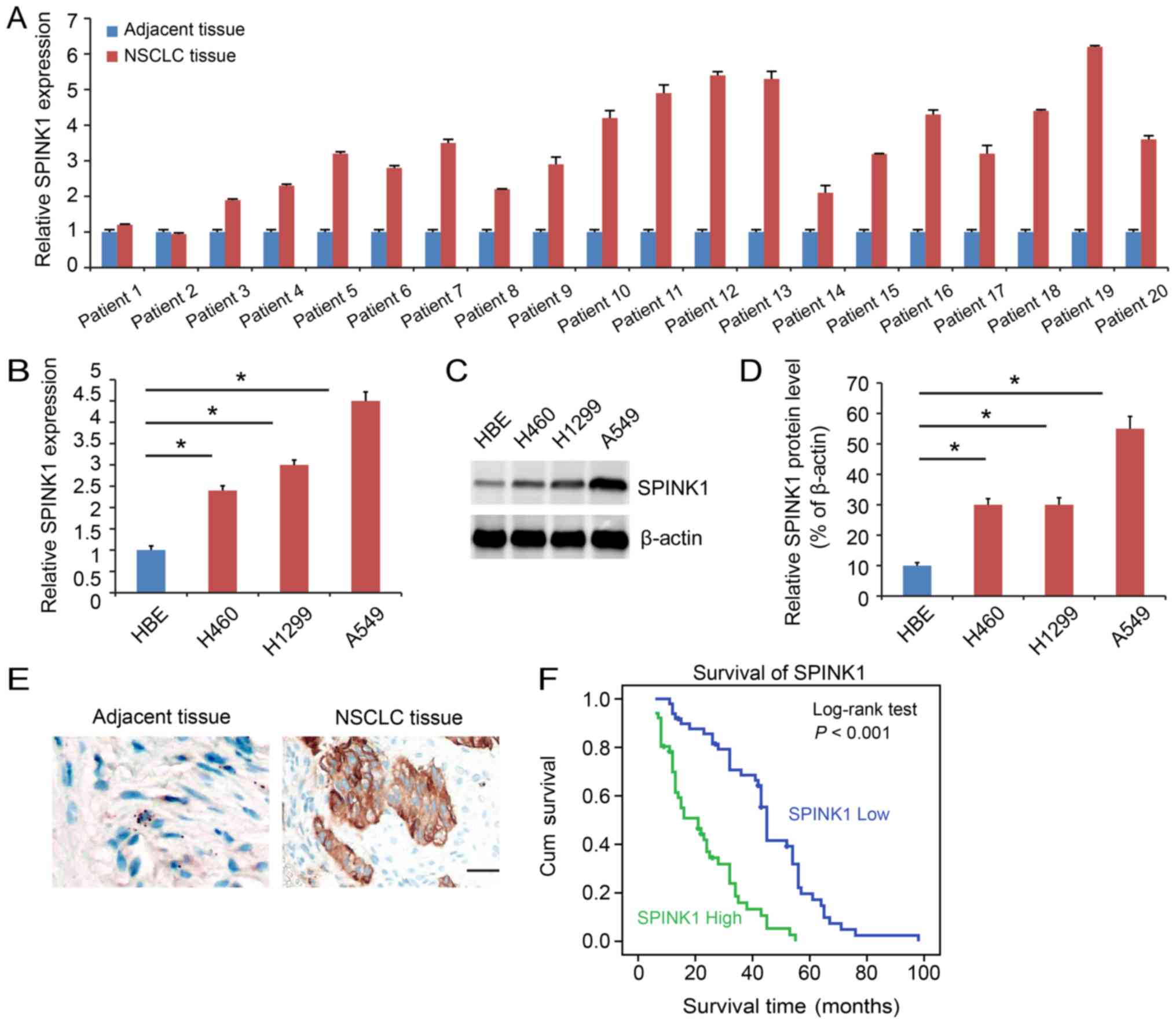
SPINK1 is expressed at a higher level
in NSCLC
SPINK1 expression was analyzed in clinical samples.
The function of SPINK1 was also analyzed. SPINK1 expression was
higher in NSCLC tissues compared with adjacent normal tissues
(P<0.05; Fig. 1A). The expression
of SPINK1 was also assessed in cell lines. SPINK1 was upregulated
in NSCLC cell lines compared with HBE at both the transcriptional
(P<0.05; Fig. 1B) and protein
levels (Fig. 1C and D).
Immunohistochemistry demonstrated a higher level of expression of
SPINK1 in NSCLC tissue samples (Fig.
