Introduction
Breast cancer is the most prevalent malignancy in
women and estimated to account for 30% of new cancer diagnoses in
women and 15.3% of all types of cancer according to a statistics
report in 2018 in the United States (1). The incidence rate varies greatly
worldwide from 19.3 per 100,000 women in Eastern Africa to 89.7 per
100,000 women in Western Europe, and the highest annual morbidity
(40,290 cases, in 2015) and mortality rate (13.8%, in 2015) of
patients is in the United States (2,3). Between
2002 and 2008, the mortality rate from breast cancer increased by
201% among urban Chinese women (4).
Triple-negative breast cancer in particular is associated with poor
prognosis due to a lack of clinically established targeted
therapies and aggressive pathological characteristics (5). Despite advanced systemic treatments
including surgical excision, local radiotherapy and adjuvant
therapy, which have successfully doubled the survival rate of
patients with breast cancer (6),
10–15% patients still experience recurrence or metastasis (7). Developing an efficient prevention
and/or treatment regimen for patients with breast cancer remains a
major challenge in clinical practice.
Carcinogenesis is the consequence of the synergistic
effects of imbalanced homeostasis and aberrant multigenetic
regulation. Accumulating experimental evidence has suggested
several molecular hallmarks in breast cancer, such as breast and
ovarian cancer susceptibility protein 1 (BRCA1) and BRCA2, which
have been used to assess the risk of developing breast cancer
(8–10). Cathepsins are ubiquitous proteinases
that serve important roles in cancer metastasis and degrade
proteins in the lysosome; based on the variation of their active
sites, cathepsins can be divided into three subgroups: Aspartate (D
and E), Cysteine (B, C, H, F, K, L, O, S, V, W and X/Z), and Serine
(G) cathepsins (11). Cathepsin D
serves a crucial biomarker in the early detection of nasopharyngeal
cancer and metastasis in patients with breast cancer (12,13).
Recently, Cathepsin L (CTSL) was demonstrated to be associated with
multiple diseases, including breast cancer metastasis (14–17).
However, the specific mechanisms and functions of CTSL in breast
cancer metastasis are still unclear. Although accumulating evidence
has revealed that CTSL expression is increased in several types of
carcinoma, including breast cancer, and that its expression pattern
reflects the degree of malignancy, the detailed mechanisms and its
interaction partners remain elusive (18).
The aim of the present study was to investigate the
expression profiles and pathological functions of CTSL and its
contribution to the prognosis of breast cancer, highlighting CTSL
as a potential novel therapeutic target for the treatment of
patients with breast cancer.
Materials and methods
Patient selection and follow-up
A total of 249 primary breast cancer tissues and 31
paired adjacent normal tissues were collected from patients
admitted to the Xiangya Hospital of Central South University
(Changsha, China) between July 2004 and July 2005. The patients
enrolled were histologically confirmed as cases of primary breast
cancer, underwent surgical resection and had an adequate size
tissue sample with no evidence of metastasis prior to the surgery.
Patients with previous or other concomitant malignancies were
excluded. The clinical information of the patients was collected
from Xiangya Hospital, including age, menstrual status, status of
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor 2 (HER-2) expression, clinical
stage (classified according to the 7th edition of the American
Joint Committee on Cancer staging manual) (19), recurrence and metastasis. The
characteristics of the patients are presented in Table I. The termination date for patient
follow-up was June 2015. Progression-free survival (PFS) was
calculated as the time between the date of resection and death,
recurrence or metastasis. Alive patients without recurrence or
metastasis at the end of the follow-up were censored. The study was
conducted in accordance with the Declaration of Helsinki (20), and the protocol was approved by The
Ethical Review Committee of Xiangya Hospital, Central South
University.
 | Table I.Patient clinicopathological
characteristics. |
Table I.
Patient clinicopathological
characteristics.
|
Characteristics | No. of patients (%)
(N=249) |
|---|
| Age, years |
|
|
≤50 | 159 (63.8) |
|
>50 | 90 (36.1) |
| Menstrual
status |
|
|
Pre-menopausal | 160 (64.2) |
|
Post-menopausal | 88 (35.3) |
|
Missing | 1 (0.4) |
| Stage |
|
|
I–II | 197 (79.1) |
|
III | 48 (19.3) |
|
Missing | 4 (1.6) |
| Lymph node
metastasis |
|
|
Positive | 126 (50.6) |
|
Negative | 122 (49.0) |
|
Missing | 1 (0.4) |
| Estrogen
receptor |
|
|
Positive | 140 (56.2) |
|
Negative | 104 (41.8) |
|
Missing | 5 (2.0) |
| Progesterone
receptor |
|
|
Positive | 131 (52.6) |
|
Negative | 112 (45.0) |
|
Missing | 6 (2.4) |
| Human endothelial
growth factor receptor 2 expression |
|
|
Positive | 76 (30.5) |
|
Negative | 162 (65.1) |
|
Missing | 1 (0.4) |
Immunohistochemistry
Immunohistochemistry was performed as described
previously (21). Briefly,
paraffin-embedded tissues were cut into 5-µm sections and mounted
on slides. The slides were deparaffinized by xylene, rehydrated
with graded ethanol series and stained with a CTSL antibody (1:200;
cat. no. TA809346; OriGene Technologies, Inc.) overnight at 4°C and
subsequently incubated with a biotinylated secondary goat
anti-mouse immunoglobulin G antibody (1:1,000; cat. no. TA130001;
OriGene Technologies, Inc.). The immunoreactions were detected
using a streptavidin-peroxidase system. The slides were evaluated
in a double-blinded manner by two independent pathologists. The
intensity of the staining was scored as follows: 0, negative/-; 1,
weak/+; 2, moderate/++; and 3, strong/+++. The proportion of
immunoreactive cells was scored as follows: 0, no cells stained; 1,
<25% of cells stained; 2, 25–50% of cells stained; 3, 51–75% of
cells stained; and 4, >75% of cells stained. The final score for
each tissue specimen was determined by multiplying the score for
the proportion of immunoreactive cells by the score for the
intensity of the staining. In order to ascertain a cut-off score to
distinguish between the expression levels of CTSL, receiver
operating characteristic (ROC) curve analysis was used. The score
closest to the point of maximum Youden's index was used as the
optimal cut-off value. CTSL low expression was defined as samples
with scores below the cut-off value (score <6.5), whereas high
expression was defined as samples with scores above the value
(score >6.5).
Protein-protein interaction (PPI)
network construction
The STRING database (string-db.org;
version 10) was searched for cyclin-dependent kinase 2-associaed
protein 1 (‘CDK2-AP1’) in ‘homo sapiens’ to identify the potential
protein-protein interactions.
