Introduction
There is mounting evidence to support the sex-biased
differences in the susceptibility, incidence and mortality of
cancer (1–3). The incidence of cancer and the
mortality rates were higher in men than in women in the USA between
2008 and 2014 (4). The
sex-specificity of cancer seems to be dependent on the type of
cancer. In certain types of cancer, such as melanoma, esophagus,
larynx, liver and bladder cancer, the incidence and mortality have
been demonstrated to be sex-specific (4–6). In
particular, there are prominent differences between men and women
in liver cancer, with almost three times higher incidence rate in
men than in women, according to the data reported in 2018 (4,7–9). On the other hand, in colorectal and
lung cancer, which are the most commonly diagnosed malignancies and
the most common leading cause of cancer-associated mortality
(7,10,11),
remarkable sex-specificity has not been reported.
In addition to lifestyle and living environments,
genetic factors also play an essential role in the wide disparity
between the sexes in terms of cancer incidence and mortality of
cancer (2,3,12–14). The
expression of cancer-associated genes was correlated with sex
differences in cancer and thus, these genes were designated as
sex-affected (3,12,14).
However, to the best of our knowledge, there is limited evidence
supporting sex differences in the correlation of gene
expression.
Cancer cell lines have been used to investigate the
underlying mechanisms, prevention, diagnosis and treatment of
cancer. The sex description of cell lines, however, is often
inadequate (15). The majority of
the animal models used for verification of in vitro results
primarily use male animal models, as the changes in hormone levels
associated with the female menstrual cycle can affect the
experimental results (16–18).
The epithelial-to-mesenchymal transition (EMT), a
cellular process involving changes in cell shape, adhesion and
movement, has been associated with cancer progression and
metastasis (19–21). The transdifferentiation of epithelial
cells into mesenchymal cells requires a number of molecular
changes, including the repression of E-cadherin, an epithelial
phenotype-associated protein (22–26).
Transcription factors, such as Snail 1 (SNAI1), Slug (SNAI2), Zinc
finger E-box binding homeobox (ZEB)1 and ZEB2, were revealed to
bind and repress the promoter region of the E-cadherin gene CDH1
(21). Vimentin (VIM), a major
constituent protein of the intermediate filament family, is known
to be overexpressed during the EMT process (27,28).
Discoidin domain receptors (DDRs) are receptor
tyrosine kinases that are activated by collagen (29). The roles of DDR1 in the EMT have been
reported in various types of cancer (30). DDR1 improved the stability of
E-cadherin via formation of the DDR1/E-cadherin complex (31) and triggered epithelial cell
differentiation by promoting cell adhesion through stabilizing
E-cadherin (32). Loss of DDR1 was
observed during the EMT in epithelial ovarian cancer (33). In contrast, DDR1 has been revealed to
induce the EMT in renal cancer cells (34). Previously, it has been demonstrated
that DDR1 was repressed by ZEB1 in breast epithelial cells
undergoing H-Ras-induced EMT (35).
In the present study, the sex differences in the
expression of EMT-associated genes CDH1, VIM, DDR1 and ZEB1 were
investigated in various cancer cell lines. Furthermore, the gender
specificity of the correlation between the expression of
EMT-associated genes was also examined.
Materials and methods
Cell culture
MCF7, MDA-MB-231 and Hs578T human breast cancer cell
lines were purchased from the Korean Cell Line Bank (KCLB; Korean
Cell Line Research Foundation) and cultured as previously described
(36). PC-3 and DU-145 human
prostate cancer cell lines, and SNU-840 and SK-OV-3 human ovarian
cancer cell lines were purchased from the KCLB; Korean Cell Line
Research Foundation and cultured in Roswell Park Memorial Institute
(RPMI) medium with L-glutamine and 25 mM HEPES, supplemented with
10% fetal bovine serum (Corning Inc.) and 100 µg/ml
penicillin-streptomycin. SNU-387, SNU-449 and SNU-878 human
hepatocellular carcinoma cell lines, SK-Hep1 human hepatic
adenocarcinoma cell line and HepG2 human hepatoblastoma cell line
were provided by Dr Sang Geon Kim (Seoul National University). The
cells used in this study were maintained in a humidified atmosphere
with 95% air and 5% CO2 at 37°C.
Comparison of mRNA expression
presented in public databases
The mRNA expression levels of CDH1, VIM, DDR1 and
ZEB1 in human cancer cell lines derived from the Broad-Novartis
Cancer Cell Line Encyclopedia (CCLE) database (portals.broadinstitute.org/ccle) were
compared in the present study. The sex origin of each cell line was
confirmed at the American Type Culture Collection website
(https://www.atcc.org) and SciCrunch (https://www.scicrunch.org). The correlation between
CDH1, VIM, DDR1 and ZEB1 was analyzed using Statistical Analysis
System (SAS 9.1.3; SAS Institute, Inc.) and represented as Pearson
correlation coefficients (r) and P-values to measure the
significance.
Reverse transcription-quantitative
(RT)-PCR
Total RNA was extracted from male- and
female-derived cancer cells using TRIsure™ (Bioline), according to
the manufacturer's protocol. The RNA was reverse-transcribed to
obtain cDNA using the Tetro Reverse Transcriptase cDNA Synthesis
kit (Bioline) according to the manufacturer's protocol. The PCR
cycling conditions and primer sequences for CDH1, VIM, DDR1, ZEB1
and β-actin were performed as previously described (35,37). The
primer sequences were as follows: Human CDH1: Forward,
5′-TCCATTTCTTGGTCTACGCC-3′, reverse, 5′-CACCTTCAGCCATCCTGTTT-3′;
VIM: Forward, 5′-GGCTCAGATTCAGGAACAGC-3′, reverse,
5′-GTCTCAACGGCAAAGTTCTC-3′; human DDR1: Forward,
5′-GGACATACCGTGGGCGGACT-3′, reverse, 5′-CCTAGGTTGTGGCGCATGG-3′;
human ZEB1: Forward, 5′-GCACAACCAAGTGCAGAAGA-3′, reverse,
5′-GAACCATTGGTGGTTGATCC-3′; and human β-actin: Forward,
5′-ACTCTTCCAGCCTTCCTT-3′, reverse, 5′-TCTCCTTCTGCATCCTGTC-3′. The
following amplification conditions were applied for the PCR of
human ZEB1 and CDH1: 94°C for 30 sec, 55°C for 30 sec and 72°C for
1 min for 27 cycles; human VIM, 94°C for 30 sec, 55°C for 30 sec
and 72°C for 45 sec for 28 cycles; and human DDR1, 94°C for 30 sec,
57°C for 30 sec, and 72°C for 1 min for 28 cycles. PCR product (10
µl) synthesized using the MyTaq™ kit (Bioline) were analyzed by
electrophoresis using gel with 1% agarose and 0.1 µg/ml ethidium
bromide (EtBr) and bands of DDR1 (320 bp), ZEB1 (284 bp), CDH1 (361
bp), VIM (327 bp) and β-actin (175 bp) were detected and quantified
using Image Lab™ Software (version 5.2; Bio-Rad Laboratories,
Inc.).
Immunoblot analysis
Immunoblot analysis was performed as previously
described (38). The antibodies used
in the present study included polyclonal rabbit anti-ZEB1 (cat. no.
sc-25388; 1:1,000), polyclonal rabbit anti-CDH1 (cat. no. sc-7870;
1:1,000), polyclonal rabbit anti-DDR1 (cat. no. sc-532; 1:1,000)
primary antibodies obtained from Santa Cruz Biotechnology, Inc. The
monoclonal rabbit anti-VIM primary antibody (cat. no. 5741S;
1:1,000) were obtained from Cell Signaling Technology, Inc. The
monoclonal mouse anti-β-actin primary antibody (cat. no. A2228;
1:4,000; Sigma-Aldrich; Merck KGaA) was also used. Goat anti-rabbit
(cat. no. 65-6120; 1:4,000) and goat anti-mouse (cat. no. 62-6520;
1:4,000) were purchased from Invitrogen (Thermo Fisher Scientific,
Inc.). Individual membranes were incubated with appropriately
diluted primary antibodies (1:1,000) overnight at 4°C. Horseradish
peroxidase (HRP)-conjugated secondary antibodies (1:4,000) were
then applied for 1.5 h at room temperature, following three
intensive washes in phosphate buffered saline with Tween 20 (PBST).
The protein expression levels were examined using enhanced
chemiluminescence (WesternBright™ ECL; Advansta Inc, Menlo Park,
CA, USA) and detected using the Gel Doc™ XR+ system (Bio-Rad
Laboratories, Inc.). Relative band intensities were determined by
the quantification of each band using Image Lab software (version
5.2; Bio-Rad Laboratories, Inc.).
