Introduction
Currently, glioma is the most common and aggressive
primary brain tumor in the central nervous system of adults
worldwide (1). The yearly incidence
of glioma is ~5 cases per 100,000 individuals and it is also has
one of the highest mortality rates worldwide (2–4). Glioma
remains a major public health problem in China and worldwide.
Therefore, it is important to elucidate its underlying mechanisms,
and identify novel diagnostic biomarkers and therapeutic targets
for glioma.
Long non-coding RNA (lncRNA) is a class of ncRNAs of
>200 nucleotides that are poorly conserved in different species
(5). lncRNAs were once considered to
be transcriptional ‘noise’ without biological functions, as the
RNAs have no evident protein-coding capacity (6). However, increasing evidence has
demonstrated that lncRNAs have pivotal roles in the initiation and
progression of human malignancies, including glioma, involving
transcriptional and post-transcriptional regulation (7,8).
lncRNA metallothionein 1J, pseudogene (MT1JP) is
located on chromosome 16 in a cluster consisting of several
homologous protein-coding genes. Liu et al (9) reported the tumor suppressive function
of MJ1JP and revealed that MT1JP exerts its roles by regulating the
expression of p53 and, therefore, regulating the activity of the
p53 signalling pathway. Subsequently, Xu et al (10) reported that MT1JP is downregulated in
gastric cancer tissues and cell lines. Additionally, low expression
of MT1JP was significantly associated with advanced Tumor, Necrosis
and Metastasis stage and lymphatic metastasis (10). Furthermore, MT1JP overexpression
suppresses gastric cancer cell proliferation, migration and
invasion, and promoted apoptosis in vitro and in vivo
via an MT1JP/microRNA (miRNA)-214-3p/runt related transcription
factor 3 axis (10); however, the
biological functions and potential mechanisms of MT1JP in glioma
remain unclear.
In the present study, the expression of MT1JP was
investigated in glioma tissues and cell lines, and its biological
functions and underlying mechanisms were explored. The present
results suggested that MT1JP was downregulated in glioma and its
overexpression inhibited glioma cell proliferation, migration and
invasion by negatively regulating miRNA-24 (miR-24). The present
data suggested that MT1JP may have an important tumor-suppressing
role in glioma and may serve as a novel diagnostic biomarker and
therapeutic target for glioma.
Materials and methods
Human tissue samples
Between January 2006 and December 2011, 40 pairs of
glioma tissues and paired adjacent normal tissues were collected
from patients (23 males, 17 females; range, 37–68 years) undergoing
resection surgery at the Department of Neurosurgery, The First
Affiliated Hospital of Gannan Medical University (Ganzhou, China).
The inclusion criteria were that patients were aged >18 years,
and had a pathological diagnosis of glioma in the brain based on
the WHO classification (11). The
exclusion criteria were that the patient had another type of tumor,
or an infection or non-surgical removal of the tumor. The adjacent
normal brain tissues were obtained 2–3 cm away from the glioma
during surgery. Histological examination was performed in order to
verify the normal brain tissues and glioma tissues. No patients
received preoperative anti-cancer treatment, including chemotherapy
or radiation, prior to the collection of specimens. All specimens
from resection surgery were frozen and stored in liquid nitrogen
(−196°C) until required. The present study was performed with the
approval of the Ethic and Research Committees of the First
Affiliated Hospital of Gannan Medical University. Informed consent
was obtained from all participants.
Cell culture
Human glioma cell lines (SHG-44 and U251),
glioblastoma of unknown origin (U87; cat. no. HTB14) and human
astroglia cells (HA) were used in the present study. All cell lines
were purchased from American Type Culture Collection and cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS, 50 U/ml penicillin and 0.1 mg/ml streptomycin (Biowest).
All the cell cultures were incubated at 37°C in 5%
CO2.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from glioma tissues and
glioma cell lines (80–90% confluence) using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Following extraction, the RNA and
miRNA samples were reverse transcribed into cDNA using a RevertAid
RT kit (Thermo Fisher Scientific, Inc.) or miRNA RT kit (Tiangen
Biotech Co., Ltd.), respectively. The reverse transcription
reaction condition was as follows: 37°C for 15 min, 85°C for 5 sec
and 4°C for 2 min. The cDNA templates were amplified by qPCR using
SYBR Green Mix (Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for 55 sec, 40 cycles of 95°C for 22 sec and
60°C for 44 sec. mRNA levels were quantified using the
2−ΔΔCq method (12).