Oncomine database
To determine the expression of CTSL in human breast
cancer, data mining was performed using the Oncomine database
(www.oncomine.org). ‘Sorlie breast’, ‘Sorlie
breast 2’, ‘Perou breast’, ‘Ma breast 4’, ‘Zhao breast’ and
‘Richardson breast 2’ databases were selected (22–27). The
gene expression of CTSL between cancer tissues and normal tissues
was compared. In addition, the expression of CTSL in different
histological grades and clinical stages of breast cancer were also
determined.
PROGgene database
The prognostic significance of CTSL was assessed
using the PROGgene database (genomics.jefferson.edu/proggene/). GSE10893-GPL887,
GSE6130-GLP1390 and GSE9893 (28–30) were
the datasets used for further analysis. Kaplan-Meier analysis was
used to evaluate the prognostic significance of CTSL.
Yeast two-hybrid (Y2H) screening
Y2H library screening was performed as described in
the Matchmaker® Gold Y2H System User Manual (Clontech
Laboratories, Inc.) with minimal modifications. Briefly, the
pGBKT7-CDK2-AP1 BD-Bait plasmid was generated and transformed into
the Y2H Gold Yeast Strain, and the bait expression, autoactivation
and cytotoxicity were assessed prior to any application.
Subsequently, concentrated Y2H Gold [pGBKT7-CDK2-AP1] recombinants
were mixed with 1 ml library strains [Mate&Plate™
Libraries-Universal Human (Normalized), Clontech Laboratories,
Inc.] and incubated at 30°C with gentle agitation until the zygotes
were visible under a phase-contrast microscope (CKX41; Olympus
Corporation). The desired colonies were screened from the DDO/X/A
plates (double synthetic dropout medium lacking tryptophan and
leucine supplemented with X-α-Gal and Aureobasidin A) dependent on
the color, and the plasmids were extracted from the positive
colonies, sequenced and analyzed using NCBI BLAST (http://www.ncbi.nlm.nih.gov) and the results were
obtained. Then, protein interaction maps were generated using
Cytoscape (https://cytoscape.org/) (31) and interaction networks of the 13
putative CDK2-AP1-interacting proteins were examined using Search
Tool for the Retrieval of Interacting Genes and Proteins (STRING)
(32).
Cell cultures
The human breast cancer cell lines T-47D, MCF-7,
MDA-MB-231 and BT-474, as well as 293T cells were purchased from
the Type Culture Collection of the Chinese Academy of Sciences; all
cell lines were authenticated by American Type Culture Collection.
The T-47D, MDA-MB-231 and 293T cells were routinely cultured in
DMEM (HyClone; GE Healthcare Life Sciences) supplemented with 10%
fetal bovine serum (FBS; Lonsera Science); the BT-474 cells were
cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences)
supplemented with 10% FBS; and the MCF-7 cells were cultured in
DMEM supplemented with 10% FBS, 1% sodium glutamate and 0.01 mg/ml
bovine insulin. All the cells were incubated at 37°C in a
humidified atmosphere with 5% CO2.
Lentivirus packaging and
infection
CTSL small interfering (si)RNAs (s1,
5′-CCAAAGACCGGAGAAACCATT-3′; s2, 5′-AGGCGATGCACAACAGATTAT-3′; and
s3, 5′-TGCCTCAGCTACTCTAACATT-3′) were designed against the open
reading frame of the human CTSL gene (NM_145918.2) and a random
non-targeting siRNA (5′-TTCTCCGAACGTGTCACGT-3′) was designed as a
negative control. Fragments of the siRNA were cloned into the
pGreenPuro™ short hairpin (sh)RNA Cloning and Expression
Lentivector (System Biosciences, LLC) and recombinant lentiviruses
were produced by co-infecting 293T cells with the siRNA expression
plasmid and pHelper plasmids (Lentiviral Packaging mix;
Sigma-Aldrich; Merck KGaA). For lentiviral infection,
1×106 T-47D cells were seeded in six-well plates and
infected with the specific recombinant lentivirus (Lv-shCTSL or
Lv-shCon) at a multiplicity of infection of 25 for 96 h. Knockdown
of endogenous CTSL was confirmed using western blot analysis with a
monoclonal antibody (1:1,000; cat. no. TA809346; OriGene
Technologies, Inc.), and infection efficiency was determined by
counting the number of green fluorescent protein (GFP)-positive
cells under a fluorescent microscope (magnification, ×400).
Western blot analysis
The total cell proteins were extracted using RIPA
buffer supplemented with a protease inhibitor cocktail (Roche
Diagnostics). Total protein concentration was measured by BCA
protein assay kit (Beyotime Institute of Biotechnology). Equal
quantities of protein (30 µg) were separated using 10% SDS-PAGE and
transferred onto PVDF membranes. The membranes were blocked in 5%
skimmed milk for 2 h at room temperature. Subsequently, the blots
were probed with one of the following primary antibodies: Rabbit
anti-CTSL (1:1,000; cat. no. TA809346; OriGene Technologies, Inc.);
rabbit anti-CDK2AP1 (1:1,000; cat. no. 13060-2-AP; Proteintech
Group, Inc.); or mouse anti-GAPDH (1:500,000; cat. no. HRP-60004;
Proteintech Group, Inc.) overnight at 4°C, followed by incubation
with the secondary anti-rabbit horseradish peroxidase
(HRP)-conjugated antibody (1:5,000; cat. no. SC-2054; Proteintech
Group, Inc.) or anti-mouse HRP (1:5,000; cat. no. SC-2005; Santa
Cruz Biotechnology, Inc.). The blots were visualized using an
enhanced chemiluminescence reagent (Beyotime Institute of
Biotechnology) and scanned using the Tanon 5200 (Tanon Science and
Technology Co., Ltd.) and analyzed using the Image J software v1.48
(National Institutes of Health).
Glutathione S-transferase (GST)
pull-down assay
Direct physical interactions between CTSL and
CDK2-AP1 in eukaryotic cells were examined using a GST pull-down
assay. A GST or GST-CDK2-AP1 fusion protein was expressed and
purified as bait from Escherichia coli BL21/DE3 recombinants
induced with isopropyl β-D-1-thiogalactopyranoside, whereas the
GFP-CTSL fusion protein was overexpressed in 293T cells as prey. A
total of 5 ng purified GST-labeled proteins were incubated with 25
µl glutathione sepharose 4B beads for 3 h at 4°C and incubated with
the cell lysate from the 293T recombinants overnight at 4°C. The
bead-bound protein complexes were detected using western blotting
(rabbit anti-GFP; 1:8,000; cat. no. 50430-2-AP; rabbit anti-GST;
1:6,000; cat. no. 10,000-0-AP; incubated overnight at 4°C;
antibodies from Proteintech Group, Inc.).