In vitro transwell invasion assay
In vitro transwell invasion assays were
performed as previously described (39). Using a 24-well transwell insert with
membranes of 8.0-µm pores (Falcon; Corning Inc.), the lower part of
the membrane was covered with 0.5 mg/ml type I collagen (Corning
Inc.), and the upper part was covered in reconstituted basement
membrane material, 0.5 mg/ml matrigel (BD Biosciences) and then
dried. Complete medium (600 µl) was inserted into the bottom of the
well and 100 µl serum-free medium containing 5×104 cells
was placed in the upper chamber of the transwell. Cells were
incubated for 24 h in a humidified atmosphere with 5%
CO2 at 37°C. Following incubation, the invaded cells
attached to the membrane were stained by 0.5% crystal violet for 20
min at room temperature. For cell imaging, the filter membrane was
cut and fixed with Canada balsam (Junsei Chemical Co., Ltd.) on the
glass slide. The invaded cells in the lower side of the filter were
viewed under the by microscope (Olympus CK2) at ×100 magnification
and the images were captured with the camera attached to a
microscope (Olympus U-PMTVC). For quantitative measurements, the
membrane filter stained with crystal violet was cut out and eluted
with 30% methanol for 5 min. The absorbance was measured at a
wavelength of 595 nm.
In order to illustrate the correlation between the
invasiveness of each cell line and mRNA expression of
EMT-associated molecules, the levels of mRNA expression of
EMT-associated molecules were converted into logarithmic values.
The DDR1 index was defined as the logarithmic value of DDR1 divided
by ZEB1, based on the mRNA data of ZEB1, CDH1, VIM and DDR1
obtained from the RT-qPCR analysis. Similarly, CDH1 index and VIM
index were defined as the logarithmic value of CDH1 divided by
ZEB1, and that of VIM divided by ZEB1, respectively.
Kaplan-Meier survival curves
Analyses including the Kaplan-Meier survival curves
were calculated and plotted by The Kaplan Meier Plotter (http://kmplot.com) using the transcriptomic datasets,
the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), the European
Genome-phenome Archive (EGA; http://ega-archive.org/) and The Cancer Genome Atlas
(TCGA; version 2016_01_28; http://cancergenome.nih.gov/) repositories (40,41). The
data of female and male patients with liver cancer were split using
the auto select best cutoff criteria. Median mRNA expression levels
of DDR1 (female, 16.3903; male, 15.9004) and ZEB1 (female, 15.9808;
male, 15.9159) were used to differentiate between high expression
and low expression).
Statistical analysis
The correlations of gene expression were analyzed
using the Pearson and Spearman correlation with GraphPad Prism
(version 6.0; GraphPad Software, Inc.). Its R2 value
(the coefficient of determination), r value (Pearson correlation
coefficient), ρ value (Spearman correlation coefficient) and the
significance (two-tailed P-value) of each correlation analysis are
represented in Figures and listed in Table I. The significance of differences in
the invasive ability of cancer cell lines from in vitro
transwell invasion assay was analyzed using one-way analysis of
variance and with Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
 | Table I.Relative correlation of mRNA
expression between EMT-associated genes and ZEB1. |
Table I.
Relative correlation of mRNA
expression between EMT-associated genes and ZEB1.
|
|
|
| Correlation
coefficients (P-value) |
|---|
|
|
|
|
|
|---|
| Sex origin of cell
lines | Organ | Correlation | ZEB1 vs. Log
CDH1 | ZEB1 vs. Log
VIM | ZEB1 vs. Log
DDR1 |
|---|
| Female | Breast | Pearson (r) | −0.8154
(P<0.0010) | 0.6947
(P<0.0010) | −0.8210
(P<0.0010) |
|
|
| Spearman (ρ) | −0.5738
(P<0.0010) | 0.6144
(P<0.0010) | −0.6072
(P<0.0010) |
|
| Ovary | Pearson (r) | −0.7944
(P<0.0010) | 0.5823
(P<0.0010) | −0.7324
(P<0.0010) |
|
|
| Spearman (ρ) | −0.7743
(P<0.0010) | 0.6772
(P<0.0010) | −0.7703
(P<0.0010) |
|
| Liver | Pearson (r) | −0.9101
(P=0.2720) | −0.7579
(P=0.4524) | −0.8043
(P=0.4051) |
|
|
| Spearman (ρ) | −0.5000
(P>0.9999) | −1.000
(P=0.3333) | −1.000
(P=0.3333) |
|
| Colon | Pearson (r) | −0.8267
(P<0.0010) | 0.8074
(P<0.0010) | −0.6324
(P<0.0100) |
|
|
| Spearman (ρ) | −0.3168
(P=0.2002) | 0.6367
(P<0.0100) | −0.3829
(P=0.1168) |
|
| Lung | Pearson (r) | −0.8152
(P<0.0010) | 0.5817
(P<0.0010) | −0.7604
(P<0.0010) |
|
|
| Spearman (ρ) | −0.7997
(P<0.0010) | 0.6715
(P<0.0010) | −0.8132
(P<0.0010) |
| Male | Prostate | Pearson (r) | −0.7579
(P<0.0500) | 0.9089
(P<0.0100) | 0.1431
(P=0.7352) |
|
|
| Spearman (ρ) | −0.8095
(P<0.0500) | 0.8571
(P<0.0500) | 0
(P>0.9999) |
|
| Liver | Pearson (r) | −0.7583
(P<0.0010) | 0.4025
(P=0.0569) | 0.2164
(P=0.3214) |
|
|
| Spearman (ρ) | −0.5741
(P<0.0100) | 0.5474
(P<0.0100) | 0.1848
(P=0.3986) |
|
| Colon | Pearson (r) | −0.8898
(P<0.0010) | 0.7627
(P<0.0010) | −0.6699
(P<0.0010) |
|
|
| Spearman (ρ) | −0.5444
(P<0.0100) | 0.3020
(P=0.0987) | −0.1887
(P=0.3093) |
|
| Lung | Pearson (r) | −0.7730
(P<0.0010) | 0.4784
(P<0.0010) | −0.5250
(P<0.0010) |
|
|
| Spearman (ρ) | −0.7589
(P<0.0010) | 0.5000
(P<0.0010) | −0.5359
(P<0.0010) |
Results
Negative correlation between DDR1 and
ZEB1 is observed only in female-derived cancer cell lines by RT-PCR
and immunoblot analysis
In order to investigate the sex differences in the
expression and correlation of EMT-associated molecules, the mRNA
expression levels of CDH1, VIM, DDR1 and ZEB1 in human cancer cell
lines were detected (Fig. 1). RT-PCR
was performed in 7 female-derived cancer cell lines (MCF7, Hs578T,
MDA-MB-231, SK-OV-3, SNU840, SNU-387 and SNU-878; Fig. 1A) and 5 male-derived cancer cell
lines (PC-3, DU-145, SNU-449, HepG2 and SK-Hep1; Fig. 1B). In order to observe not only the
correlations between gene expression, but also the correlations
between protein expression, immunoblot analyses were conducted
using the same, aforementioned cell line samples (Fig. 1C and D).