GAPDH was used as an internal control for lncRNAs and miRNA samples
were normalized to U6 expression. The following primers were used
for the qPCR: MT1JP, forward:
5′-TACCGAGCTCGGATCCTTGCGGTCTCTCCATTTATCG-3′, reverse:
5′-TACCGAGCTCGGATCCTTGCGGTCTCTCCATTTATCG-3′; GAPDH, forward:
5′-GCACCGTCAAGGCTGAGAAC-3′, reverse: 5′-TGGTGAAGACGCCAGTGGA-3′. U6
forward, 5′-CTCGCTTCGGCAGCACA3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The aforementioned primers were
designed and synthesized by Shangai GenePharma Co., Ltd. (Shanghai,
China).
Constructs, synthesized oligos and
transfection
The MT1JP overexpression plasmid and miR-24 mimic
were purchased from Shanghai GenePharma Co., Ltd. The plasmids and
mimics were both introduced into cells at a final concentration of
50 nM. The sequence was amplified and inserted into the pcDNA3.1
(+) vector at the BamH1 sites. The pcDNA3.1 (+) empty vector
(Shanghai GenePharma Co., Ltd) was used as a negative control for
MT1JP overexpression. Lipofectamine® 3000 (Thermo Fisher
Scientific, Inc.) was used for transfection according to the
manufacturer's instructions. At 48 h after transfection, cells were
used for the subsequent experiments. The sequence of the negative
control mimic was as follows: 5′-UUCUCCGAACGUGUCACGUTT-3′.
Cell proliferation assay
The cell proliferation assay was performed using a
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.)
according to the manufacturer's protocol. Cells were placed into
the 96 well plates at a density of ~3,000 cells/well, and examined
every 24 h. The absorbance was recorded at 450 nm and analyzed
following the instructions provided.
Cell invasion assay
Cell invasion was assessed using a Matrigel assay.
Cells (~1×105) were seeded into the upper chambers of a
Transwell plate (Corning, Inc.) with Matrigel (Corning, Inc.) in
300 µl serum-free DMEM, and the lower chambers were filled with 700
µl DMEM containing 10% FBS. After 6 h of incubation at 37°C, the
filters were treated with 4% paraformaldehyde for 15 min at room
temperature and crystal violet for 10 min at room temperature.
Finally, the number of cells that invaded through the membrane was
counted and calculated in ≥5 randomly selected fields of view under
an Olympus CKX31 inverted light microscope (magnification, ×400;
Olympus Corporation). Image J (version 1.40; National Institutes
for Health) software was used to quantify cell number.
Dual-luciferase reporter assay
The potential binding sites were mutated by the
QuikChang site-directed mutagenesis kit (Agilent Technologies,
Inc.). The wild type MT1JP (WT) and mutant MT1JP (MUT) sequence
containing the seeding site of miR-24 were established and
incorporated into the Firefly luciferase expressing psiCHECK-2
vector (Promega Corporation). All the cloned sequences were
validated by DNA sequencing. U87 cells were seeded into a 24-well
plate for 24 h before transfection. Cells were transfected with 0.4
µg pMiR-report-MT1JP-WT or pMiR-report-MT1JP-MUT plasmid, and 20 µM
miR-24 mimic or control scrambled oligo (Shanghai GenePharma Co.,
Ltd.; sequence: 5′-UUCUCCGAACGUGUCACGUTT-3′), together with 0.02 µg
Renilla luciferase vector pRL-TK (Promega Corporation) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the luciferase activities were
detected consecutively using the Dual-Luciferase®
Reporter assay system (Promega Corporation) following the
manufacturer's instructions.
Statistical analysis
All results are presented as the mean ± SD and
analysed using GraphPad Prism 5 (GraphPad Software, Inc.) from ≥3
independent experiments. The χ2 test was performed to
examine the associations between MT1JP level and
clinicopathological features. The Kaplan-Meier method was used to
calculate the survival curve, and log-rank test was used to
determine statistical significance. The differences among groups
were analyzed using one-way ANOVA followed by Tukey's post-hoc
test. Expression correlations were analyzed using Pearson's
correlation coefficient. P<0.05 was considered to indicate a
statistically significant difference.