Proliferation assay
The proliferation or viability of the tumor cells
with or without CTSL-knockdown was assessed using an MTT assay
(Sigma-Aldrich; Merck KGaA). Briefly, T-47D cells transfected with
either Lv-shCon or Lv-shCTSL were plated into a 96-well plate at a
density of 3×103 cells/well and incubated with 20 µl MTT
(5 mg/ml) at 37°C for a further 4 h at the indicated time-points
(day 1, 2, 3, 4 and 5). Subsequently, the formazan crystals were
dissolved with 100 µl acidic isopropanol (10% SDS, 5% isopropanol
and 0.01 mol/l HCl) and the absorbance was measured at 570 nm using
a microplate spectrophotometer.
Colony formation assay
Lentivirus-infected T-47D cells were trypsinized and
seeded into a six-well plate at a density of 1×103
cells/well and incubated for 14 days until colonies were visible by
eye. The samples were carefully washed twice with PBS, fixed with
4% paraformaldehyde for 30 min and stained with crystal violet for
15 min at room temperature. The number of colonies in each group
was visually counted manually and the average values were
calculated.
Flow cytometric analysis of apoptosis
and cell cycle distribution
Flow cytometry was used to determine the proportion
of apoptotic cells and the cell cycle distribution in breast cancer
cells following CTSL knockdown. T-47D cells infected with Lv-shCon
or Lv-shCTSL were cultured to 80% confluency, harvested and washed
twice with PBS. For cell cycle analysis, the cells were fixed and
permeabilized with −20°C 70% ethanol for 12 h prior to treating
with DNase-free RNase and stained with propidium iodide (both
Sigma-Aldrich; Merck KGaA) for 30 min at 4°C. For the analysis of
apoptosis, the cells were resuspended in binding buffer containing
Annexin V-fluorescein isothiocyanate and 7-AAD for 15 min in the
dark according to the manufacturer's protocol (Annexin APC/V-7-AAD
apoptosis detection kit, Nanjing KeyGen Biotech Co., Ltd.). The
stained cells were subsequently subjected to flow cytometry
analysis using a FACScalibur flow cytometer (BD Biosciences). The
data was analyzed using FlowJo v7.6 software (FlowJo LLC).
Statistical analysis
Data were analyzed using either paired or unpaired
Student's t-test to identify differences between two groups.
One-way or two-way ANOVA with Dunnett's post hoc test to compare
multiple groups using GraphPad prism v5.0 software (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference. mRNA expression levels of
CTSL in cancer and normal clinical specimens in six databases were
compared using paired or unpaired Student's t-test. The prognostic
value of CTSL expression in breast cancer was determined using
Kaplan-Meier analysis of overall survival or PFS using PROGgene and
compared using the log rank test, a tool that assesses the effects
of genes on survival in patients with cancer (33). The covariates and cumulative PFS
rates were calculated using a Cox-proportional hazard model with
SPSS version 18.0 (SPSS, Inc.).
Results
CTSL directly interacts with
CDK2-AP1
Our previous study demonstrated that CDK2-AP1 serves
as a tumor suppressor in breast cancer in vitro and in
vivo (34). To identify the
proteins interacting with CDK2-AP1, Y2H screening was performed.
