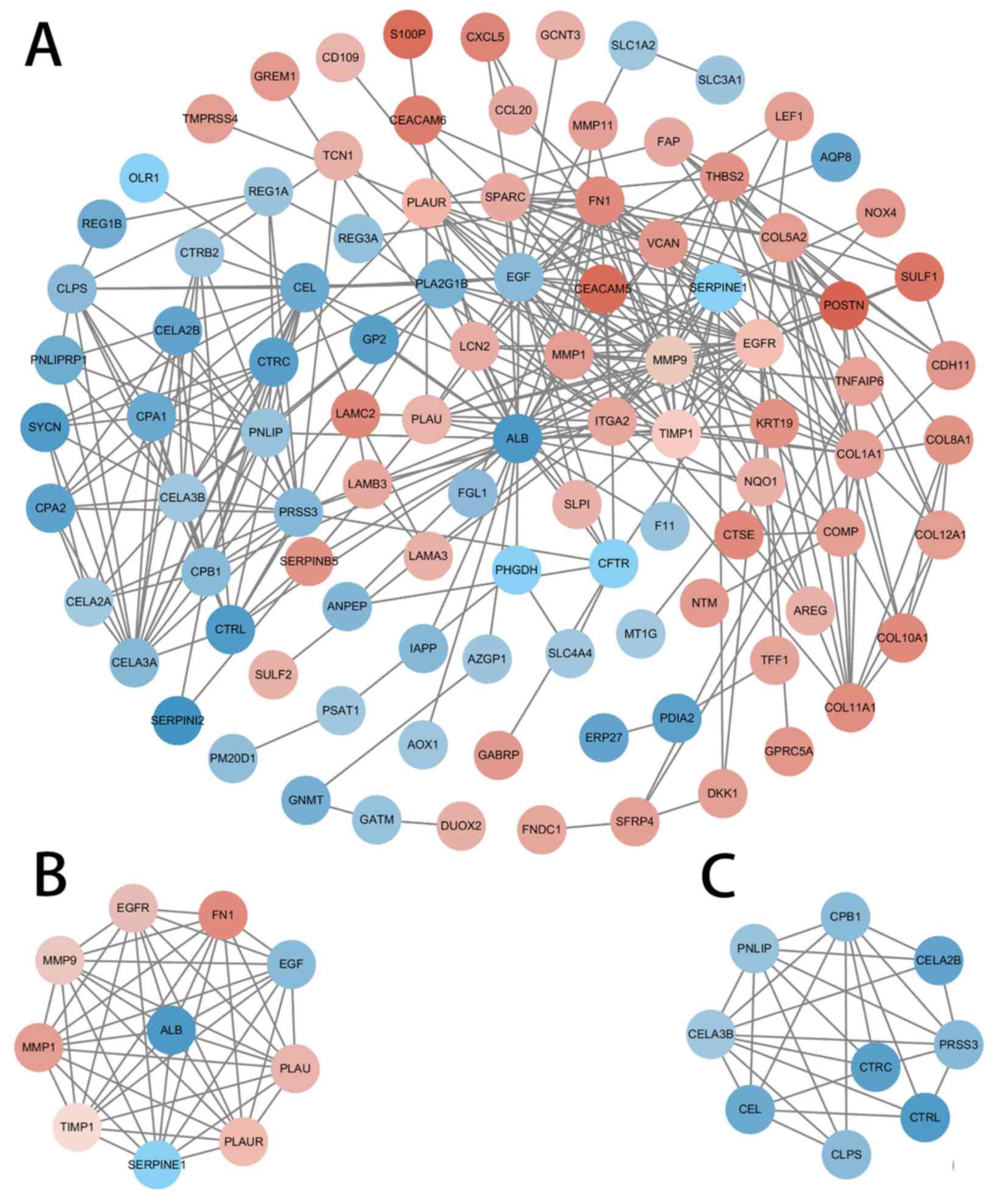
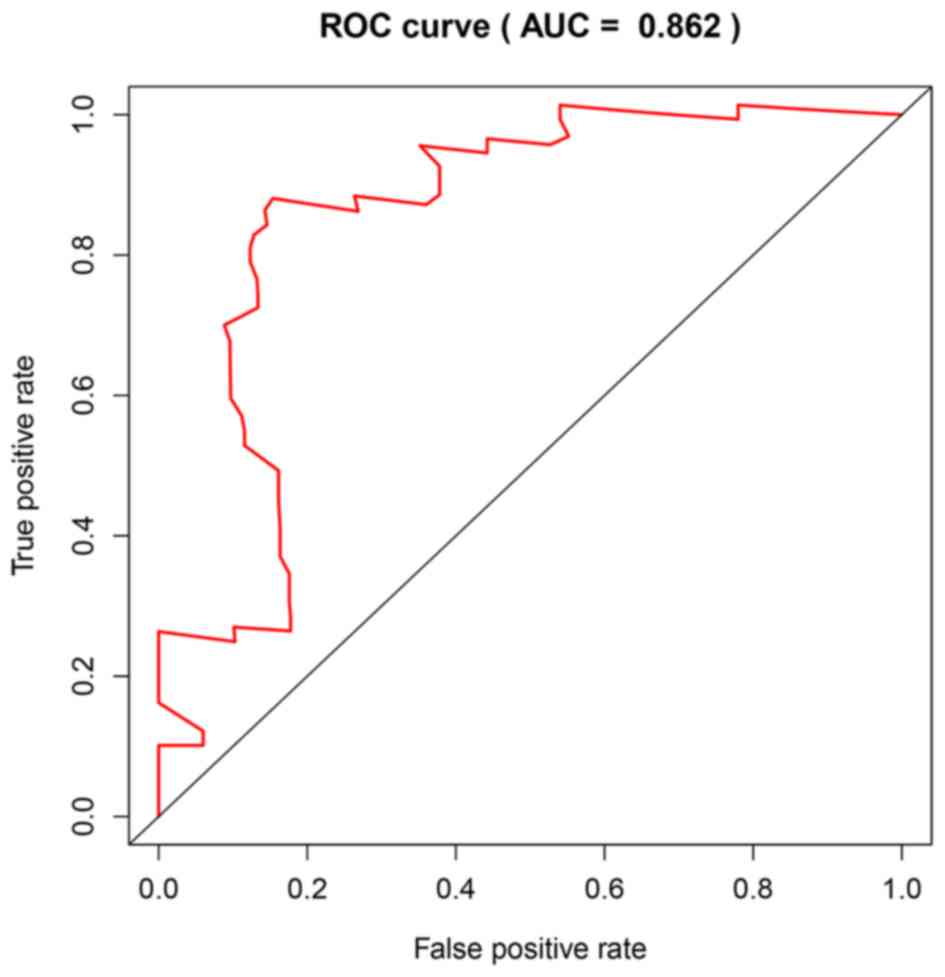
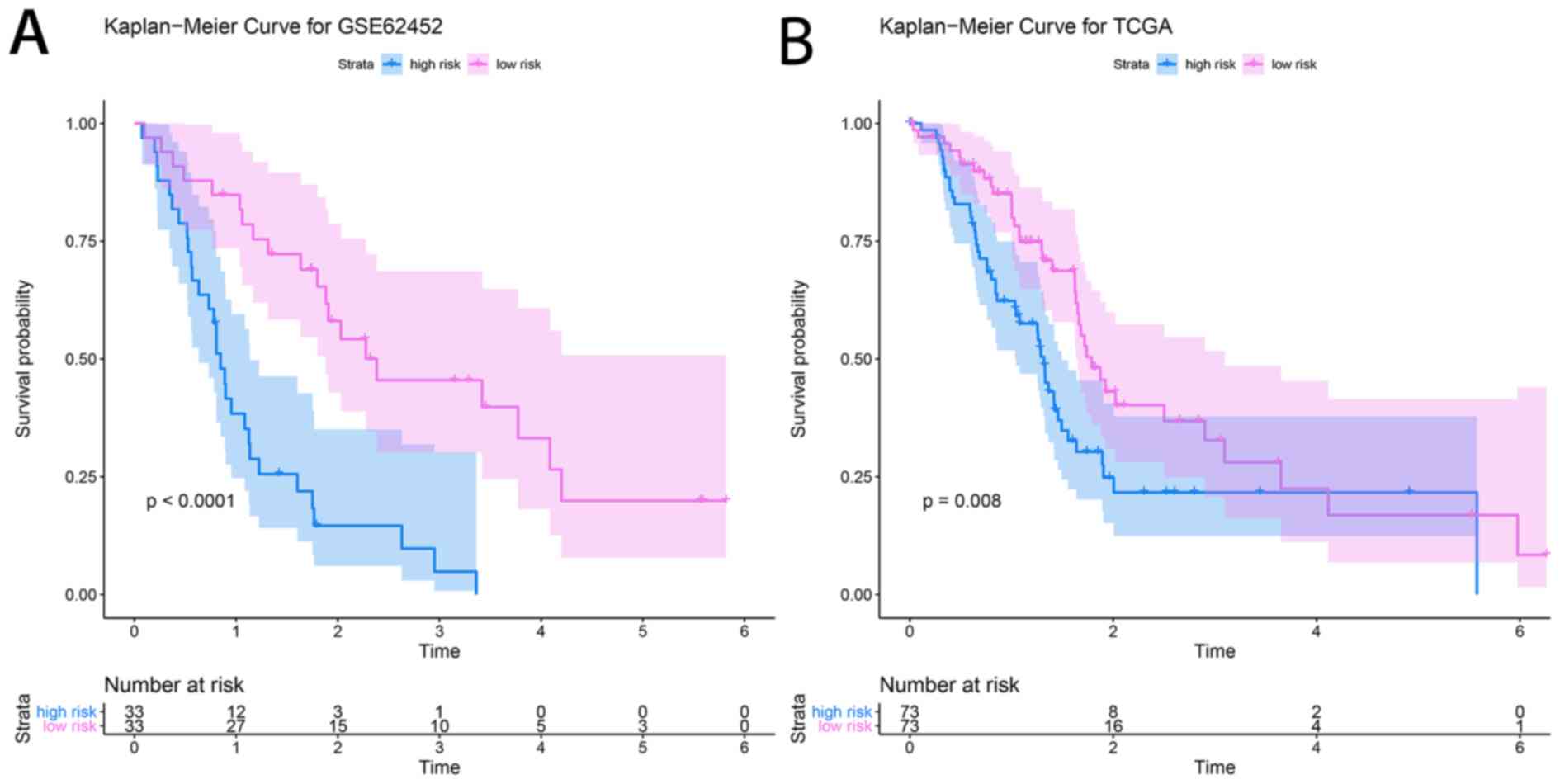
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shen Q, Yu M, Jia JK, Li WX, Tian YW and
Xue HZ: Possible Molecular markers for the diagnosis of pancreatic
ductal adenocarcinoma. Med Sci Monit. 24:2368–2376. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ger M, Kaupinis A, Petrulionis M,
Kurlinkus B, Cicenas J, Sileikis A, Valius M and Strupas K:
Proteomic identification of FLT3 and PCBP3 as potential prognostic
biomarkers for pancreatic cancer. Anticancer Res. 38:5759–5765.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang Y, Zhang Z, Tang Y, Chen X and Zhou
J: Identification of potential target genes in pancreatic ductal
adenocarcinoma by bioinformatics analysis. Oncol Lett.
16:2453–2461. 2018.PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Research Network:; and
Cancer Genome Atlas Research Network, . Integrated Genomic
Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell.
32:185–203 e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kolde R and Laur S.; RobustRankAggreg, :
Methods for robust rank aggregation. 2013.
|
|
9
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
11
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frampton AE, Castellano L, Colombo T,
Giovannetti E, Krell J, Jacob J, Pellegrino L, Roca-Alonso L, Funel
N, Gall TM, et al: MicroRNAs cooperatively inhibit a network of
tumor suppressor genes to promote pancreatic tumor growth and
progression. Gastroenterology. 146:268–277 e18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Janky R, Binda MM, Allemeersch J, Van den
Broeck A, Govaere O, Swinnen JV, Roskams T, Aerts S and Topal B:
Prognostic relevance of molecular subtypes and master regulators in
pancreatic ductal adenocarcinoma. BMC Cancer. 16:6322016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang S, He P, Wang J, Schetter A, Tang W,
Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al: A
Novel MIF signaling pathway drives the malignant character of
pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moffitt RA, Marayati R, Flate EL, Volmar
KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung
AH, et al: Virtual microdissection identifies distinct tumor- and
stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat
Genet. 47:1168–1178. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang J, Azevedo-Pouly AC, Redis RS, Lee
EJ, Gusev Y, Allard D, Sutaria DS, Badawi M, Elgamal OA, Lerner MR,
Brackett DJ, et al: Globally increased ultraconserved noncoding RNA
expression in pancreatic adenocarcinoma. Oncotarget. 7:53165–53177.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sutaria DS, Jiang J, Azevedo-Pouly ACP,
Lee EJ, Lerner MR, Brackett DJ, Vandesompele J, Mestdagh P and
Schmittgen TD: Expression Profiling Identifies the Noncoding
Processed Transcript of HNRNPU with Proliferative Properties in
Pancreatic Ductal Adenocarcinoma. Noncoding RNA.