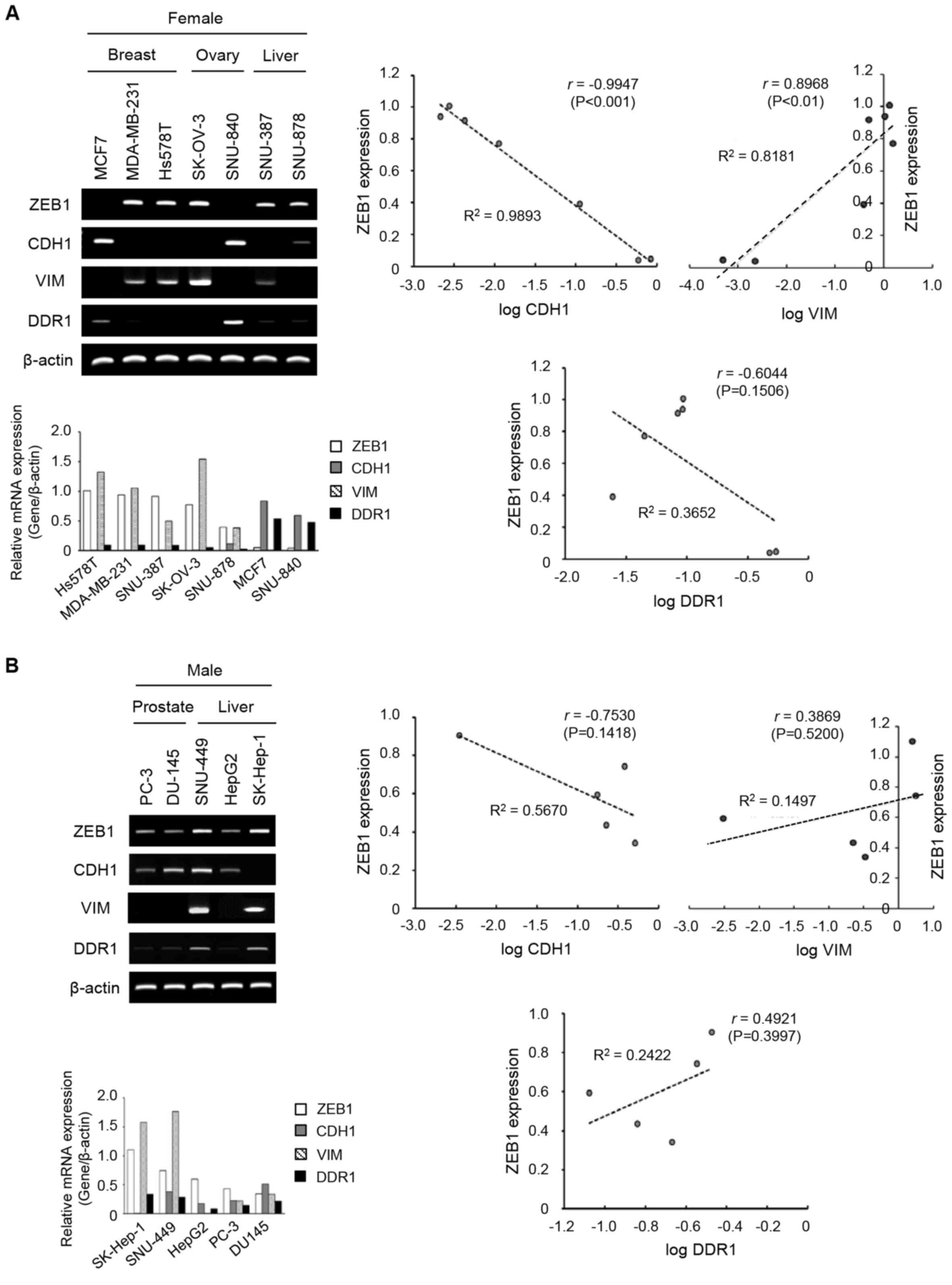 | Figure 1.mRNA and protein levels of ZEB1,
CDH1, VIM and DDR1 were detected by reverse transcription-PCR and
immunoblot analyses in cancer cell lines. Left-hand sides, the
bands of ZEB1, CDH1, VIM and DDR1 were quantified as relative
values to β-actin as a loading control. Right-hand sides, the level
of ZEB1 was plotted against the logarithmic values of CDH1, VIM or
DDR1. Each graph represents a trend line with its R2
value (the coefficient of determination) and r value (Pearson
correlation coefficient). The levels of mRNA expression in the
female- (A) and male- (B) derived cancer cell lines. The levels of
protein expression in the female- (C) and male- (D) derived cancer
cell lines. ZEB1, zing finger E-box binding homeobox; VIM,
vimentin; DDR1, discoidin domain receptor 1. |
These data were statistically analyzed using the
Pearson correlation coefficients of log CDH1, log VIM, or log DDR1
with ZEB1 and a curve was plotted. As presented in Fig. 1A, the plot of log CDH1 and ZEB1
revealed a negative linear correlation (r, −0.9947; P<0.001) and
log VIM and ZEB1 revealed a positive linear correlation (r, 0.8968;
P<0.010) in female-derived cell lines. A negative correlation
was observed between DDR1 and ZEB1 (r, −0.6044; P=0.1506) in
female-derived cell lines. In Fig.
1C, the correlation of protein expression of the EMT-associated
molecules revealed a similar pattern to that of mRNA expressions.
Expression of the same molecule was examined at the mRNA and
protein levels, with similar patterns but not significantly
consistent (Fig. 1). This can be
explained by the results of previous studies that the expression
levels of genes and proteins are not always consistent because of
cellular modification following gene expression (42).
As presented in Fig.
1B, male-derived cell lines, showed a tendency towards negative
correlation between log CDH1 and ZEB1 (r, −0.7530; P=0.1418), and a
tendency towards positive correlation between log VIM and ZEB1 (r,
0.3869 P=0.5200). The data demonstrated a trend of negative
correlation between CDH1 and DDR1 and a positive correlation
between VIM and DDR1 in the female- and male-derived cell lines
(Fig. 1A and B). Unlike
female-derived cell lines, the correlation between log DDR1 and
ZEB1 was positive in male-derived cell lines (r, 0.4921; P=0.3997;
Fig. 1B). In Fig. 1D, the correlation coefficients
between EMT-associated molecules and ZEB1 in male-derived cancer
cell lines was lower, which suggested that the correlation between
the two protein expression levels more decreased in male-derived
cancer cell lines than in female-derived cancer cell lines.
These data demonstrate that the correlation between
DDR1 and ZEB1 revealed a negative tendency specific to
female-derived cancer cell lines, although this was not
statistically significant.
Negative correlation between DDR1
index and invasive phenotype is observed in cancer cell lines in a
sex-specific manner
The differences in gene expression in regard to
their association with the invasive phenotype of cancer cell lines
was investigated in the present study. To this end, an in
vitro transwell invasion assay was performed and the invasive
ability from three independent experiments was quantified relative
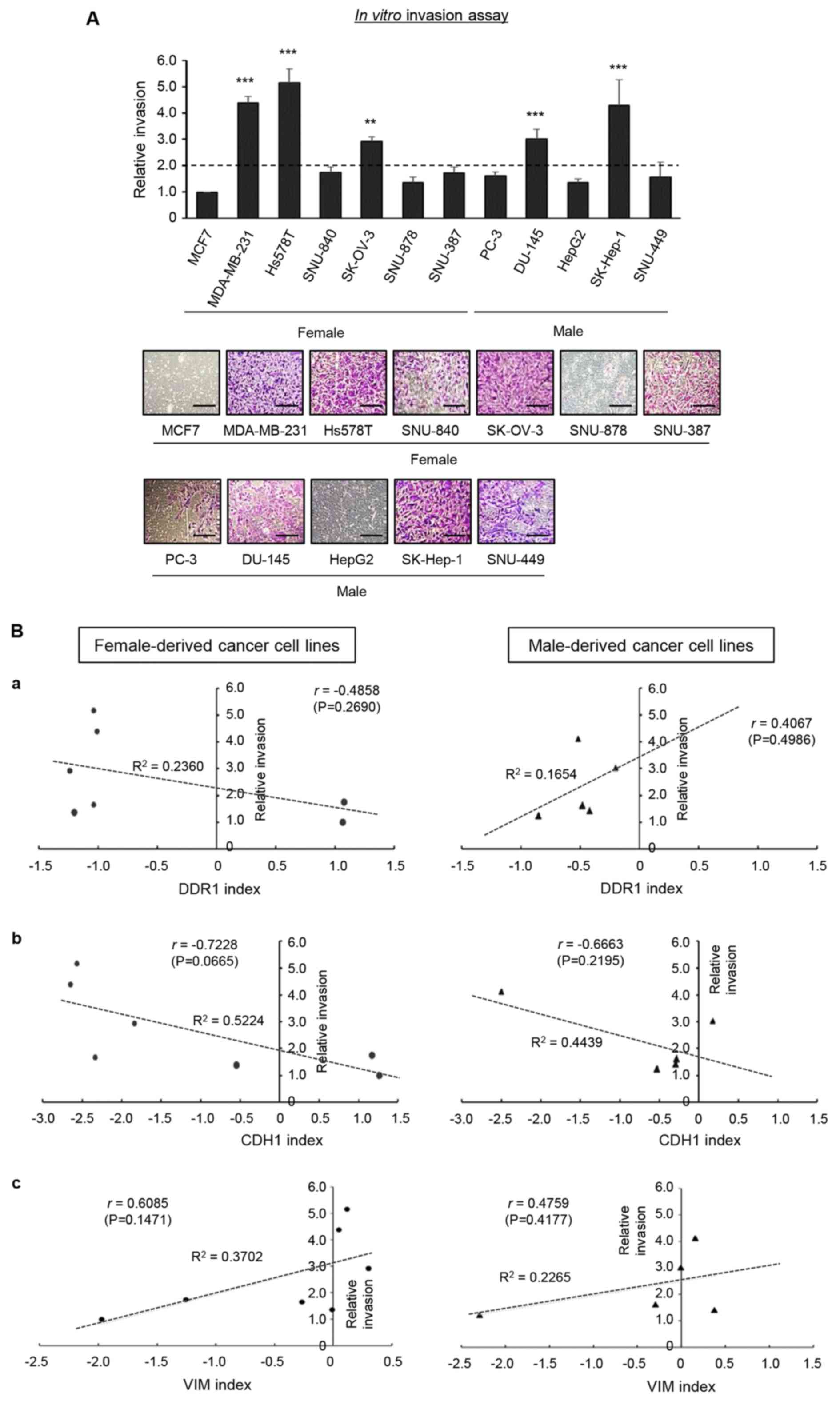
to the invasiveness of MCF7 cells (Fig.