1E). Kaplan-Meier analysis indicated that higher expression of
SPINK1 predicted a poorer prognosis for patients (P<0.001;
Fig. 1F).
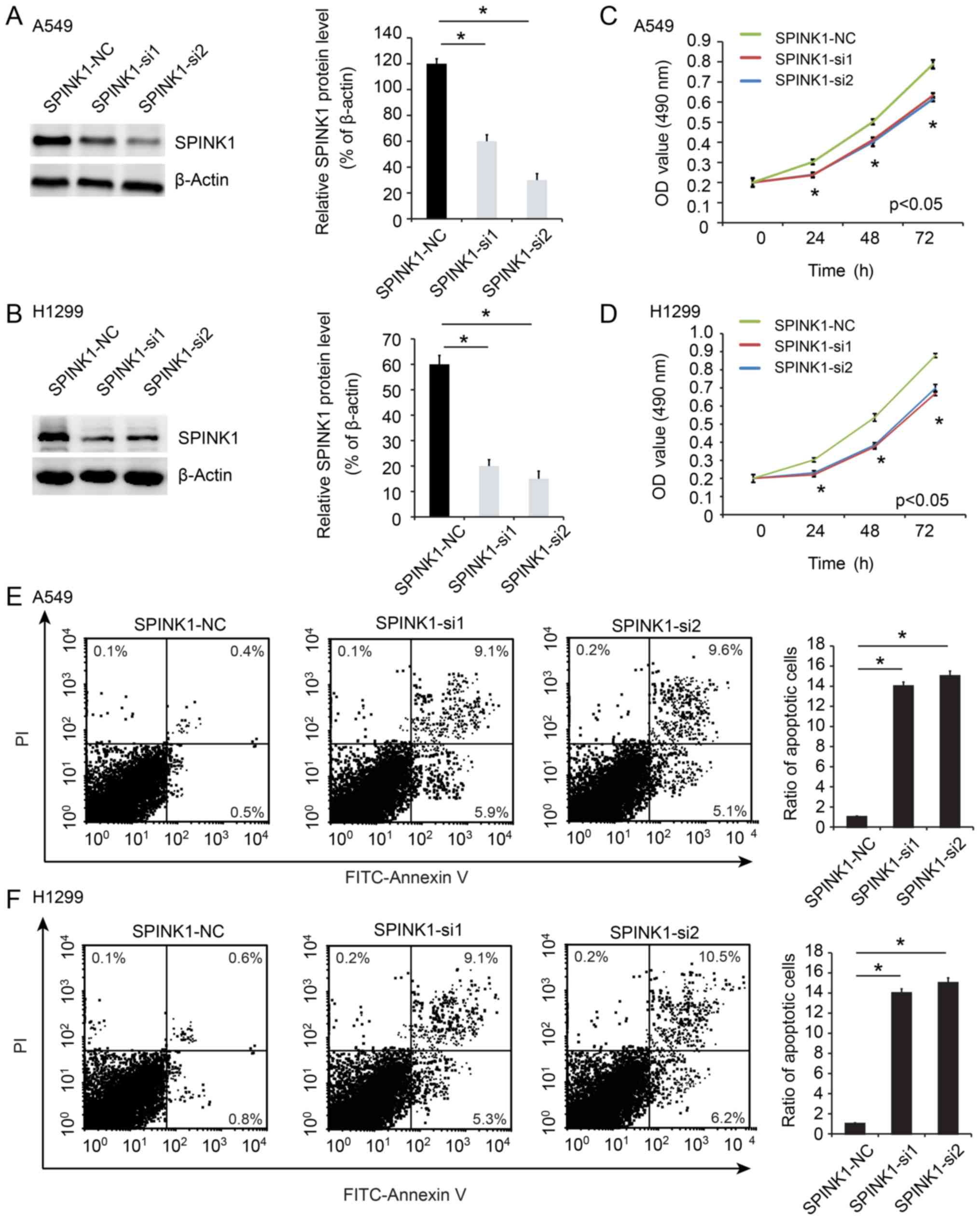
SPINK1 regulates proliferation and
apoptosis in NSCLC
In order to investigate the biological effects of
SPINK1 in NSCLC cells, SPINK1 was depleted using RNAi. The
knockdown efficiency was determined using RT-PCR and western
blotting (P<0.05; Fig. 2A and B).
The biological function of SPINK1 was examined in NSCLC cell lines.