Results
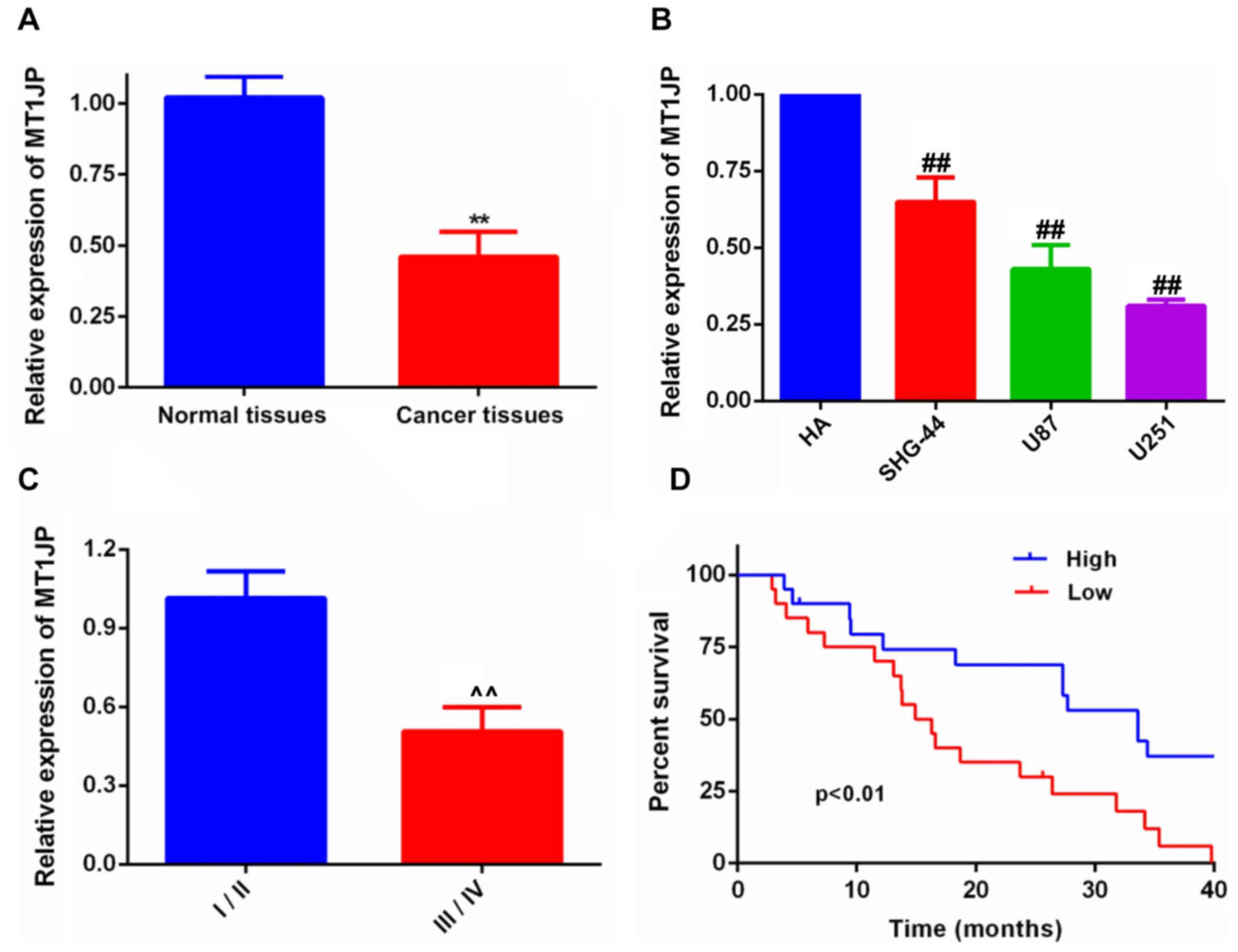
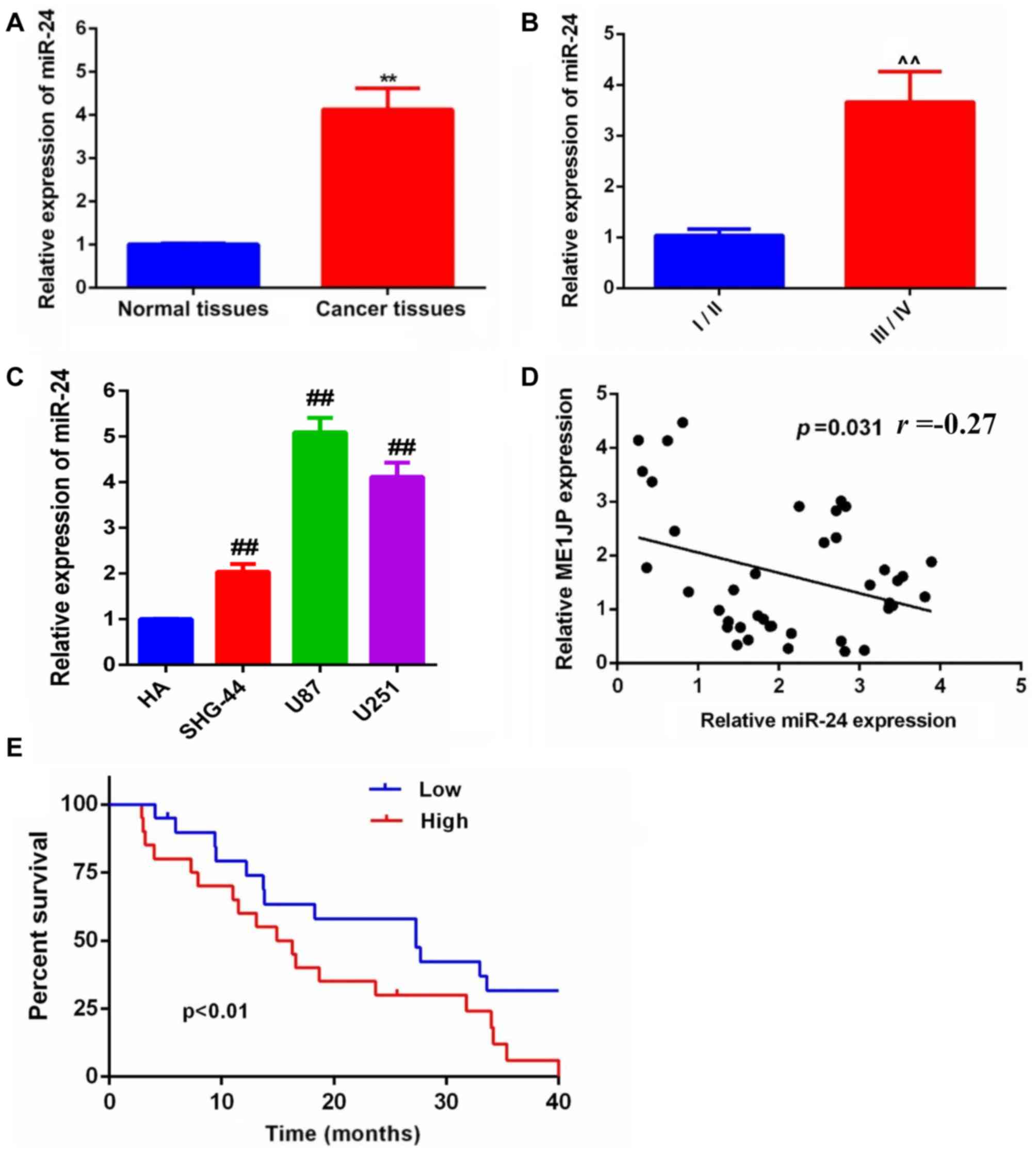
MT1JP is downregulated in glioma
MT1JP expression was determined in glioma tissues
and cell lines. The results indicated that MT1JP expression was
downregulated in glioma tissues (Fig.
1A) and cell lines (Fig. 1B).
Furthermore, the expression of MT1JP was lower in samples
presenting higher World Health Organisation (WHO) (11) tumor grades compared with lower WHO
grades (Fig. 1C). Additionally,
Kaplan-Meier survival analysis (Fig.