3:ncrna30300242017.
|
|
18
|
Kohl M, Wiese S and Warscheid B.
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao X, Sun S, Zeng X and Cui L:
Expression profiles analysis identifies a novel three-mRNA
signature to predict overall survival in oral squamous cell
carcinoma. Am J Cancer Res. 8:450–461. 2018.PubMed/NCBI
|
|
20
|
Zhu T, Gao YF, Chen YX, Wang ZB, Yin JY,
Mao XY, Li X, Zhang W, Zhou HH and Liu ZQ: Genome-scale analysis
identifies GJB2 and ERO1LB as prognosis markers in patients with
pancreatic cancer. Oncotarget. 8:21281–21289. 2017.PubMed/NCBI
|
|
21
|
Li H, Wang X, Fang Y, Huo Z, Lu X, Zhan X,
Deng X, Peng C and Shen B: Integrated expression profiles analysis
reveals novel predictive biomarker in pancreatic ductal
adenocarcinoma. Oncotarget. 8:52571–52583. 2017.PubMed/NCBI
|
|
22
|
Hynes RO and Naba A: Overview of the
matrisome-an inventory of extracellular matrix constituents and
functions. Cold Spring Harb Perspect Biol. 4:a0049032012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Humphrey JD, Dufresne ER and Schwartz MA:
Mechanotransduction and extracellular matrix homeostasis. Nat Rev
Mol Cell Biol. 15:802–812. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Venning FA, Wullkopf L and Erler JT:
Targeting ECM disrupts cancer progression. Front Oncol. 5:2242015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang M, Yuan J, Peng C and Li Y: Collagen
as a double-edged sword in tumor progression. Tumour Biol.
35:2871–2882. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miles FL and Sikes RA: Insidious changes
in stromal matrix fuel cancer progression. Mol Cancer Res.
12:297–312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pickup MW, Mouw JK and Weaver VM: The
extracellular matrix modulates the hallmarks of cancer. EMBO Rep.
15:1243–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mollenhauer J, Roether I and Kern HF:
Distribution of extracellular matrix proteins in pancreatic ductal
adenocarcinoma and its influence on tumor cell proliferation in
vitro. Pancreas. 2:14–24. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Armstrong T, Packham G, Murphy LB, Bateman
AC, Conti JA, Fine DR, Johnson CD, Benyon RC and Iredale JP: Type I
collagen promotes the malignant phenotype of pancreatic ductal
adenocarcinoma. Clin Cancer Res. 10:7427–7437. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vaquero EC, Edderkaoui M, Nam KJ, Gukovsky
I, Pandol SJ and Gukovskaya AS: Extracellular matrix proteins
protect pancreatic cancer cells from death via mitochondrial and
nonmitochondrial pathways. Gastroenterology. 125:1188–1202. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koenig A, Mueller C, Hasel C, Adler G and
Menke A: Collagen type I induces disruption of E-cadherin-mediated
cell-cell contacts and promotes proliferation of pancreatic
carcinoma cells. Cancer Res. 66:4662–4671. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Grzesiak JJ and Bouvet M: The alpha2beta1
integrin mediates the malignant phenotype on type I collagen in
pancreatic cancer cell lines. Br J Cancer. 94:1311–1319. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Topalovski M and Brekken RA: Matrix
control of pancreatic cancer: New insights into fibronectin
signaling. Cancer Lett. 381:252–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Glasner A, Levi A, Enk J, Isaacson B,
Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, et al:
NKp46 receptor-mediated interferon-ү production by natural killer
cells increases fibronectin 1 to alter tumor architecture and
control metastasis. Immunity. 48:107–119 e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu D, Ansari D, Zhou Q, Sasor A, Said
Hilmersson K and Andersson R: Stromal fibronectin expression in
patients with resected pancreatic ductal adenocarcinoma. World J
Surg Oncol. 17:292019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shibata K, Kikkawa F, Nawa A, Thant AA,
Naruse K, Mizutani S and Hamaguchi M: Both focal adhesion kinase
and c-Ras are required for the enhanced matrix metalloproteinase 9
secretion by fibronectin in ovarian cancer cells. Cancer Res.