Sequencing from the positive colonies identified 13 potential
putative CDK2-AP1-interacting proteins, and the interaction
networks were reconstructed using Cytoscape and STRING (Fig. 1A and B). To determine whether CTSL
interacted with CDK2-AP1, a GST pull-down assay was performed with
GST-CDK2-AP1 fusion protein as bait to capture GFP-CTSL from
lysates of engineered 293T cells. The results demonstrated that
GFP-CTSL could be pulled down from the cell lysates by
GST-CDK2-AP1, but not by GST, suggesting a direct interaction
between CTSL and CDK2-AP1 (Fig. 1C and
D). Western blot analysis revealed that in cell lines
exhibiting high expression of CTSL, CDK2-AP1 expression was low,
whereas low level of CTSL corresponded to high CDK2-AP1 expression
(Fig. 1E-G), suggesting that a
balance between CTSL and CDK2-AP1 may be involved in the regulation
of tumorigenesis.
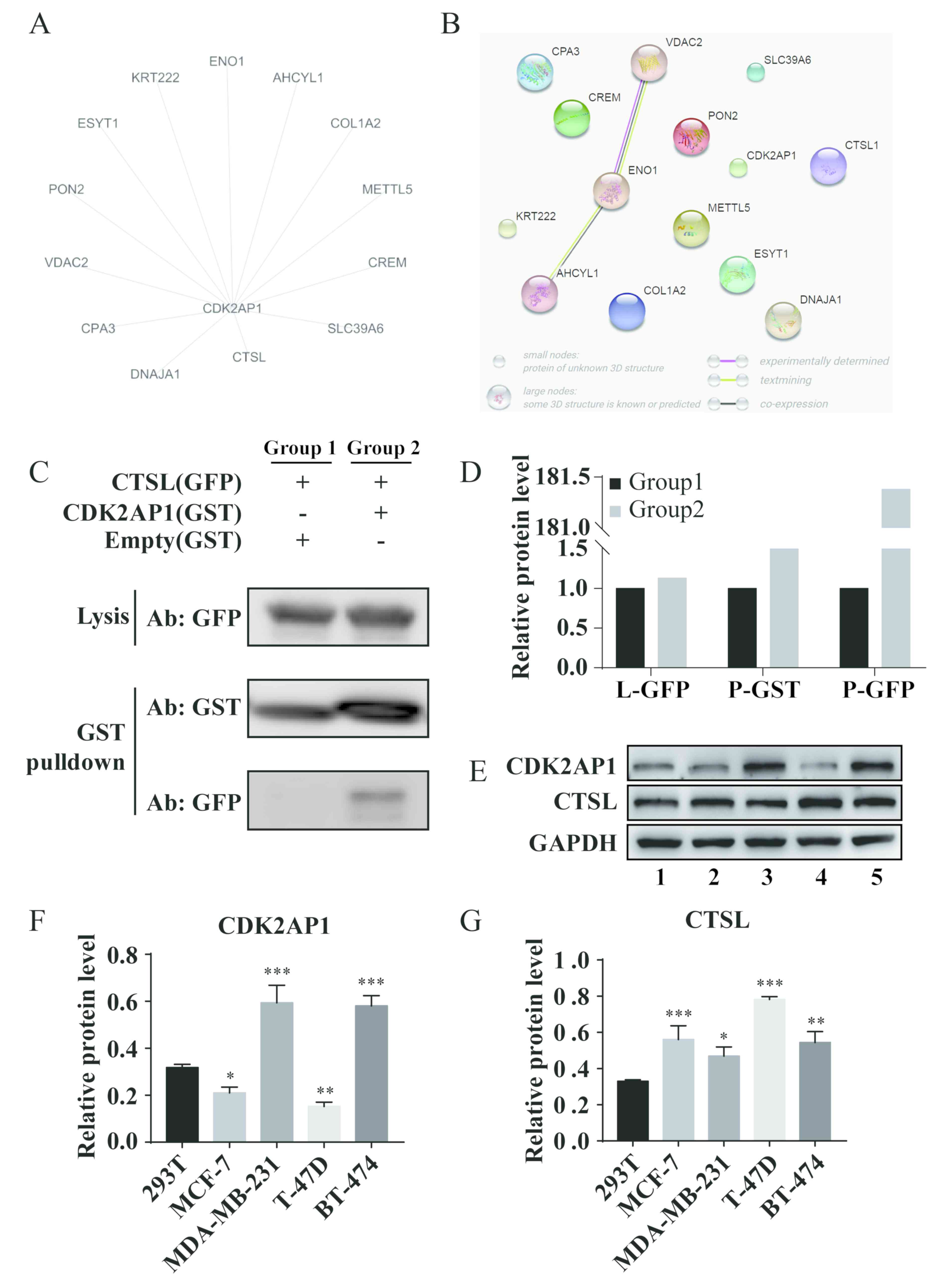 | Figure 1.CTSL directly interacts with
CDK-2AP1. (A) Protein interaction map identified by yeast
two-hybrid screening indicated putative CDK2-AP1-interacting
proteins using Cytoscape. (B) Protein-protein interaction networks
of the indicated candidates were constructed using Search Tool for
the Retrieval of Interacting Genes/Proteins database. (C) GST
pull-down assay was performed in 293T cells to identify the
interaction of CTSL and CDK2-AP1. (D) The densitometry analysis of
the protein bands of GST pull-down assay. (E) Western blot analysis
was used to determine the endogenous expression of CDK2AP1 and CTSL
in breast cancer cell lines. 1, 2, 3, 4 and 5 represents 293T,
MCF-7, MDA-MB-231, T-47D and BT-474, respectively. (F) CDK2-AP1
protein expression in breast cancer cell lines. (G) CTSL protein
expression in breast cancer cell lines. Statistical analyses were
performed using one-way ANOVA with Dunnett's multiple comparisons
test: *P<0.05 and **P<0.01, ***P<0.001 vs. 293T. CTSL,
Cathepsin L; CDK2-AP1, cyclin-dependent kinase 2-associated protein
1; GST, glutathione S-transferase; GFP, green fluorescent
protein. |
CTSL is upregulated and associated
with poor prognosis in patients with breast cancer
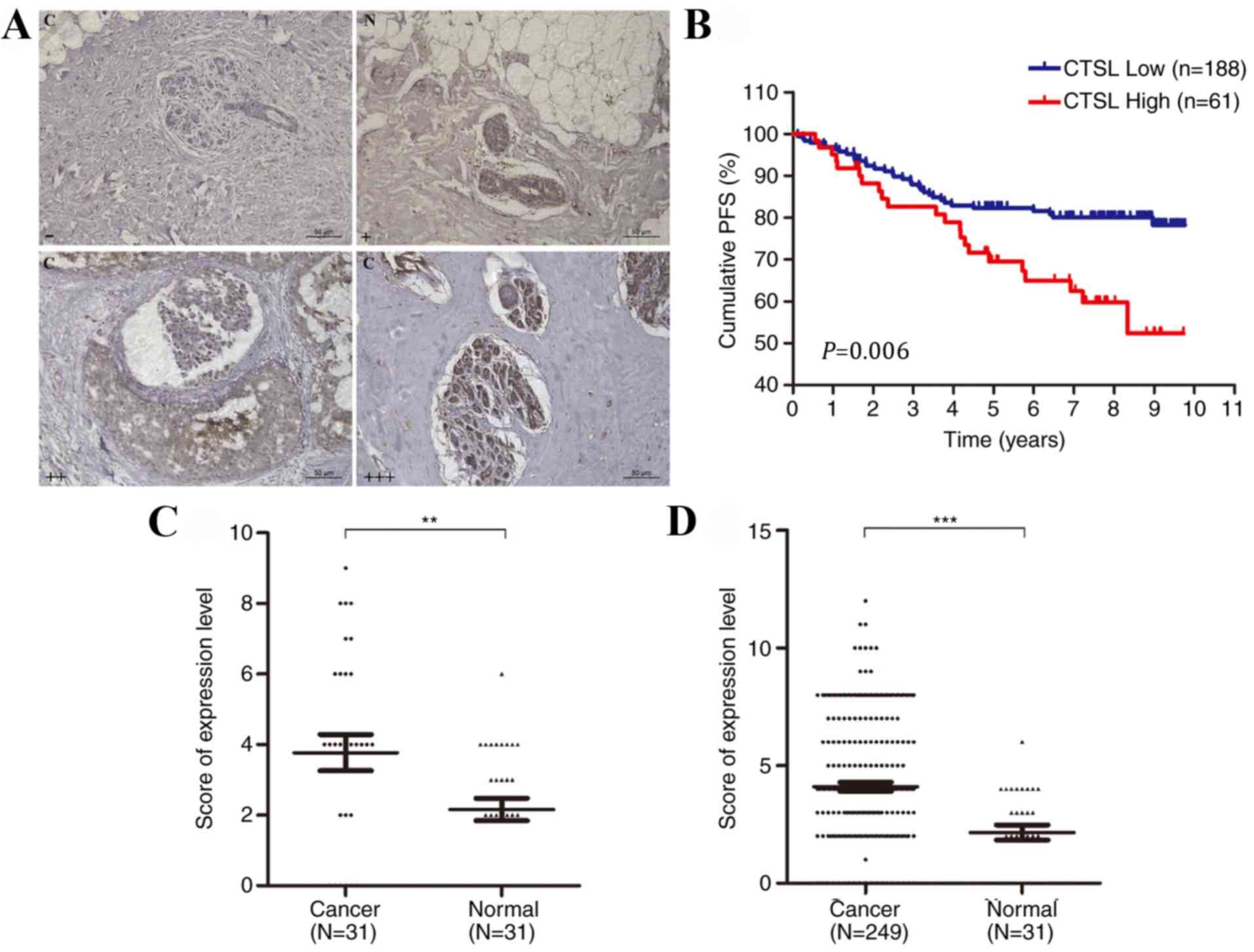
Immunohistochemistry was performed to determine CTSL
expression levels in patients with breast cancer, and PFS analysis
was performed to determine its prognostic value (Fig. 2A and B). CTSL was upregulated in
breast cancer tissues compared with adjacent normal tissues
(Fig. 2C and D). For PFS analysis,
the median follow-up time was 7.59 years; CTSL expression levels
(P=0.006), menstrual (P=0.007), lymph node metastasis (P<0.001),
ER (P=0.004) and PR (P=0.014) statuses were significantly
associated with PFS in the univariate Cox-proportional hazard model
(Table II). Patients with low CTSL
expression levels exhibited a higher PFS rate (Fig. 2B). The parameters significantly
associated with PFS were analyzed using multivariate Cox regression
analysis. By adjusting menstrual status, lymph node metastasis, ER
and PR status as covariates, CTSL expression level was not
significantly associated with PFS in patients with breast cancer
(hazard ratio, 0.61; 95% confidence interval, 0.35–1.09; P=0.096;
Table II).