2A). Since the MCF7 cell line is widely known to be
non-invasive (43), the invasiveness
of each cell line was compared to this cell line. The DDR1, CDH1
and VIM index was defined as the logarithmic value of each molecule
divided by ZEB1, based on the mRNA data of ZEB1, CDH1, VIM and DDR1
obtained from the RT-PCR analysis, respectively (Fig. 1A and B). A plot of the relative
invasion vs. DDR1 index revealed a negative correlation only in
female-derived cell lines, and not in male-derived cell lines
(Fig. 2B-a). A plot of the relative
invasion vs. CDH1 index or VIM index revealed that the relative
invasion was negatively correlated with CDH1 index and positively
correlated with VIM index in both female- and male-derived cancer
cell lines (Fig. 2B-b and B-c,
respectively). Although the results were not of statistical
significance, due to the limited number of samples, these results
suggest that DDR1 index may have the potential to serve as an
indicator of invasive phenotype of female-derived cancer cell
lines. However, as there were no sex-specific differences observed
in the correlations between invasion and CDH1 and VIM indices,
these would not be suitable as invasive phenotype indicators.
Analysis of CCLE database reveals a
sex-biased difference in the correlation between DDR1 and ZEB1 in
liver-derived cell lines
In order to expand the experimental results that
demonstrated a negative correlation of sex difference between DDR1
and ZEB1, the Broad-Novartis CCLE database was analyzed. As the
expression levels of DDR1 and ZEB1 in cancer cell lines were not
sex-biased, the correlation between the expression levels of DDR1
and ZEB1 was investigated. As presented in Fig. 3A, the gene expression profiles from
the CCLE database revealed that the mRNA levels of ZEB1 were
negatively correlated with DDR1 with a clear, negative linear
correlation in female-derived cancer cell lines. The results were
as follows: 58 breast (r, −0.8210; P<0.001), 52 ovary (r,
−0.7324; P<0.001), 3 female liver (r, −0.8043; P=0.4051), 18
female colon (r, −0.6324; P<0.01) and 48 female lung cancer cell
lines (r, −0.7604; P<0.001).
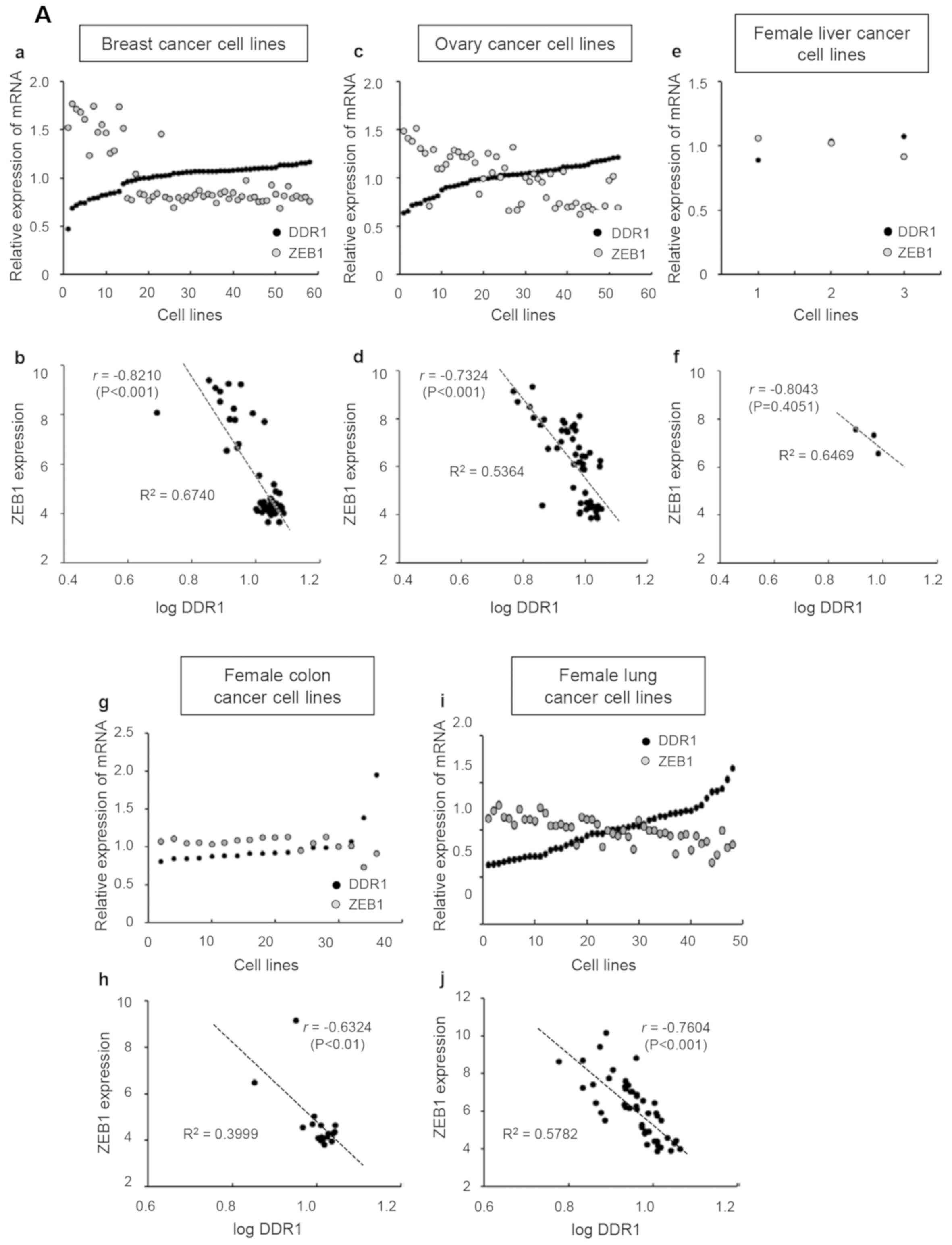 | Figure 3.Broad-Novartis Cancer Cell Line
Encyclopedia database analysis for mRNA levels of DDR1 and ZEB1 in
cancer cell lines. Each graph presents a trend line with its
R2 value (the coefficient of determination) and r value
(Pearson correlation coefficient). DDR1, discoidin domain receptor;
ZEB1, zinc finger E-box binding homeobox. (A) Female-derived cancer
cell lines. (a, c, e, g and i) mRNA value of DDR1 or ZEB1 in each
cell line was divided by the average value of mRNA expression and
plotted. Lower plots (b, d, f, h and j), ZEB1 expression levels
were plotted against the logarithmic values of DDR1 levels. DDR1,
discoidin domain receptor; ZEB1, zinc finger E-box binding
homeobox. (B) Male-derived cancer cell lines. (a, c, e and g) mRNA
value of DDR1 or ZEB1 in each cell line was divided by the average
value of mRNA expression and plotted. Lower plots (b, d, f and h),
ZEB1 expression levels were plotted against the logarithmic values
of DDR1 levels. . DDR1, discoidin domain receptor; ZEB1, zinc
finger E-box binding homeobox. |
In male-derived cancer cell lines (8 prostate, 23
male liver, 31 male colon and 119 male lung cancer cell lines),
however, the negative correlation between ZEB1 and DDR1 was
observed only in male colon-derived (r, −0.6699; P<0.001) and
male lung-derived cell lines (r, −0.5250; P<0.001) (Fig. 3B). There was a positive correlation
between ZEB1 and DDR1 in male prostate- and liver-derived cell
lines (r, 0.1431; r, 0.2163, respectively) but these values were
not significant as follows: Prostate (r, 0.1431; P=0.7352) and male
liver (r, 0.2164; P=0.3214). Among the analyzed cancer types
between the two sexes (liver, colon and lung cancers), only
liver-derived cell lines revealed a sex-biased difference between
DDR1 and ZEB1. Given that the marked difference between men and
women were observed in liver cancer, but not in colon and lung
cancers, the results of this analysis suggest that the correlation
between DDR1 and ZEB1 may be used for investigating the types of
cancer that are sex-biased.