1D) suggested that the overall survival time of patients with
glioma was improved in the higher MT1JP expression group compared
with that in the lower expression group. The mean value was used to
determine high and low expression of MT1JP.
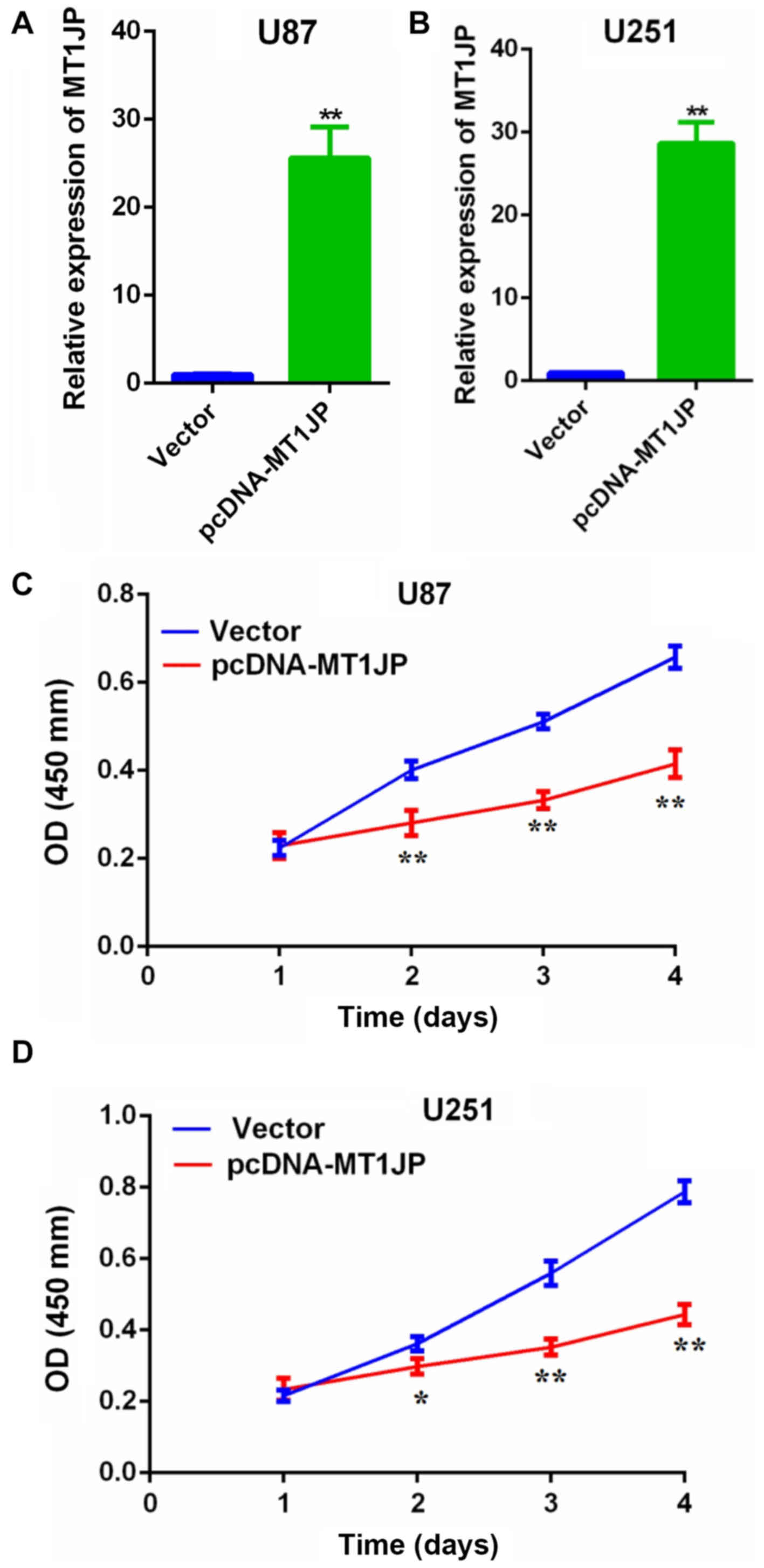
Overexpression of MT1JP suppresses
glioma cell proliferation and invasion
RT-qPCR revealed that the expression of MT1JP was
significantly upregulated in U87 (Fig.