 | Table II.Univariate and multivariate Cox
regression analyses of CTSL expression and PFS in patients with
breast cancer. |
Table II.
Univariate and multivariate Cox
regression analyses of CTSL expression and PFS in patients with
breast cancer.
|
| PFS |
|---|
|
|
|
|---|
| Variables | Univariate HR (95%
CI) | P-value | Multivariate HR
(95% CI) | P-value |
|---|
| Age (≤50 vs.
>50) | 0.65
(0.38–1.09) | 0.104 |
|
|
| Menstrual status
(pre-menopausal vs. post-menopausal) | 2.05
(1.21–3.46) | 0.007a | 2.36
(1.36–4.12) | 0.002a |
| Stage (I–II vs.
III) | 1.06
(0.52–2.18) | 0.862 |
|
|
| Lymph node
metastasis (positive vs. negative) | 0.24
(0.13–0.46) |
<0.001a | 0.21
(0.11–0.40) |
<0.001a |
| ER status (negative
vs. positive) | 2.21
(1.29–3.79) | 0.004a | 2.51
(1.32–4.80) | 0.005a |
| PR status (negative
vs. positive) | 1.96
(1.15–3.38) | 0.014a | 0.93
(0.48–1.80) | 0.831 |
| HER-2 status
(negative vs. positive) | 0.83
(0.47–1.46) | 0.512 |
|
|
| CTSL expression
(high vs. low) | 0.47
(0.27–0.80) | 0.006a | 0.61
(0.35–1.09) | 0.096 |
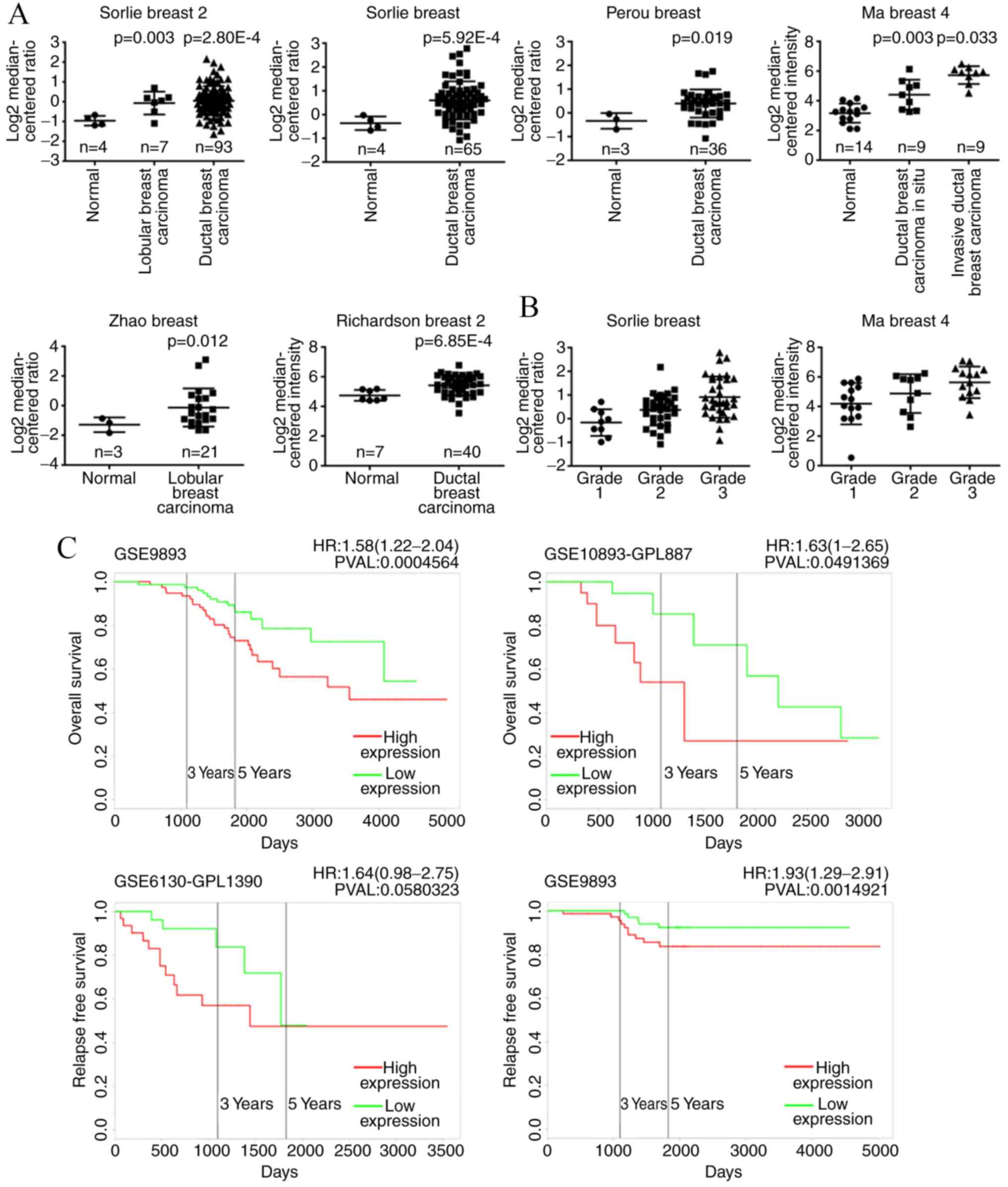
To further investigate the role of CTSL in breast
cancer, six independent microarray datasets from the ONCOMINE
database were analyzed, which exhibited statistically higher CTSL
expression levels in the majority of breast cancer tissues compared
with adjacent non-cancerous tissues (Fig. 3A). In addition, increased expression
of CTSL was associated with histological grade and clinical stage
of breast cancer (Fig. 3B). To
further investigate the prognostic significance of CTSL expression
in patients with breast cancer, Kaplan-Meier analysis was performed
using the PROGgene database (33).