The viability of SPINK1-siRNA treated cells was significantly
decreased compared with the control group (P<0.05; Fig. 2C and D). In addition, the level of
apoptosis was higher in SPINK1-siRNA treated cells (P<0.05;
Fig. 2E and F). These results
indicated a role for SPINK1 in promoting cell growth and inhibiting
apoptosis.
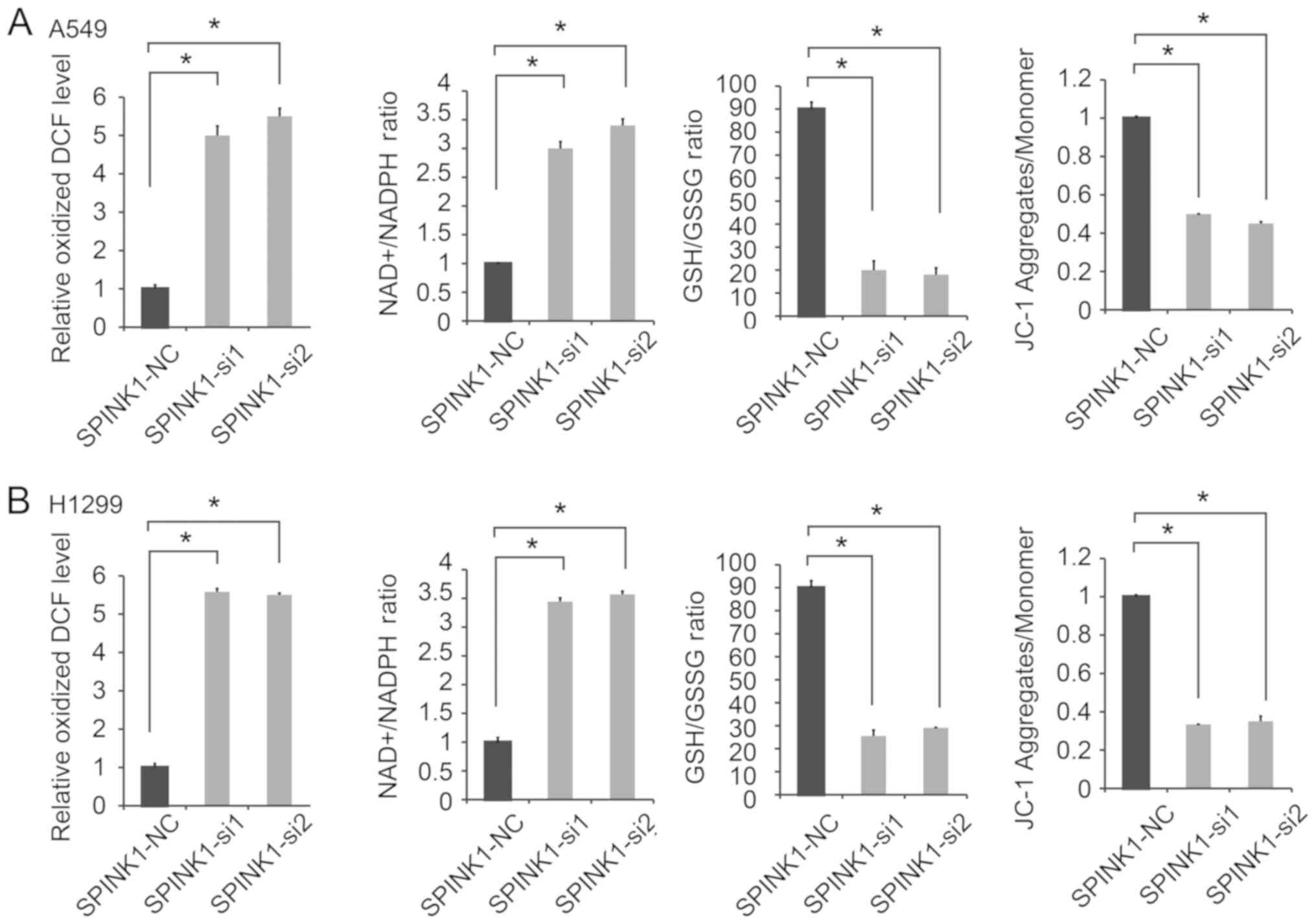
SPINK1 regulates the redox state of
NSCLC cells
The potential molecular mechanisms of SPINK1 in
NSCLC were investigated in the present study. Redox homeostasis is
an important factor in tumor progression (23,24).