Both female- and male-derived cell
lines from CCLE database present a negative correlation between
CDH1 and ZEB1, and a positive correlation between VIM and ZEB1
In female-derived cancer cell lines, the mRNA levels
of ZEB1 were negatively correlated with CDH1 (Fig. 4A-a, A-c, A-e, A-g and A-i). The plot
of log CDH1 and ZEB1 in these cell lines revealed a negative linear
correlation (Fig. 4A-b, A-d, A-f, A-h
and A-j). The r values of log CDH1 with ZEB1 in female breast-,
ovary-, liver-, colon- and lung-derived cell lines were −0.8154
(P<0.001), −0.7944 (P<0.001), −0.9101 (P=0.2720), −0.8267
(P<0.001) and −0.8152 (P<0.001), respectively. The mRNA
levels of ZEB1 in male-derived cancer cell lines were also
negatively correlated with CDH1 (Fig.
4B-a, B-c, B-e and B-g). The plot of log CDH1 and ZEB1 in these
cell lines revealed a negative linear association (Fig. 4B-b, B-d, B-f and B-h). The r values
of the male prostate-, liver-, colon- and lung-derived cell lines
were −0.7579 (P<0.05), −0.7583 (P<0.001), −0.8898
(P<0.001) and −0.7730 (P<0.001), respectively.
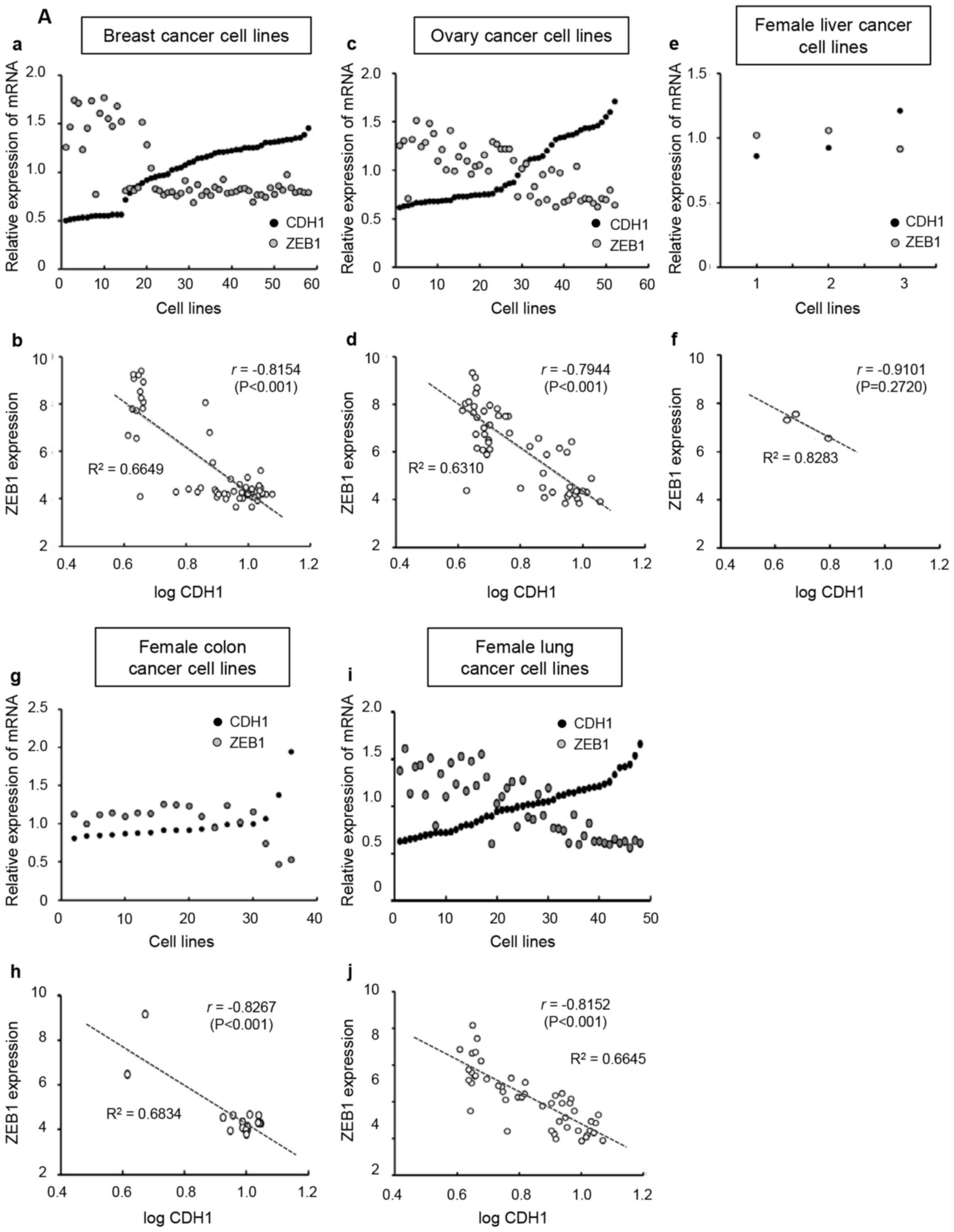 | Figure 4.Broad-Novartis Cancer Cell Line
Encyclopedia database analysis for mRNA levels of CDH1 and ZEB1 in
cancer cell lines. Each graph presents a trend line with its R2
value (the coefficient of determination) and r value (Pearson
correlation coefficient). (A) Female-derived cancer cell lines. (a,
c, e, g and i) mRNA value of CDH1 or ZEB1 in each cell line was
divided by the average value of mRNA expression and plotted. Lower
plots (b, d, f, h and j), ZEB1 expression levels were plotted
against the logarithmic values of CDH1 levels. CDH1, E-cadherin;
ZEB1, zinc finger E-box binding homeobox. (B) Male-derived cancer
cell lines. (a, c, e and g) mRNA value of CDH1 or ZEB1 in each cell
line was divided by the average value of mRNA expression and
plotted. Lower plots (b, d, f and h), ZEB1 expression levels were
plotted against the logarithmic values of CDH1 levels. |
The correlation between VIM and ZEB1 revealed a
positive linear relationship in the female-derived (Fig. 5A), except female liver cancer cell
lines, and male-derived (Fig. 5B)
cell lines. The r values of log VIM with ZEB1 in female breast-,
ovary-, liver-, colon- and lung-derived cell lines were 0.6947
(P<0.001), 0.5823 (P<0.001), −0.7579 (P=0.4524), 0.8074
(P<0.001) and 0.5817 (P<0.001), respectively. The r values of
log VIM and ZEB1 in male prostate-, liver-, colon- and lung-derived
cell lines were 0.9089 (P<0.01), 0.4025 (P=0.0569), 0.7627
(P<0.001) and 0.4784 (P<0.001), respectively. Taken together,
the data from the CCLE databases revealed that there were no
sex-specific differences in the correlation between CDH1 and ZEB1,
as well as in that between VIM and ZEB1.