Patients with high CTSL expression exhibited worse overall survival
compared with patients with low CTSL expression levels, suggesting
that CTSL served as a promoting factor during the tumorigenesis and
malignant progression of breast cancer, which may be regarded as a
valuable hallmark for prognosis.
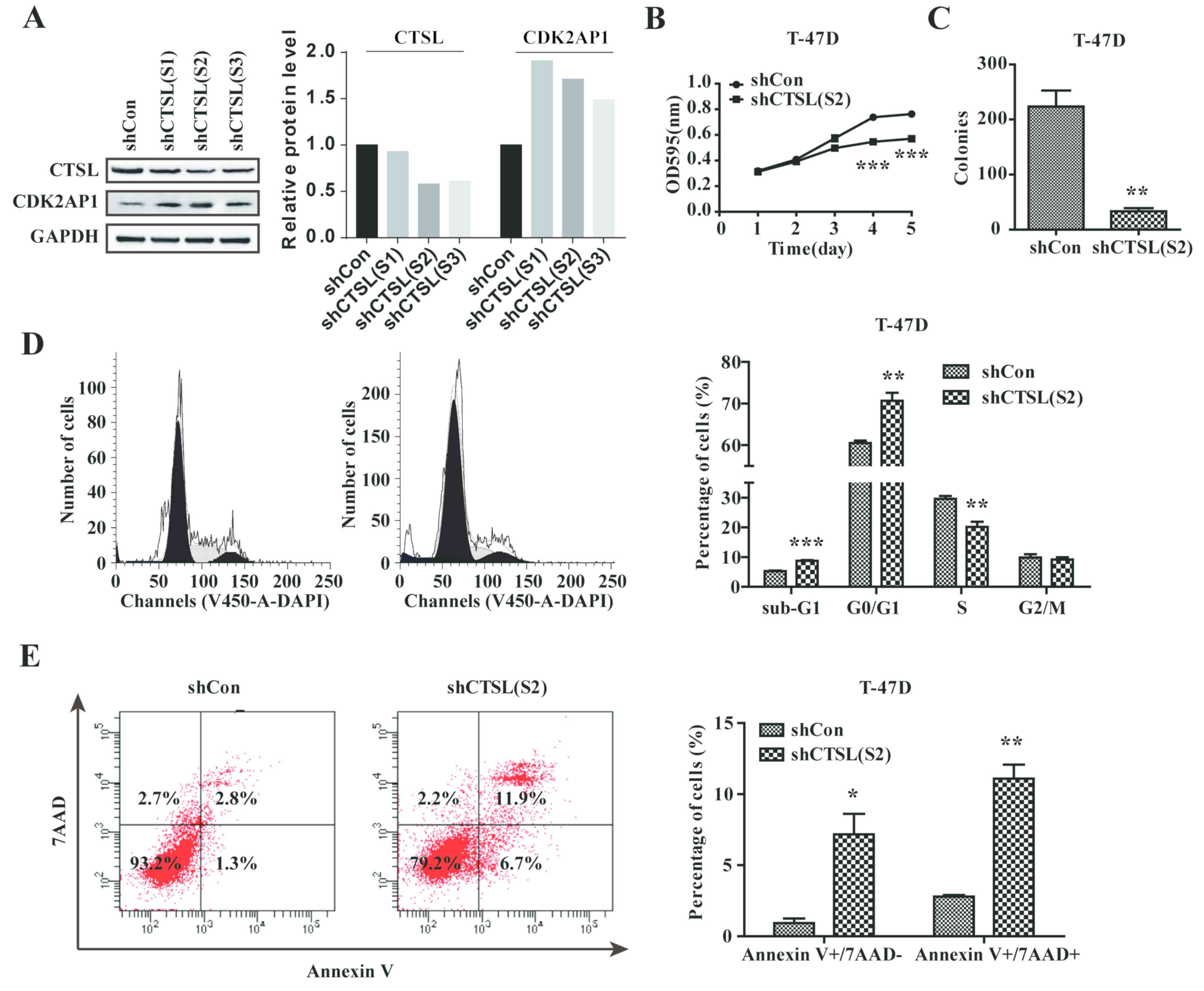
CTSL knockdown reduces the progression
of breast cancer cells in vitro
As upregulation of CTSL was significantly associated
with the progression and poor prognosis in breast cancer, the
pathological functions of CTSL were studied in vitro. T-47D
cells were selected due to high expression levels of CTSL (Fig. 1D). Endogenous CTSL was knocked down
using the specific Lv-shCTSL and CDK2-AP1 was increased in
CTSL-downregulated cells (Fig. 4A).
An MTT assay demonstrated that the proliferation of T-47D cells
transfected with shCTSL was decreased significantly compared with
those transfected with shCon (Fig.
4B). Colony formation of T-47D cells was significantly
decreased following CTSL downregulation compared with the negative
control (Fig. 4C), suggesting that
CTSL may contribute to the proliferation and colony formation of
breast cancer cells. In addition, flow cytometry revealed that
knockdown of CTSL expression significantly induced G0/G1-phase
arrest in T-47D cells and increased the apoptotic index compared
with the control cells (Fig. 4D and
E), suggesting that CTSL knockdown may decelerate the
progression of breast cancer by arresting the cell cycle and
inducing apoptosis.
Discussion
Accumulating evidence has demonstrated that
proteolytic enzymes serve a contributing role in cancer metastasis
(35,36). Proteases are involved in several
stages of tumorigenesis or malignant progression, including
proteolytic activation of latent growth factors and pro-angiogenic
factors, degradation of the extracellular and interstitial
matrices, as well as intravasation or extravasation across the
capillary/lymphatic system (37,38). In
light of the crucial functions of proteases in tumor development
and progression, numerous inhibitors have been developed and
applied in clinical trials, such as MPP inhibitors. A number of
protease-targeted drugs have been successful, although adverse side
effects hinder their application in oncotherapy (39). Therefore, it is important to identify
novel potential proteolytic targets and antagonists to impair tumor
metastasis and progression.