58:900–903. 1998.PubMed/NCBI
|
|
37
|
Himelstein BP, Canete-Soler R, Bernhard
EJ, Dilks DW and Muschel RJ: Metalloproteinases in tumor
progression: The contribution of MMP-9. Invasion Metastasis.
14:246–258. 1994-1995.
|
|
38
|
Bloomston M, Zervos EE and Rosemurgy AS
II: Matrix metalloproteinases and their role in pancreatic cancer:
A review of preclinical studies and clinical trials. Ann Surg
Oncol. 9:668–674. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Grunwald B, Vandooren J, Locatelli E,
Fiten P, Opdenakker G, Proost P, Kruger A, Lellouche JP, Israel LL,
Shenkman L and Comes Franchini M: Matrix metalloproteinase-9
(MMP-9) as an activator of nanosystems for targeted drug delivery
in pancreatic cancer. J Control Release. 239:39–48. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agarwal A, Tressel SL, Kaimal R, Balla M,
Lam FH, Covic L and Kuliopulos A: Identification of a
metalloprotease-chemokine signaling system in the ovarian cancer
microenvironment: Implications for antiangiogenic therapy. Cancer
Res. 70:5880–5890. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Grunwald B, Schoeps B and Kruger A:
Recognizing the molecular multifunctionality and interactome of
TIMP-1. Trends Cell Biol. 29:6–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hirsch FR, Varella-Garcia M and Cappuzzo
F: Predictive value of EGFR and HER2 overexpression in advanced
non-small-cell lung cancer. Oncogene. 28((Suppl 1)): S32–S37. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Normanno N, De Luca A, Bianco C, Strizzi
L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F and
Salomon DS: Epidermal growth factor receptor (EGFR) signaling in
cancer. Gene. 366:2–16. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Olayioye MA, Neve RM, Lane HA and Hynes
NE: The ErbB signaling network: Receptor heterodimerization in
development and cancer. EMBO J. 19:3159–3167. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chong CR and Janne PA: The quest to
overcome resistance to EGFR-targeted therapies in cancer. Nat Med.
19:1389–1400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rivera F, Lopez-Tarruella S, Vega-Villegas
ME and Salcedo M: Treatment of advanced pancreatic cancer: From
gemcitabine single agent to combinations and targeted therapy.
Cancer Treat Rev. 35:335–339. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang F, Xiao W, Sun J, Han D and Zhu Y:
MiRNA-181c inhibits EGFR-signaling-dependent MMP9 activation via
suppressing Akt phosphorylation in glioblastoma. Tumour Biol.
35:8653–8658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qiu Q, Yang M, Tsang BK and Gruslin A:
EGF-induced trophoblast secretion of MMP-9 and TIMP-1 involves
activation of both PI3K and MAPK signalling pathways. Reproduction.
128:355–363. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang T, Yamashita K, Iwata K and Hayakawa
T: Both tissue inhibitors of metalloproteinases-1 (TIMP-1) and
TIMP-2 activate Ras but through different pathways. Biochem Biophys
Res Commun. 296:201–205. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tarpgaard LS, Orum-Madsen MS, Christensen
IJ, Nordgaard C, Noer J, Guren TK, Glimelius B, Sorbye H, Ikdahl T,
Kure EH, et al: TIMP-1 is under regulation of the EGF signaling
axis and promotes an aggressive phenotype in KRAS-mutated
colorectal cancer cells: A potential novel approach to the
treatment of metastatic colorectal cancer. Oncotarget.
7:59441–59457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Curry S, Mandelkow H, Brick P and Franks
N: Crystal structure of human serum albumin complexed with fatty
acid reveals an asymmetric distribution of binding sites. Nat
Struct Biol. 5:827–835. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mueller TC, Burmeister MA, Bachmann J and
Martignoni ME: Cachexia and pancreatic cancer: Are there treatment
options? World J Gastroenterol. 20:9361–9373. 2014.PubMed/NCBI
|
|
53
|
Bachmann J, Buchler MW, Friess H and
Martignoni ME: Cachexia in patients with chronic pancreatitis and
pancreatic cancer: Impact on survival and outcome. Nutr Cancer.