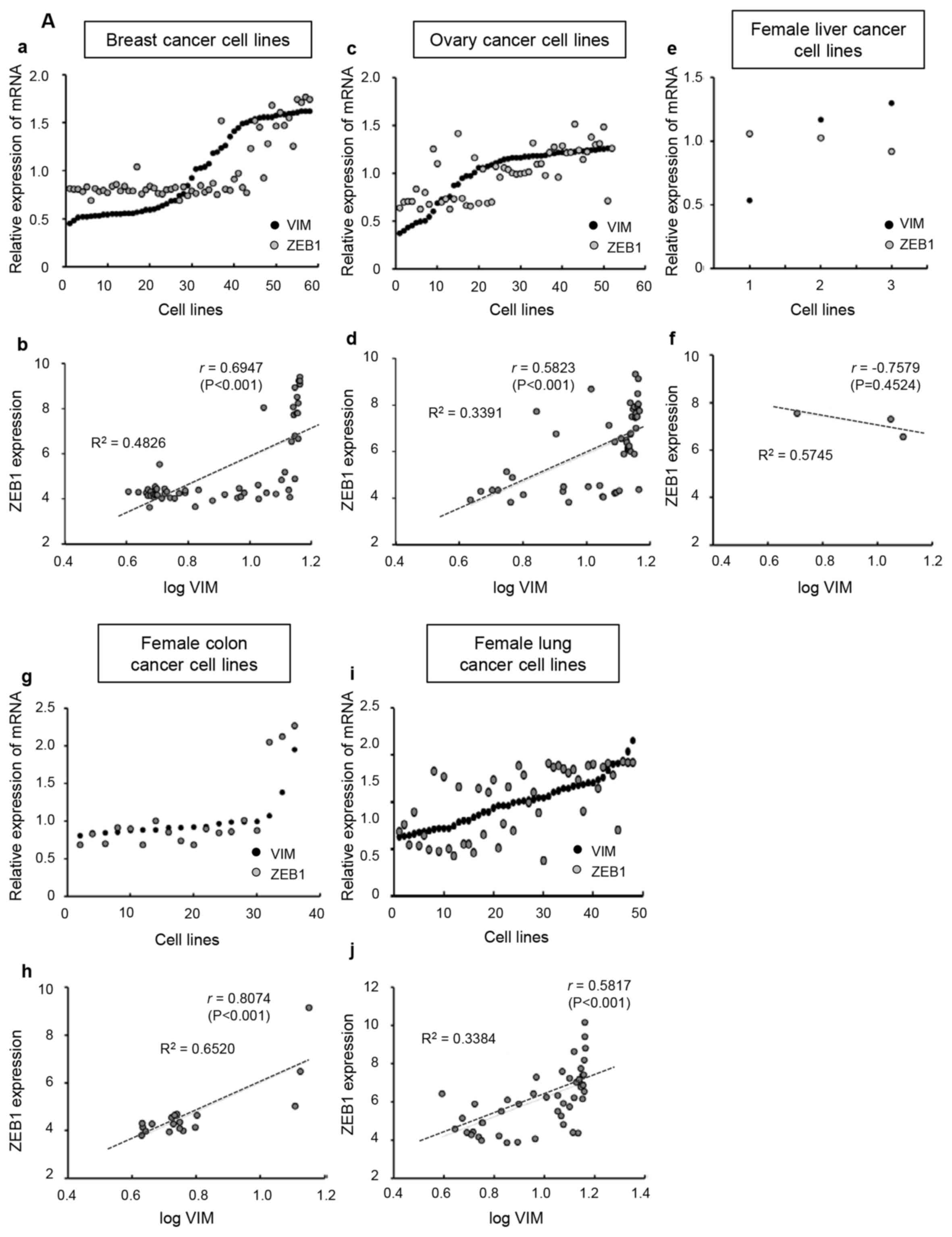 | Figure 5.Broad-Novartis Cancer Cell Line
Encyclopedia database analysis for mRNA levels of VIM and ZEB1 in
cancer cell lines. Each graph presents a trend line with its
R2 value (the coefficient of determination) and r value
(Pearson correlation coefficient). (A) Female-derived cancer cell
lines. (a, c, e, g and i) the mRNA value of VIM or ZEB1 in each
cell line was divided by the average value of mRNA expression and
plotted. (b, d, f, h and j) ZEB1 expression levels were plotted
against the logarithmic values of VIM levels. VIM, vimentin; ZEB1,
zinc finger E-box binding homeobox. (B) Male-derived cancer cell
lines. (a, c, e and g) the mRNA value of VIM or ZEB1 in each cell
line was divided by the average value of mRNA expression and
plotted. (b, d, f and h) ZEB1 expression levels were plotted
against the logarithmic values of VIM levels. VIM, vimentin; ZEB1,
zinc finger E-box binding homeobox. |
Relative correlation of mRNA
expression between EMT-associated genes and ZEB1 by Pearson and
Spearman correlation coefficients
In order to evaluate the relative correlation of
mRNA expression between EMT-associated genes and ZEB1, both the
Pearson and Spearman correlation coefficients were calculated
(Table I). Out of the total 27
correlation sets, 5 sets revealed differences in the correlation
coefficients of Pearson and Spearman. The expression levels of CDH1
and ZEB1 were negatively correlated, and those of VIM and ZEB1 were
positively correlated, regardless of the sexual origin of cancer
cell lines (Table I). In contrast,
the negative correlation between DDR1 and ZEB1 in cell lines was
observed in a female-specific manner in cancer cell lines such as
female-specific organ cancers and liver cancer. These data revealed
that the negative or positive correlation between DDR1 and ZEB1
indicated a sex-biased correlation in liver cancer cell lines
specifically.
Analysis of TCGA database reveals a
sex-biased difference in the correlation between DDR1 expression
level and overall survival in female or male patients with liver
cancer
The transcriptomic datasets, such as GEO, EGA and
TCGA, were analyzed for the gene expression levels of DDR1/ZEB1 and
overall survival time in female and male patients with liver cancer
(40,41). The Kaplan-Meier survival curves were
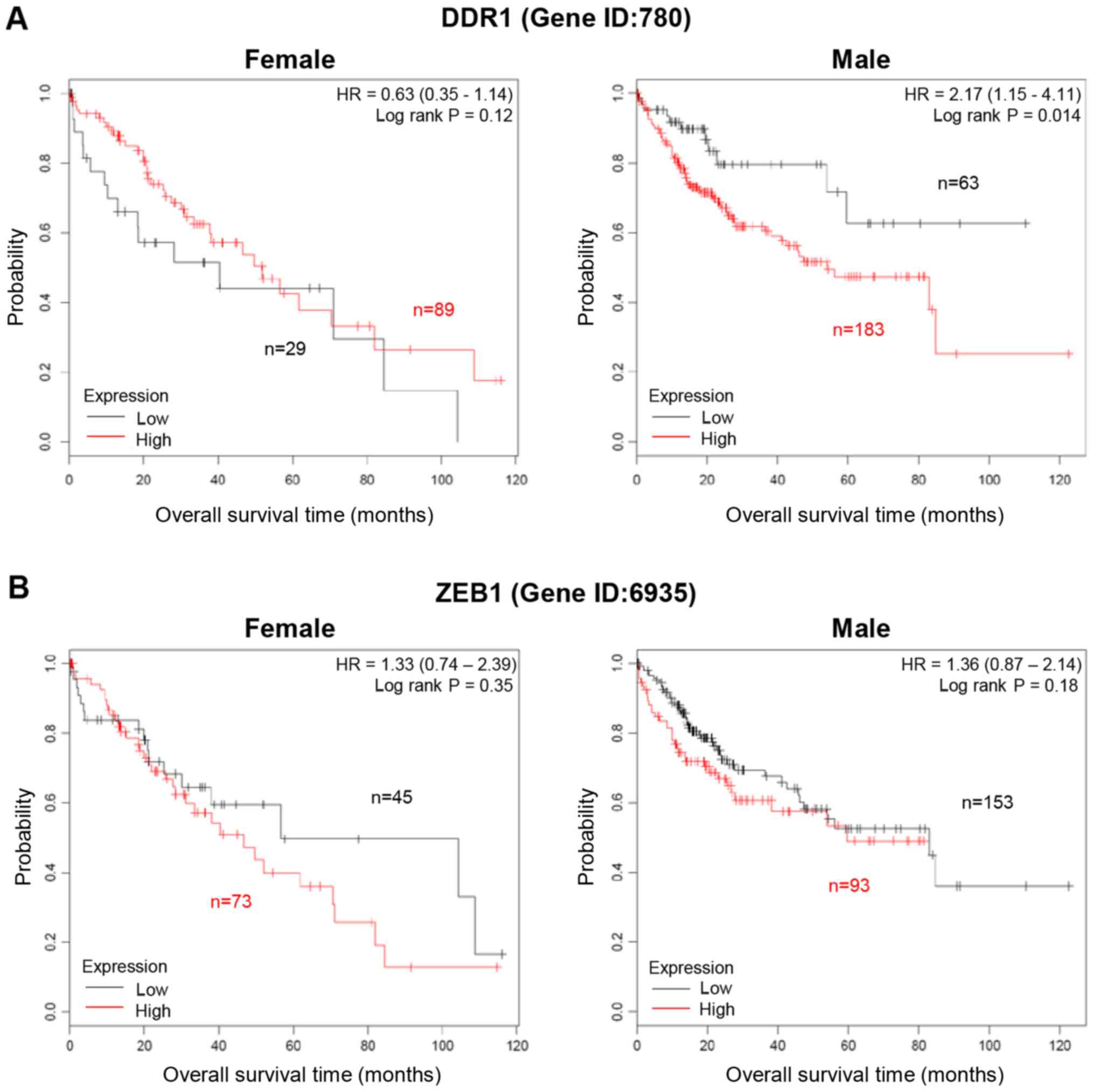
obtained using the Kaplan-Meier Plotter program (Fig. 6). In Fig.