In the present study, CTSL was identified as a
potential target for treating breast cancer. CTSL is a member of
the papain superfamily of cysteine proteases, is overexpressed in
multiple cancers and associated with cancer metastasis (40–42), and
perceived as a potential facilitator of tumorigenesis and
neoplastic progression (18,43–45). For
example, CTSL ablation led to a significant reduction in MDA-MB-231
tumor cell-induced angiogenesis in vitro and in vivo
by affecting cell cycle-associated genes, including cyclin D1-D3,
E2, A2, B2 and H (15). In addition,
CTSL blocked TGF-β-induced cell migration via the PI3K-AKT and Wnt
signaling pathways in A549 and MCF-7 cell lines (46). Qin et al (47) demonstrated that CTSL affected
proliferation of breast cancer; downregulation of CTSL
significantly inhibited the proliferation of MCF-7 cells, which was
similar to the results of the present study.
As previously reported (34), CDK2-AP1 serves as a tumor suppressor
in breast cancer in vitro and in vivo. In the present
study, Y2H screening further revealed 13 putative
CDK2-AP1-interacting proteins, including CTSL; a direct physical
interaction between CDK2-AP1 and CTSL was demonstrated using a GST
pull-down assay. Analysis using the Oncomine and PROGgene databases
revealed increased expression of CTSL in breast cancer tissues
compared with normal tissues, and the expression profile of CTSL in
tumor tissues exhibited a negative association with clinical
outcomes and overall survival, suggesting a potential role for CTSL
as a drug target in the clinical oncotherapy of breast cancer.
To delineate how CTSL functioned in breast cancer,
the endogenous expression pattern of CTSL in a range of established
breast cancer cell lines was initially determined. In agreement
with the bioinformatics analysis, CTSL was upregulated in breast
cancer cells compared with 293T cells and exhibited an inverse
association with CDK2-AP1 expression. In addition, knockdown of
CTSL in T-47D cells induced by lentiviral shRNA notably suppressed
cell proliferation and colony formation in vitro, and T-47D
cells exhibited G0/G1-phase arrest and increased apoptosis when
CTSL was knocked down compared with the negative control, which was
in agreement with our previous study (34). Knockdown of CTSL may thus interrupt
the cell cycle and promote apoptosis. In addition, as T-47D is
invasive, whether CTSL may affect the migration and invasion of
breast cancer requires further study.
As CDK2-AP1 functions as a specific negative
regulator for cyclin-dependent kinase 2 (CDK2) and CDK2 is a
checkpoint in the cellular G1/S-phase (48,49),
CTSL may be an upstream negative regulator of CDK2-AP1 that
proteolytically inactivates CDK2-AP1 and weakens or reverses its
inhibition of CDK2. This mechanism may be explained by the
observation that CTSL was occasionally detected in normal tissues,
but upregulated in the majority of breast cancer tissues associated
with malignant progression and poor prognosis. Additional studies
are required to validate this hypothesis.
In conclusion, results of the present study
demonstrated that CTSL may act as a tumor promoter by interfering
with the tumor suppressor CDK2-AP1, leading to the progression of
breast cancer. These findings suggest a novel potential drug target
for the treatment of breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported The National Natural
Science Foundation of China (grant nos. 81572612, 81803640 and
81372842), the Hunan Provincial Natural Science Foundation (grant
no. 2015JJ2183), Youth Science Foundation of Xiangya Hospital,
Central South University (grant no. 2017Q02), The National Key
Clinical Specialist Construction Programs of China, the Research
Innovation Program for Graduate Students of Central South
University (grant no. 2018zzts912) and The Program of China
Scholarships Council (grant no. 201706370123 to ZW).
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW, ZX, LQS and WBZ conceived and designed the
experiments. ZW and TZ performed the experiments, ZW and JC
analyzed the data. MZZ, JH, KSW, and LL contributed to the
collection of clinical samples, performing experiments and data
analysis. ZW wrote the manuscript. ZW, TZ, LQS and WBZ reviewed and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by The Ethical Review
Committee of Xiangya Hospital, Central South University (Changsha,
China). All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gansler T, Ganz PA, Grant M, Greene FL,
Johnstone P, Mahoney M, Newman LA, Oh WK, Thomas CR Jr, Thun MJ, et
al: Sixty years of CA: A cancer journal for clinicians. CA Cancer J
Clin. 60:345–350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Fedewa SA, Goding Sauer A,
Kramer JL, Smith RA and Jemal A: Breast cancer statistics, 2015:
Convergence of incidence rates between black and white women. CA
Cancer J Clin. 66:31–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He M, Guo Q and Hu G: Reversed urban-rural
differences in breast cancer mortality (China, 2002–2008). Breast
Cancer Res Treat. 126:231–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mahamodhossen YA, Liu W and Rong-Rong Z:
Triple-negative breast cancer: New perspectives for novel
therapies. Med Oncol. 30:6532013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bilal E, Dutkowski J, Guinney J, Jang IS,
Logsdon BA, Pandey G, Sauerwine BA, Shimoni Y, Moen Vollan HK,
Mecham BH, et al: Improving breast cancer survival analysis through
competition-based multidimensional modeling. PLoS Comput Biol.
9:e10030472013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Freedman GM and Fowble BL: Local
recurrence after mastectomy or breast-conserving surgery and
radiation. Oncology (Williston Park). 14:1561–1581. 2000.PubMed/NCBI
|
|
8
|
Duffy MJ: Biochemical markers in breast
cancer: Which ones are clinically useful? Clin Biochem. 34:347–352.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mohammadzadeh F, Mosayebi G, Montazeri V
and Darabi M, Fayezi S, Shaaker M, Rahmati M, Baradaran B,
Mehdizadeh A and Darabi M: Fatty acid composition of tissue
cultured breast carcinoma and the effect of stearoyl-coa desaturase
1 inhibition. J Breast Cancer. 17:136–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo Y, Wang P, Li X, Zhu S, Xu H, Li S,
Deng H and Yuan L: Identifying a BRCA2 c.5722_5723del mutation in a
Han-Chinese family with breast cancer. Biosci Rep. 39:2019.
View Article : Google Scholar
|
|
11
|
Rawlings ND, Barrett AJ and Bateman A:
MEROPS: The database of proteolytic enzymes, their substrates and
inhibitors. Nucleic Acids Res. 40:D343–D350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kabel AM: Tumor markers of breast cancer:
New prospectives. J Oncol Sci. 3:5–11. 2017.
|
|
13
|
Tan G, Liu Q, Tang X, Kang T, Li Y, Lu J,
Zhao X and Tang F: Diagnostic values of serum cathepsin B and D in
patients with nasopharyngeal carcinoma. BMC Cancer. 16:2412016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uhlman A, Folkers K, Liston J, Pancholi H
and Hinton A: Effects of vacuolar H+-ATPase inhibition on
activation of cathepsin B and Cathepsin L secreted from MDA-MB231
breast cancer cells. Cancer Microenviron. 10:49–56. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sudhan DR, Rabaglino MB, Wood CE and
Siemann DW: Cathepsin L in tumor angiogenesis and its therapeutic
intervention by the small molecule inhibitor KGP94. Clin Exp
Metastasis. 33:461–473. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Li X, Peng D, Tan Z, Liu H, Qing Y,
Xue Y and Shi GP: Usefulness of serum Cathepsin L as an independent
biomarker in patients with coronary heart disease. Am J Cardiol.