Therefore, whether SPINK1 is as an oxidative factor involved in
protecting basal mitochondrial respiration in NSCLC cells was
investigated in the present study. Since A549 and H1299 have
relatively higher endogenous SPINK1 expression, these cell lines
were chosen for SPINK1 silencing experiments. The results revealed
that the inhibition of SPINK1 significantly increased ROS
production. Furthermore, the knockdown of SPINK1 increased the
NADP/NADPH ratio, decreased the GSH/GSSG ratio and JC-1 aggregates
compared with the control cells (P<0.05; Fig. 3A and B). These results suggested that
SPINK1 played an important role in the redox balance of NSCLC
cells.
SPINK1 transcriptionally activates
NRF2 expression in NSCLC
Next, the potential underlying molecular mechanism
of SPINK1 regulating redox homeostasis was investigated.
Antioxidant response elements (AREs) are in the regulatory regions
of several genes that encode for enzymes that contribute to the
regulation of antioxidants (21).
The results of the present study revealed that ARE activity was
increased in a dose-dependent manner after cloning the ARE
sequences into a luciferase vector and transfecting cells with a
SPINK1 expression vector, indicating that ARE-driven gene were
significantly decreased in SPINK1 knockdown cells (P<0.05;
Fig. 4A and B). The knockdown of
SPINK1 decreased NRF2 expression and further decreased the mRNA
levels of ARE-driven genes, including NADP-dependent malic enzyme
(ME1), thioredoxin reductase 1 (TXNRD), glutamate-cysteine ligase
catalytic subunit (GCLC) and glutamate-cysteine ligase regulatory
subunit (GCLM) (P<0.05; Fig. 4C and
D). Furthermore, the expression levels of NRF2, ME1, TXNRD 1,
GCLC and GCLM were validated using 20 NSCLC specimens frozen in
liquid nitrogen. The results revealed that the expression levels of
NRF2, ME1, TXNRD 1, GCLC and GCLM were increased in NSCLC tissues
(in 18 of the 20 cases; P<0.05; Fig.
4E). SPINK1 maintained the redox balance by activating NRF2,
upregulating antioxidant response element-driven gene expression,
leading to increased cell proliferation and decreased cell
apoptosis (Fig. 4F).
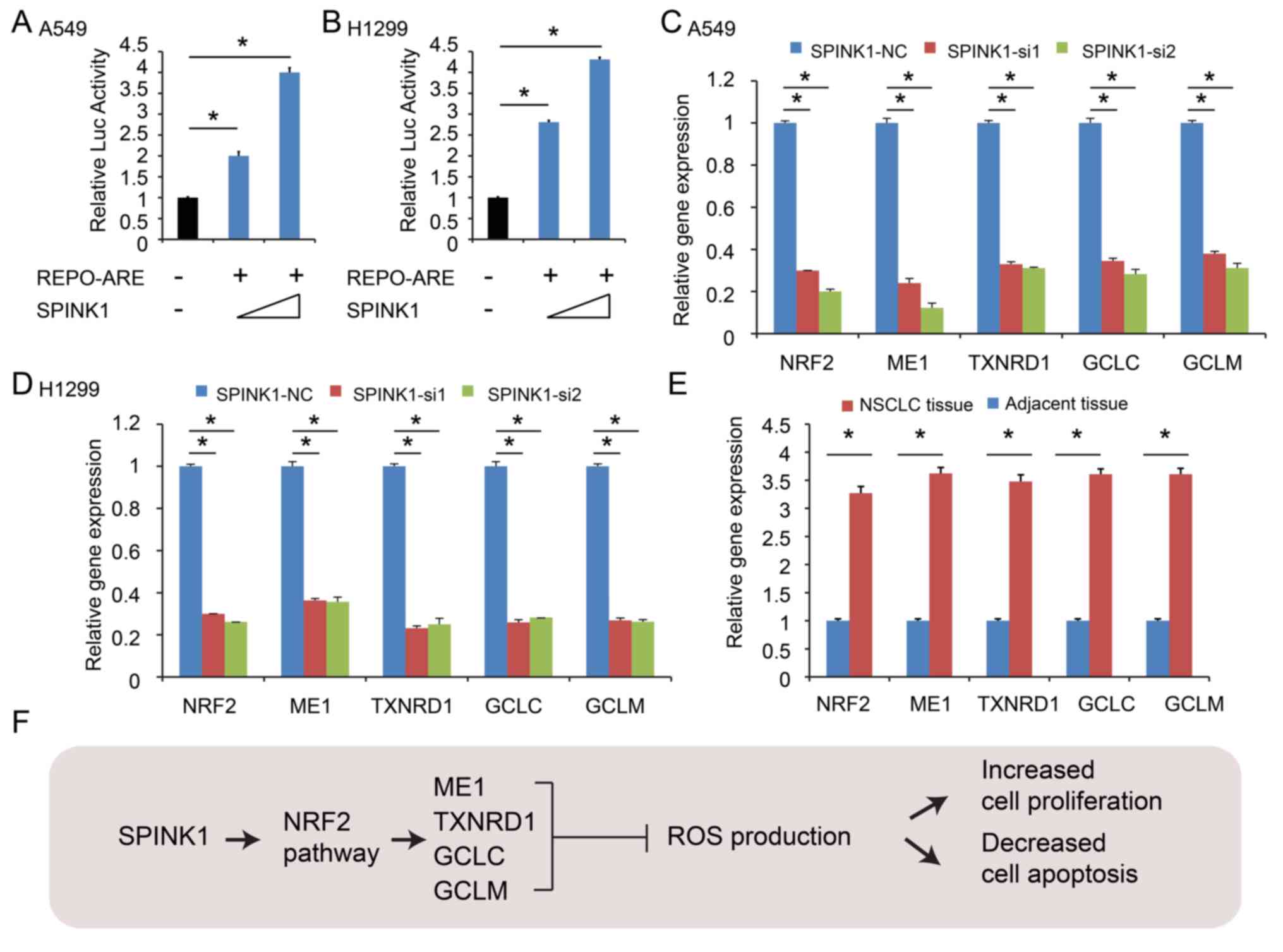 | Figure 4.SPINK1 activates the transcription of
NRF2 in NSCLC. (A and B) ARE activity was increased in a
dose-dependent manner after cloning of the ARE sequences into a