6A, high expression level of DDR1 revealed an improved survival
time in female patients with liver cancer (Hazard ratio, HR=0.63,
logrank P=0.12). In male patients with liver cancer however, high
levels of DDR1 revealed a worse survival time (HR, 2.17; logrank
P=0.014). High levels of ZEB1 revealed a worse overall survival
time in both female and male patients with liver cancer although
not specific, logrank P=0.35 and 0.18, respectively. (Fig. 6B). These data suggest a sex-biased
difference in correlation between DDR1 expression pattern and
overall survival time in patients with liver cancer although not
all of the KM plots analyzed showed demonstrated significant
results.
Discussion
The differences in incidence, progression and
mortality rates of cancer between each sex have been well
documented (1–3). Sex differences in cancer have recently
been characterized at the molecular level (14). The present study aimed to elucidate
the molecular basis for sex-biased gene expression in cancer. The
mRNA expression of EMT-associated genes including CDH1, VIM, DDR1
and ZEB1 in female- and male-derived cancer cell lines were
investigated experimentally (Fig. 1)
and using CCLE database analysis (Figs.
3–5) of human cancer cell lines
and Kaplan-Meier survival curves of female or male patients with
liver cancer (Fig. 6). The present
study revealed that the expression levels of these genes in cancer
cell lines were not sex-biased, which was consistent with a
previous report (14).
In order to evaluate the relative correlation of
mRNA expression between EMT-associated genes and ZEB1, the Pearson
and Spearman correlation coefficients were calculated (Table I). As presented in Table I, the expression levels of CDH1 and
ZEB1 were negatively correlated, and those of VIM and ZEB1 were
positively correlated, regardless of sexual origin of cancer cell
lines (Table I). However, two
correlation coefficients of ZEB1 and logVIM were calculated as a
negative value only in female liver cancer cell lines. This may be
due to the low number of cell lines used in the study because there
are relatively few studies on female liver cancer compared to many
studies on the risk of male liver cancer. Therefore, further
studies on female liver cancer are needed.
In contrast, the negative correlation between DDR1
and ZEB1 in cancer cell lines was observed in a female-specific
manner in cell lines of female-specific organs, such as breast and
ovary cancer, and liver cancer, where the significant sex
difference was reported (4–6). The correlation between DDR1 and ZEB1
were negative in both male- and female-derived cancer cell lines of
colon and lung cancer. Notably, recent reports have revealed that
the incidence and mortality of colon and lung cancer were more
likely to be associated with socioeconomic development, and not
with sex (44,45). The sex-biased correlation between
DDR1 and ZEB1 in liver cancer cell lines suggests that the
regulatory mechanism of gene expression, rather than the expression
itself, may be affected by sex.
Invasiveness is an important step in the malignant
transformation of cells (46). CDH1,
VIM, DDR1 and ZEB1 are associated with the invasive phenotype of
cancer cells (27,30,36,47–49). To
simplify the CDH1-, VIM- or DDR1-to-ZEB1 ratios, the CDH1, VIM, or
the DDR1 indices were determined in the present study. The data
suggest that the DDR1 index may be a sex-specific indicator of the
invasive potential of cancer cell lines, while the correlation
between the CDH1 index or the VIM index and relative invasion was
not affected by the origin of sex, and is therefore not a suitable
indicator (Fig. 2).
The correlation between DDR1 index and invasiveness
may be associated with sex. Genetic factors, environmental causes
and sex hormones may contribute to sex disparity in susceptibility
to cancer (1,3,12,13). As
all cell lines used in research can be assigned to a sex (50), cells derived from females or males
are different in cellular biochemistry and physiology due to the
presence of sex chromosomes (51).
The effect of hormones on the sex-specific gene expression of a
cell line has been controversial. Several studies have suggested
that hormones affect the gene expression profiles of a cell line
(12,13), while other studies have demonstrated
that the sex-specific genetic differences in cell lines existed
prior to hormone exposure (52–54). It
has been demonstrated in a number of genes that sexually dimorphic
gene expression was associated with the presence of the X or Y
chromosomes, and not with hormones (55,56). As
the cell lines were not treated with hormones in the present study,
it can be suggested that the sex-specific difference of correlation
between DDR1 and ZEB1 expression may not be due to the effects of
hormones. The human clinical data from GEO, EGA and TCGA databases
were also analyzed for the expressions of DDR1 and ZEB1 in patients
with liver cancer. These human data reflect the effect of hormones.
Although the databases do not directly address the cause of these
differences, several studies using the databases mentioned hormones
as the cause of these differences in liver cancer (57,58).
A recent study revealed the sex-biased molecular
signatures affecting cancer (14).
The present study demonstrated sex-biased differences in the
correlation of EMT-associated gene expression. Comparative studies
using gene-expression data of various types of cancer reported that
the expression patterns of certain genes were specific to
individual cancers depending on the type (4,59,60). The
results from the present study demonstrate that the negative
correlation of DDR1 and ZEB1 expression was observed only in
certain types of cancer, which may be explained by the
cancer-specific expression patterns of certain genes. A
sex-specific correlation was observed between DDR1 and ZEB1 in cell
lines derived from liver cancer, which is already known to be
sex-biased (4,5), whereas it was not detected in cell
lines from colon or lung cancers in which the obvious sex
dependence has not previously been observed (10,11). The
results of the present study imply that the sex-biased regulation
of gene expression may contribute to the sex differences in the
susceptibility and progression of cancer. The present study
suggests a potential application of the correlation between DDR1
and ZEB1 for investigating sex-biased cancers, such as liver
cancer. These results may provide useful information for
establishing sex-specific strategies for diagnosis and therapy of
cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Bio &
Medical Technology Development Program (grant no. 2015M3A9B6074045)
of the National Research Foundation funded by the Korean
government.
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SYK and SL designed and conducted the study and
wrote the manuscript. EL, HL and JYS conducted the study and helped
to write the manuscript. JJ and SGK designed the study and helped
to analyze the data. AM conceived and designed the study as
corresponding author of the manuscript. All authors reviewed and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zahm SH and Fraumeni JF Jr: Racial,
ethnic, and gender variations in cancer risk: Considerations for
future epidemiologic research. Environ Health Perspect. 103 (Suppl
8):283–286. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kirsch-Volders M, Bonassi S, Herceg Z,
Hirvonen A, Möller L and Phillips DH: Gender-related differences in
response to mutagens and carcinogens. Mutagenesis. 25:213–221.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dorak MT and Karpuzoglu E: Gender
differences in cancer susceptibility: An inadequately addressed
issue. Front Genet. 3:2682012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127:S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oh CM, Cho H, Won YJ, Kong HJ, Roh YH,
Jeong KH and Jung KW: Nationwide trends in the incidence of
melanoma and non-melanoma skin cancers from 1999 to 2014 in South
Korea. Cancer Res Treat. 50:729–737. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jung KW, Won YJ, Kong HJ and Lee ES:
Prediction of cancer incidence and mortality in Korea, 2018. Cancer
Res Treat. 50:317–323. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryerson AB, Eheman CR, Altekruse SF, Ward
JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM,
et al: Annual Report to the Nation on the Status of Cancer,
1975–2012, featuring the increasing incidence of liver cancer.
Cancer. 122:1312–1337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao RN, Neutel CI and Wai E: Gender
differences in colorectal cancer incidence, mortality,
hospitalizations and surgical procedures in Canada. J Public Health
(Oxf). 30:194–201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thun MJ, Hannan LM, Adams-Campbell LL,
Boffetta P, Buring JE, Feskanich D, Flanders WD, Jee SH, Katanoda
K, Kolonel LN, et al: Lung cancer occurrence in never-smokers: An
analysis of 13 cohorts and 22 cancer registry studies. PLoS Med.