103:476–481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sui H, Shi C, Yan Z and Wu M:
Overexpression of Cathepsin L is associated with chemoresistance
and invasion of epithelial ovarian cancer. Oncotarget.
7:45995–46001. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sudhan DR and Siemann DW: Cathepsin L
targeting in cancer treatment. Pharmacol Ther. 155:105–116. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
World Medical Association, . World Medical
Association declaration of helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Chen J, Zhong MZ, Huang J, Hu YP,
Feng DY, Zhou ZJ, Luo X, Liu ZQ, Jiang WZ and Zhou WB:
Overexpression of ANLN contributed to poor prognosis of
anthracycline-based chemotherapy in breast cancer patients. Cancer
Chemother Pharmacol. 79:535–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sorlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Prolc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar
|
|
24
|
Perou CM, Sorlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11:R72009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao H, Langerod A, Ji Y, Nowels KW,
Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D,
Børresen-Dale AL and Jeffrey SS: Different gene expression patterns
in invasive lobular and ductal carcinomas of the breast. Mol Biol
Cell. 15:2523–2536. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Richardson AL, Wang ZC, De Nicolo A, Lu X,
Brown M, Miron A, Liao X, Iglehart JD, Livingston DM and Ganesan S:
X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weigman VJ, Chao HH, Shabalin AA, He X,
Parker JS, Nordgard SH, Grushko T, Huo D, Nwachukwu C, Nobel A, et
al: Basal-like breast cancer DNA copy number losses identify genes
involved in genomic instability, response to therapy, and patient
survival. Breast Cancer Res Treat. 133:865–880. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mullins M, Perreard L, Quackenbush JF,
Gauthier N, Bayer S, Ellis M, Parker J, Perou CM, Szabo A and
Bernard PS: Agreement in breast cancer classification between
microarray and quantitative reverse transcription PCR from
fresh-frozen and formalin-fixed, paraffin-embedded tissues. Clin
Chem. 53:1273–1279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chanrion M, Negre V, Fontaine H, Salvetat
N, Bibeau F, Mac Grogan G, Mauriac L, Katsaros D, Molina F,
Theillet C and Darbon JM: A gene expression signature that can
predict the recurrence of tamoxifen-treated primary breast cancer.
Clin Cancer Res. 14:1744–1752. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goswami CP and Nakshatri H: PROGgeneV2:
Enhancements on the existing database. BMC Cancer. 14:9702014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou W, Guan X, Wang L, Liao Y and Huang
J: p12(CDK2-AP1) inhibits breast cancer cell proliferation and in
vivo tumor growth. J Cancer Res Clin Oncol. 138:2085–2093. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Herszenyi L, Barabas L, Hritz I, Istvan G
and Tulassay Z: Impact of proteolytic enzymes in colorectal cancer
development and progression. World J Gastroenterol. 20:13246–13257.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Herszenyi L, Lakatos G, Hritz I, Varga MZ,
Cierny G and Tulassay Z: The role of inflammation and proteinases
in tumor progression. Dig Dis. 30:249–254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mason SD and Joyce JA: Proteolytic
networks in cancer. Trends Cell Biol. 21:228–237. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tan GJ, Peng ZK, Lu JP and Tang FQ:
Cathepsins mediate tumor metastasis. World J Biol Chem. 4:91–101.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Turk B: Targeting proteases: Successes,
failures and future prospects. Nat Rev Drug Discov. 5:785–799.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tholen M, Wolanski J, Stolze B, Chiabudini
M, Gajda M, Bronsert P, Stickeler E, Rospert S and Reinheckel T:
Stress-resistant translation of Cathepsin L mRNA in breast cancer
progression. J Biol Chem. 290:15758–15769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sudhan DR, Pampo C, Rice L and Siemann DW:
Cathepsin L inactivation leads to multimodal inhibition of prostate
cancer cell dissemination in a preclinical bone metastasis model.
Int J Cancer. 138:2665–2677. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sigloch FC, Tholen M, Gomez-Auli A,
Biniossek ML, Reinheckel T and Schilling O: Proteomic analysis of
lung metastases in a murine breast cancer model reveals divergent
influence of CTSB and CTSL overexpression. J Cancer. 8:4065–4074.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao Y, Liu X, Li Y, Lu Y, Zhong H, Jiang
W, Chen AF, Billiar TR, Yuan H and Cai J: Cathepsin L activity
correlates with proteinuria in chronic kidney disease in humans.
Int Urol Nephrol. 49:1409–1417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fei Y, Xiong Y, Zhao Y, Wang W, Han M,
Wang L, Tan C and Liang Z: Cathepsin L knockdown enhances
curcumin-mediated inhibition of growth, migration, and invasion of
glioma cells. Brain Res. 1646:580–588. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Han ML, Zhao YF, Tan CH, Xiong YJ, Wang
WJ, Wu F, Fei Y, Wang L and Liang ZQ: Cathepsin L
upregulation-induced EMT phenotype is associated with the
acquisition of cisplatin or paclitaxel resistance in A549 cells.
Acta Pharmacol Sin. 37:1606–1622. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qingqing Z, Meiling H, Wenjuan W, Song Y,
Chen G, Wang Z and Liang Z: Downregulation of Cathepsin L
suppresses cancer invasion and migration by inhibiting transforming
growth factor?β?mediated epithelial?mesenchymal transition. Oncol
Rep. 33:1851–1859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qin G, Cai Y, Long J, Zeng H, Xu W, Li Y,
Liu M, Zhang H, He ZL and Chen WG: Cathepsin L is involved in
proliferation and invasion of breast cancer cells. Neoplasma.
63:30–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He X, Xiang H, Zong X, Yan X, Yu Y, Liu G,
Zou D and Yang H: CDK2-AP1 inhibits growth of breast cancer cells
by regulating cell cycle and increasing docetaxel sensitivity in
vivo and in vitro. Cancer Cell Int. 14:1302014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chai J, Ju J, Zhang SW, Shen ZY, Liang L,
Yang XM, Ma C, Ni QW and Sun MY: p12CDK2-AP1 interacts with CD82 to
regulate the proliferation and survival of human oral squamous cell
carcinoma cells. Oncol Rep. 36:737–744. 2016. View Article : Google Scholar : PubMed/NCBI
|