luciferase vector and co-transfection with a SPINK1 expression
vector. (C and D) SPINK1 increased the expression of NRF2, ME1,
TXNRD, GCLC and GCLM. (E) NRF2, ME1, TXNRD1, GCLC and GCLM were
upregulated in NSCLC tissues. (F) The outline of the mechanism for
SPINK1 mediated redox regulation in NSCLC. *P<0.05 vs. control.
ARE, antioxidant response element; SPINK1, serine protease
inhibitor Kazal-type 1; NSCLC, non-small cell lung cancer; ARE,
antioxidant response element; NRF2, nuclear factor erythroid
2-related factor; ME1, NADP-dependent malic enzyme; TXNRD,
thioredoxin reductase 1; GCLC, glutamate-cysteine ligase catalytic
subunit; GCLM, glutamate-cysteine ligase regulatory subunit. |
Discussion
The serine protease inhibitor SPINK1 is a trypsin
inhibitor, which is known to play an important role in a number of
physiological and pathological processes (25). SPINK1 has an established function in
protecting the pancreas from premature activation of trypsinogen
(26). SPINK1 functions as an acute
phase reactant in severe inflammatory disease and tissue injury,
contributing to normal tissue maintenance and repair (14). Furthermore, SPINK1 promotes the
initiation and progression of cancer. Changes in SPINK1 expression
levels are associated with the prognosis of a number of different
types of cancer (17); the high
expression of SPINK1 in liver tissue is associated with liver
metastasis (27), and SPINK1 levels
in the serum of patients with colorectal cancer are associated with
OS (28).
In the present study, SPINK1 expression levels were
determined in NSCLC. The expression of SPINK1 was higher in tumor
samples and lower in the adjacent normal tissue.
Clinicopathological analysis revealed that SPINK1 expression was
associated with TNM stage. The univariate and multivariate analyses
indicated that SPINK1 may be a prognostic factor in patients with
NSCLC. The Kaplan-Meier analysis suggested that higher expression
levels of SPINK1 predicted a poorer OS time.
The biological functions of SPINK1 were also
investigated in the present study. SPINK1 was highly expressed in
NSCLC cells and tissue samples. The viability of SPINK1-siRNA
treated cells was significantly attenuated compared with the
control group. In addition, an increased level of apoptosis was
observed in SPINK1-siRNA treated cells.