5:e1852008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cook MB, Dawsey SM, Freedman ND, Inskip
PD, Wichner SM, Quraishi SM, Devesa SS and McGlynn KA: Sex
disparities in cancer incidence by period and age. Cancer Epidemiol
Biomarkers Prev. 18:1174–1182. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klein SL: The effects of hormones on sex
differences in infection: From genes to behavior. Neurosci Biobehav
Rev. 24:627–638. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao
H, Li J, Mills GB, Shu Y, Li L and Liang H: Comprehensive
characterization of molecular differences in cancer between male
and female patients. Cancer Cell. 29:711–722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park MN, Park JH, Paik HY and Lee SK:
Insufficient sex description of cells supplied by commercial
vendors. Am J Physiol Cell Physiol. 308:C578–C580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Becker JB, Arnold AP, Berkley KJ,
Blaustein JD, Eckel LA, Hampson E, Herman JP, Marts S, Sadee W,
Steiner M, et al: Strategies and methods for research on sex
differences in brain and behavior. Endocrinology. 146:1650–1673.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Keitt SK, Fagan TF and Marts SA:
Understanding sex differences in environmental health: A thought
leaders' roundtable. Environ Health Perspect. 112:604–609. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zucker I and Beery AK: Males still
dominate animal studies. Nature. 465:6902010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat (Basel). 154:8–20.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalluri R and Neilson EG:
Epithelial-mesenchymal transition and its implications for
fibrosis. J Clin Invest. 112:1776–1784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grooteclaes ML and Frisch SM: Evidence for
a function of CtBP in epithelial gene regulation and anoikis.
Oncogene. 19:3823–3828. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Comijn J, Berx G, Vermassen P, Verschueren
K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D and van Roy
F: The two-handed E box binding zinc finger protein SIP1
downregulates E-cadherin and induces invasion. Mol Cell.
7:1267–1278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guaita S, Puig I, Franci C, Garrido M,
Dominguez D, Battle E, Sancho E, Dedhar S, De Herreros AG and
Baulida J: Snail induction of epithelial to mesenchymal transition
in tumor cells is accompanied by MUC1 repression and ZEB1
expression. J Biol Chem. 277:39209–39216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hajra KM, Chen DY and Fearon ER: The SLUG
zinc-finger protein represses E-cadherin in breast cancer. Cancer
Res. 62:1613–1618. 2002.PubMed/NCBI
|
|
27
|
Gilles C and Thompson EW: The epithelial
to mesenchymal transition and metastatic progression in carcinoma.
Breast J. 2:83–96. 1996. View Article : Google Scholar
|
|
28
|
Thompson EW, Newgreen DF and Tarin D:
Carcinoma invasion and metastasis: A role for
epithelial-mesenchymal transition? Cancer Res. 65:5991–5995,
discusion 5995. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu HL, Valiathan RR, Arkwright R, Sohail
A, Mihai C, Kumarasiri M, Mahasenan KV, Mobashery S, Huang P,
Agarwal G and Fridman R: Discoidin domain receptors: Unique
receptor tyrosine kinases in collagen-mediated signaling. J Biol
Chem. 288:7430–7437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valiathan RR, Marco M, Leitinger B, Kleer
CG and Fridman R: Discoidin domain receptor tyrosine kinases: New
players in cancer progression. Cancer Metastasis Rev. 31:295–321.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang CZ, Yeh YC and Tang MJ:
DDR1/E-cadherin complex regulates the activation of DDR1 and cell
spreading. Am J Physiol Cell Physiol. 297:C419–C429. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yeh YC, Wu CC, Wang YK and Tang MJ: DDR1
triggers epithelial cell differentiation by promoting cell adhesion
through stabilization of E-cadherin. Mol Biol Cell. 22:940–953.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chung VY, Tan TZ, Huang RL, Lai HC and
Huang RY: Loss of discoidin domain receptor 1 (DDR1) via CpG
methylation during EMT in epithelial ovarian cancer. Gene.
635:9–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song J, Chen X, Bai J, Liu Q, Li H, Xie J,
Jing H and Zheng J: Discoidin domain receptor 1 (DDR1), a promising
biomarker, induces epithelial to mesenchymal transition in renal
cancer cells. Tumour Biol. 37:11509–11521. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koh M, Woo Y, Valiathan RR, Jung HY, Park
SY, Kim YN, Kim HR, Fridman R and Moon A: Discoidin domain receptor
1 is a novel transcriptional target of ZEB1 in breast epithelial
cells undergoing H-Ras-induced epithelial to mesenchymal
transition. Int J Cancer. 136:E508–E520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jeon YR, Kim SY, Lee EJ, Kim YN, Noh DY,
Park SY and Moon A: Identification of annexin II as a novel
secretory biomarker for breast cancer. Proteomics. 13:3145–3156.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cha Y, Kang Y and Moon A: HER2 induces
expression of leptin in human breast epithelial cells. BMB Rep.
45:719–723. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shin I, Kim S, Song H, Kim HR and Moon A:
H-Ras-specific activation of Rac-MKK3/6-p38 pathway: Its critical
role in invasion and migration of breast epithelial cells. J Biol
Chem. 280:14675–14683. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim MS, Lee EJ, Kim HR and Moon A: p38
kinase is a key signaling molecule for H-Ras-induced cell motility
and invasive phenotype in human breast epithelial cells. Cancer
Res. 63:5454–5461. 2003.PubMed/NCBI
|
|
40
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. Royal Soc Open
Sci. 5:1810062018. View Article : Google Scholar
|
|
41
|
Cancer Genome Atlas Research Network.
Electronic address, . simplewheeler@bcm.edu; Cancer
Genome Atlas Research Network: Comprehensive and integrative
genomic characterization of hepatocellular carcinoma. Cell.
169:1327.e23–1341.e23. 2017.
|
|
42
|
Gygi SP, Rochon Y, Franza BR and Aebersold
R: Correlation between protein and mRNA abundance in yeast. Mol
Cell Biol. 19:1720–1730. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sommers CL, Byers SW, Thompson EW, Torri
JA and Gelmann EP: Differentiation state and invasiveness of human
breast cancer cell lines. Breast Cancer Res Treat. 31:325–335.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wong MCS, Lao XQ, Ho KF, Goggins WB and
Tse SLA: Incidence and mortality of lung cancer: Global trends and
association with socioeconomic status. Sci Rep. 7:143002017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fidler IJ: Critical factors in the biology
of human cancer metastasis: Twenty-eighth G.H.A. Clowes memorial
award lecture. Cancer Res. 50:6130–6138. 1990.PubMed/NCBI
|
|
47
|
Vleminckx K, Vakaet L Jr, Mareel M, Fiers
W and Van Roy F: Genetic manipulation of E-cadherin expression by
epithelial tumor cells reveals an invasion suppressor role. Cell.
66:107–119. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Perl AK, Wilgenbus P, Dahl U, Semb H and
Christofori G: A causal role for E-cadherin in the transition from
adenoma to carcinoma. Nature. 392:190–193. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Spaderna S, Schmalhofer O, Wahlbuhl M,
Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A,
et al: The transcriptional repressor ZEB1 promotes metastasis and
loss of cell polarity in cancer. Cancer Res. 68:537–544. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wizemenn TM and Pardue ML: Exploring the
biological contributions to human health:Does sex matter?
Washington DC: Natl Acad Press; 2001
|
|
51
|
Miller VM: In pursuit of scientific
excellence: Sex matters. Adv Physiol Educ. 36:83–84. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Avery B, Jørgensen CB, Madison V and Greve
T: Morphological development and sex of bovine in vitro-fertilized
embryos. Mol Reprod Dev. 32:265–270. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pergament E, Fiddler M, Cho N, Johnson D
and Holmgren WJ: Sexual differentiation and preimplantation cell
growth. Hum Reprod. 9:1730–1732. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu KP, Yadav BR, King WA and Betteridge
KJ: Sex-related differences in developmental rates of bovine
embryos produced and cultured in vitro. Mol Reprod Dev. 31:249–252.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang X, Schadt EE, Wang S, Wang H, Arnold
AP, Ingram-Drake L, Drake TA and Lusis AJ: Tissue-specific
expression and regulation of sexually dimorphic genes in mice.
Genome Res. 16:995–1004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shah K, McCormack CE and Bradbury NA: Do
you know the sex of your cells? Am J Physiol Cell Physiol.
306:C3–C18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ge H, Yan Y, Tian F, Wu D and Huang Y:
Prognostic value of estrogen receptor α and estrogen receptor β in
gastric cancer based on a meta-analysis and The Cancer Genome Atlas
(TCGA) datasets. Int J Surg. 53:24–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang X, Shu C, Chen L and Yao B: Impact
of sex, body mass index and initial pathologic diagnosis age on the
incidence and prognosis of different types of cancer. Oncol Rep.
40:1359–1369. 2018.PubMed/NCBI
|
|
59
|
Xu K, Cui J, Olman V, Yang Q, Puett D and
Xu Y: A comparative analysis of gene-expression data of multiple
cancer types. PLoS One. 5:e136962010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Martinez-Ledesma E, Verhaak RG and Treviño
V: Identification of a multi-cancer gene expression biomarker for
cancer clinical outcomes using a network-based algorithm. Sci Rep.
23:119662015. View Article : Google Scholar
|