65:827–833. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mengele K, Napieralski R, Magdolen V,
Reuning U, Gkazepis A, Sweep F, Brunner N, Foekens J, Harbeck N and
Schmitt M: Characteristics of the level-of-evidence-1 disease
forecast cancer biomarkers uPA and its inhibitor PAI-1. Expert Rev
Mol Diagn. 10:947–962. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Asuthkar S, Stepanova V, Lebedeva T,
Holterman AL, Estes N, Cines DB, Rao JS and Gondi CS:
Multifunctional roles of urokinase plasminogen activator (uPA) in
cancer stemness and chemoresistance of pancreatic cancer. Mol Biol
Cell. 24:2620–2632. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xue A, Xue M, Jackson C and Smith RC:
Suppression of urokinase plasminogen activator receptor inhibits
proliferation and migration of pancreatic adenocarcinoma cells via
regulation of ERK/p38 signaling. Int J Biochem Cell Biol.
41:1731–1738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Botla SK, Savant S, Jandaghi P, Bauer AS,
Mucke O, Moskalev EA, Neoptolemos JP, Costello E, Greenhalf W,
Scarpa A, et al: Early epigenetic downregulation of microRNA-192
expression promotes pancreatic cancer progression. Cancer Res.
76:4149–4159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ding J, Yang C and Yang S: LINC00511
interacts with miR-765 and modulates tongue squamous cell carcinoma
progression by targeting LAMC2. J Oral Pathol Med. 47:468–476.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kinoshita T, Nohata N, Hanazawa T, Kikkawa
N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto
Y and Seki N: Tumour-suppressive microRNA-29s inhibit cancer cell
migration and invasion by targeting laminin-integrin signalling in
head and neck squamous cell carcinoma. Br J Cancer. 109:2636–2645.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jung SN, Lim HS, Liu L, Chang JW, Lim YC,
Rha KS and Koo BS: LAMB3 mediates metastatic tumor behavior in
papillary thyroid cancer by regulating c-MET/Akt signals. Sci Rep.
8:27182018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang XM, Li J, Yan MX, Liu L, Jia DS, Geng
Q, Lin HC, He XH, Li JJ and Yao M: Integrative analyses identify
osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for
lung cancer. PLoS One. 8:e557142013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ohike N, Maass N, Mundhenke C, Biallek M,
Zhang M, Jonat W, Luttges J, Morohoshi T, Kloppel G and Nagasaki K:
Clinicopathological significance and molecular regulation of maspin
expression in ductal adenocarcinoma of the pancreas. Cancer Lett.
199:193–200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kashima K, Ohike N, Mukai S, Sato M,
Takahashi M and Morohoshi T: Expression of the tumor suppressor
gene maspin and its significance in intraductal papillary mucinous
neoplasms of the pancreas. Hepatobiliary Pancreat Dis Int. 7:86–90.
2008.PubMed/NCBI
|
|
64
|
Cao D, Zhang Q, Wu LS, Salaria SN, Winter
JW, Hruban RH, Goggins MS, Abbruzzese JL, Maitra A and Ho L:
Prognostic significance of maspin in pancreatic ductal
adenocarcinoma: Tissue microarray analysis of 223 surgically
resected cases. Mod Pathol. 20:570–578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang L, Wu H, Wang L, Lu J, Duan H, Liu X
and Liang Z: Expression of amphiregulin predicts poor outcome in
patients with pancreatic ductal adenocarcinoma. Diagn Pathol.
11:602016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Busser B, Sancey L, Brambilla E, Coll JL
and Hurbin A: The multiple roles of amphiregulin in human cancer.
Biochim Biophys Acta. 1816:119–131. 2011.PubMed/NCBI
|
|
67
|
Pawar NM and Rao P: Secreted frizzled
related protein 4 (sFRP4) update: A brief review. Cell Signal.
45:63–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Deshmukh A, Kumar S, Arfuso F, Newsholme P
and Dharmarajan A: Secreted Frizzled-related protein 4 (sFRP4)
chemo-sensitizes cancer stem cells derived from human breast,
prostate, and ovary tumor cell lines. Sci Rep. 7:22562017.
View Article : Google Scholar : PubMed/NCBI
|