The molecular mechanism of SPINK1 in NSCLC was
investigated in the present study. ROS have several roles in the
development of tumors; the physiological effects of ROS vary with
concentration, duration and cell changes (29). Low concentrations of ROS can
accelerate cell division and proliferation, while moderate levels
cause the arrest of cell growth and high levels induce apoptosis or
necrosis (30). A large increase in
ROS leads to the opening of the mitochondrial membrane permeability
transport pores, activation of the caspase cascade, DNA breaks and
tumor cell apoptosis mediated by Fas-FasL through activating the
p38 mitogen-activated protein kinase (MAPK) signaling pathway
(31,32). ROS cause DNA damage (33), affecting mitochondrial DNA (mtDNA)
and impeding oxidative phosphorylation (34). Previous studies have demonstrated
that tumors, such as lung cancer, stomach cancer, breast cancer,
colon cancer and lymphoma, have mtDNA mutations (35–38). In
addition, low levels of ROS act as signaling molecules to regulate
key proteins and promote cell proliferation. In proliferation, ROS
leads to the decreased expression of p53 and p21, and increased
activity of the cyclin-cyclin dependent kinase complex, leading to
cell cycle progression (39,40). In NSCLC, the inhibition of SPINK1
significantly increased ROS production, increased the NADPH/NADPH
ratio and decreased the GSH/GSSG ratio, and decreased JC-1
aggregates. These results suggested that SPINK1 played an important
role in the redox balance of NSCLC cells.
Previous studies have revealed that the NRF2 pathway
is associated with ROS (41–44). AREs are found in the regulatory
regions of antioxidant regulating enzymes (45). Following the transfection of the
luciferase vector with ARE sequences and the SPINK1 vector in the
present study, ARE activity increased in a dose-dependent manner.
In addition, SPINK1 increased the expression of NRF2 and increased
the mRNA levels of ARE-driven genes, including ME1, TXNRD, GCLC and
GCLM. Taken together, SPINK1 participated in the redox balance by
activating NRF2 in NSCLC cells. However, how SPINK1 regulates NRF2
requires further investigation. Structurally, SPINK1 was identified
as having a structure with ~50% similarity to epidermal growth
factor (EGF), which indicates the ability of SPINK1 to induce the
dimerization and phosphorylation of the EGF receptor (EGFR), and
promote signal transduction and cell proliferation through the MAPK
pathway (46). Wang et al
(47) reported that SPINK1 promotes
the epithelial-to-mesenchymal transition through the EGFR signaling
pathway in prostate cancer. The concomitant expression of SPINK1 at
high levels and EGFR was also identified to be associated with poor
prognosis in colorectal cancer (48). Ateeq et al (49) suggested that SPINK1 functions, at
least in part, to stimulate EGFR signaling in an autocrine loop. A
direct interaction between SPINK1 and EGFR was also identified in
prostate cancer. Future research in the field should focus on
this.
In conclusion, the present study revealed a novel
regulatory function for SPINK1 through the activation of the
NRF2-mediated antioxidant regulation. In addition, SPINK1 may be a
prognostic factor for NSCLC. Thus, developing an inhibitor for
SPINK1 is expected to provide therapeutic benefits for NSCLC.
Acknowledgements
The authors would like to thank Dr Ting Wang and Dr
Quanyi Wang (Department of Pathology, Affiliated Hospital of Jining
Medical University), for their contributions to the scoring of
immunohistochemistry.
Funding
The present study was supported by the Science and
Technology Development Plan of Jining [grant no. (2016)56-086].
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MG, XZ, and XH performed the experiments and wrote
the manuscript. XH and LJ made substantial contributions to the
conception and design of the manuscript. XH and YZ analyzed the
experimental data. MG, XZ, XH and YZ assisted with the statistical
analysis. MG and XH critically revised the manuscript and provided
final approval of the version to be published. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from the
patients and the study was approved by The Affiliated Hospital of
Jining Medical University (Jining, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ciencewicki J, Trivedi S and Kleeberger
SR: Oxidants and the pathogenesis of lung diseases. J Allergy Clin
Immunol. 122:456–468, 469-470. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ghio AJ, Carraway MS and Madden MC:
Composition of air pollution particles and oxidative stress in
cells, tissues, and living systems. J Toxicol Environ Health B Crit
Rev. 15:1–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Höhn A, Jung T and Grune T:
Pathophysiological importance of aggregated damaged proteins. Free
Radic Biol Med. 71:70–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barouki R: Ageing free radicals and
cellular stress. Med Sci (Paris). 22:266–272. 2006.(In French).
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ziech D, Franco R, Georgakilas AG,
Georgakila S, Malamou-Mitsi V, Schoneveld O, Pappa A and
Panayiotidis MI: The role of reactive oxygen species and oxidative
stress in environmental carcinogenesis and biomarker development.
Chem Biol Interact. 188:334–339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kryston TB, Georgiev AB, Pissis P and
Georgakilas AG: Role of oxidative stress and DNA damage in human
carcinogenesis. Mutat Res. 711:193–201. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reuter S, Gupta SC, Chaturvedi MM and
Aggarwal BB: Oxidative stress, inflammation, and cancer: How are
they linked? Free Radic Biol Med. 49:1603–1616. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kardeh S, Ashkani-Esfahani S and Alizadeh
AM: Paradoxical action of reactive oxygen species in creation and
therapy of cancer. Eur J Pharmacol. 735:150–168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Azimi I, Petersen RM, Thompson EW,
Roberts-Thomson SJ and Monteith GR: Hypoxia-induced reactive oxygen
species mediate N-cadherin and SERPINE1 expression, EGFR signalling
and motility in MDA-MB-468 breast cancer cells. Sci Rep.
7:151402017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chiarugi P, Pani G, Giannoni E, Taddei L,
Colavitti R, Raugei G, Symons M, Borrello S, Galeotti T and Ramponi
G: Reactive oxygen species as essential mediators of cell adhesion:
The oxidative inhibition of a FAK tyrosine phosphatase is required
for cell adhesion. J Cell Biol. 161:933–944. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Poillet-Perez L, Despouy G,
Delage-Mourroux R and Boyer-Guittaut M: Interplay between ROS and
autophagy in cancer cells, from tumor initiation to cancer therapy.
Redox Biol. 4:184–192. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Itkonen O and Stenman UH: TATI as a
biomarker. Clin Chim Acta. 431:260–269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stenman UH: Role of the tumor-associated
trypsin inhibitor SPINK1 in cancer development. Asian J Androl.
13:628–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Räsänen K, Itkonen O, Koistinen H and
Stenman UH: Emerging Roles of SPINK1 in Cancer. Clin Chem.
62:449–457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stenman UH: SPINK1: A new therapeutic
target in cancer? Clin Chem. 57:1474–1475. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiao F, Hu H, Han T, Zhuo M, Yuan C, Yang
H and Wang L and Wang L: Aberrant expression of nuclear HDAC3 and
cytoplasmic CDH1 predict a poor prognosis for patients with
pancreatic cancer. Oncotarget. 7:16505–16516. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peirson SN, Butler JN and Foster RG:
Experimental validation of novel and conventional approaches to
quantitative real-time PCR data analysis. Nucleic Acids Res.
31:e732003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kwak MK, Kensler TW and Casero RJ Jr:
Induction of phase 2 enzymes by serum oxidized polyamines through
activation of Nrf2: Effect of the polyamine metabolite acrolein.
Biochem Biophys Res Commun. 305:662–670. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kong H and Chandel NS: Regulation of redox
balance in cancer and T cells. J Biol Chem. 293:7499–7507. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang C, Shi S, Liu M, Qin Y, Meng Q, Hua
J, Ji S, Zhang Y, Yang J, Xu J, et al: PIN1 maintains redox balance
via the c-Myc/NRF2 axis to counteract Kras-induced mitochondrial
respiratory injury in pancreatic cancer cells. Cancer Res.
79:133–145. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Masamune A, Mizutamari H, Kume K, Asakura
T, Satoh K and Shimosegawa T: Hereditary pancreatitis as the
premalignant disease: A Japanese case of pancreatic cancer
involving the SPINK1 gene mutation N34S. Pancreas. 28:305–310.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohmuraya M, Hirota M, Araki M, Mizushima
N, Matsui M, Mizumoto T, Haruna K, Kume S, Takeya M, Ogawa M, et
al: Autophagic cell death of pancreatic acinar cells in serine
protease inhibitor Kazal type 3-deficient mice. Gastroenterology.
129:696–705. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ying HY, Gong CJ, Feng Y, Jing DD and Lu
LG: Serine protease inhibitor Kazal type 1 (SPINK1) downregulates
E-cadherin and induces EMT of hepatoma cells to promote
hepatocellular carcinoma metastasis via the MEK/ERK signaling
pathway. J Dig Dis. 18:349–358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gaber A, Stene C, Hotakainen K, Nodin B,
Palmquist I, Bjartell A, Stenman UH, Jeppsson B, Johnson LB and
Jirström K: Effects of radiation therapy on tissue and serum
concentrations of tumour associated trypsin inhibitor and their
prognostic significance in rectal cancer patients. Radiat Oncol.
6:1002011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moloney JN and Cotter TG: ROS signalling
in the biology of cancer. Semin Cell Dev Biol. 80:50–64. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prasad S, Gupta SC and Tyagi AK: Reactive
oxygen species (ROS) and cancer: Role of antioxidative
nutraceuticals. Cancer Lett. 387:95–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bapat S, Verkleij A and Post JA:
Peroxynitrite activates mitogen-activated protein kinase (MAPK) via
a MEK- independent pathway: A role for protein kinase C. Febs Lett.
499:21–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Farrukh MR, Nissar UA, Afnan Q, Rafiq RA,
Sharma L, Amin S, Kaiser P, Sharma PR and Tasduq SA: Oxidative
stress mediated Ca(2+) release manifests endoplasmic reticulum
stress leading to unfolded protein response in UV-B irradiated
human skin cells. J Dermatol Sci. 75:24–35. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Panayiotidis M: Reactive oxygen species
(ROS) in multistage carcinogenesis. Cancer Lett. 266:3–5. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miller JH, Jin S, Morgan WF, Yang A, Wan
Y, Aypar U, Peters JS and Springer DL: Profiling mitochondrial
proteins in radiation-induced genome-unstable cell lines with
persistent oxidative stress by mass spectrometry. Radiat Res.
169:700–706. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zong WX, Rabinowitz JD and White E:
Mitochondria and cancer. Mol Cell. 61:667–676. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Zhang L, Yu X, Zhou H, Luo Y, Wang
W and Wang L: Clinical application of plasma mitochondrial DNA
content in patients with lung cancer. Oncol Lett. 16:7074–7081.
2018.PubMed/NCBI
|
|
37
|
Sansone P, Savini C, Kurelac I, Chang Q,
Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly
L, et al: Packaging and transfer of mitochondrial DNA via exosomes
regulate escape from dormancy in hormonal therapy-resistant breast
cancer. Proc Natl Acad Sci USA. 114:E9066–E9075. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin CS, Liu LT, Ou LH, Pan SC, Lin CI and
Wei YH: Role of mitochondrial function in the invasiveness of human
colon cancer cells. Oncol Rep. 39:316–330. 2018.PubMed/NCBI
|
|
39
|
DeGraff WG, Krishna MC, Kaufman D and
Mitchell JB: Nitroxide-mediated protection against X-ray- and
neocarzinostatin-induced DNA damage. Free Radic Biol Med.
13:479–487. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou D, Shao L and Spitz DR: Reactive
oxygen species in normal and tumor stem cells. Adv Cancer Res.
122:1–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu J, Sun W, Song Y, Liu J, Xue F, Gong K,
Yang X and Kang Q: SIRT6 protects retinal ganglion cells against
hydrogen peroxide-induced apoptosis and oxidative stress by
promoting Nrf2/ARE signaling via inhibition of Bach1. Chem Biol
Interact. 300:151–158. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sajadimajd S and Khazaei M: Oxidative
stress and cancer: The role of Nrf2. Curr Cancer Drug Targets.
18:538–557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Deshmukh P, Unni S, Krishnappa G and
Padmanabhan B: The Keap1-Nrf2 pathway: Promising therapeutic target
to counteract ROS-mediated damage in cancers and neurodegenerative
diseases. Biophys Rev. 9:41–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Taguchi K and Yamamoto M: The KEAP1-NRF2
system in cancer. Front Oncol. 7:852017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hayes AJ, Skouras C, Haugk B and Charnley
RM: Keap1-Nrf2 signalling in pancreatic cancer. Int J Biochem Cell
Biol. 65:288–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ozaki N, Ohmuraya M, Hirota M, Ida S, Wang
J, Takamori H, Higashiyama S, Baba H and Yamamura K: Serine
protease inhibitor Kazal type 1 promotes proliferation of
pancreatic cancer cells through the epidermal growth factor
receptor. Mol Cancer Res. 7:1572–1581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang C, Wang L, Su B, Lu N, Song J, Yang
X, Fu W, Tan W and Han B: Serine protease inhibitor Kazal type 1
promotes epithelial-mesenchymal transition through EGFR signaling
pathway in prostate cancer. Prostate. 74:689–701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen YT, Tsao SC, Yuan SS, Tsai HP and
Chai CY: Serine protease inhibitor Kazal type 1 (SPINK1) promotes
proliferation of colorectal cancer through the epidermal growth
factor as a prognostic marker. Pathol Oncol Res. 21:1201–1208.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ateeq B, Tomlins SA, Laxman B, Asangani
IA, Cao Q, Cao X, Li Y, Wang X, Feng FY, Pienta KJ, et al:
Therapeutic targeting of SPINK1-positive prostate cancer. Sci
Transl Med. 3:72ra172011. View Article : Google Scholar : PubMed/NCBI
|