2A) and U251 cells (Fig. 2B)
transfected with pcDNA-MT1JP compared with control. The CCK-8 assay
suggested that overexpression of MT1JP significantly inhibited
glioma cell proliferation (Fig. 2C and
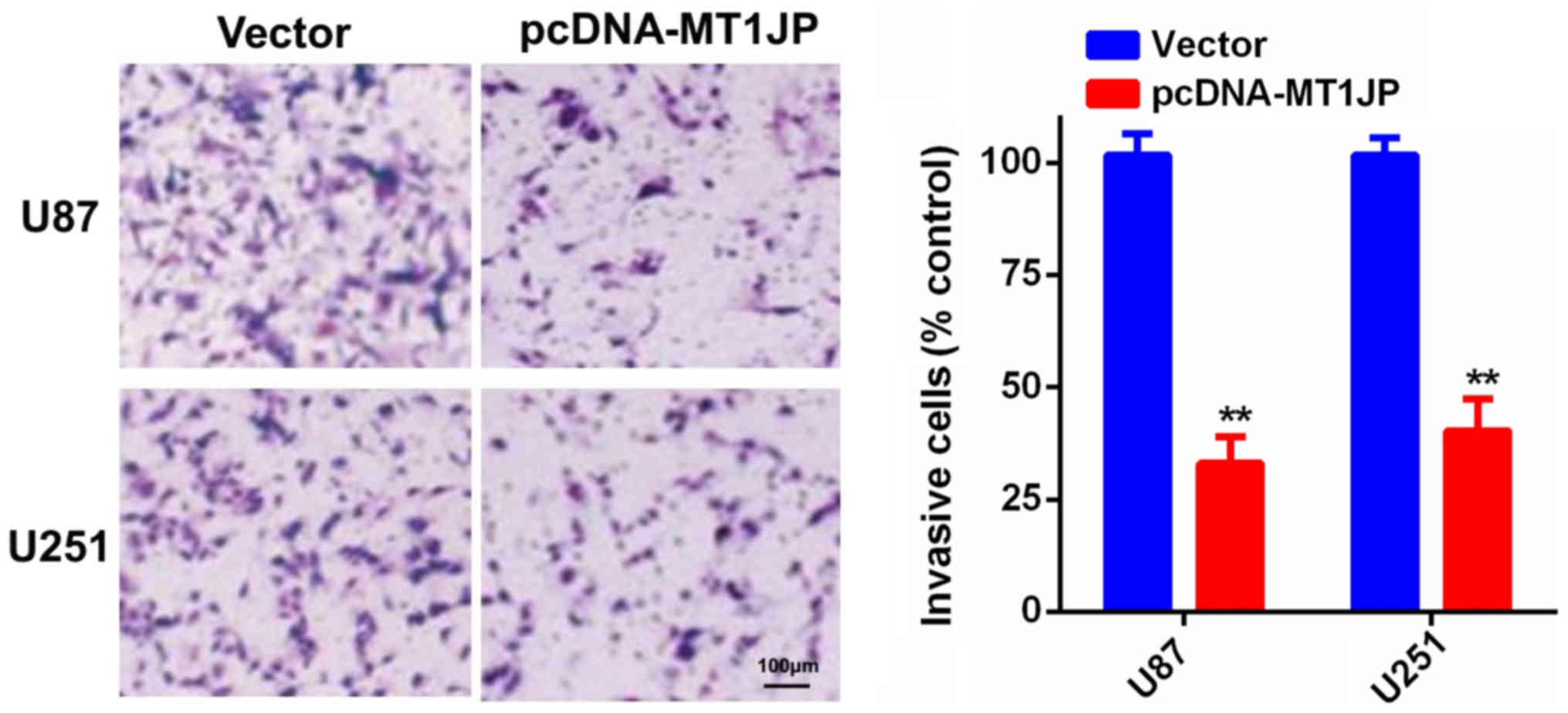
D). Additionally, the Matrigel invasion assay indicated that
overexpression of MT1JP significantly inhibited glioma cell
invasion (Fig. 3).
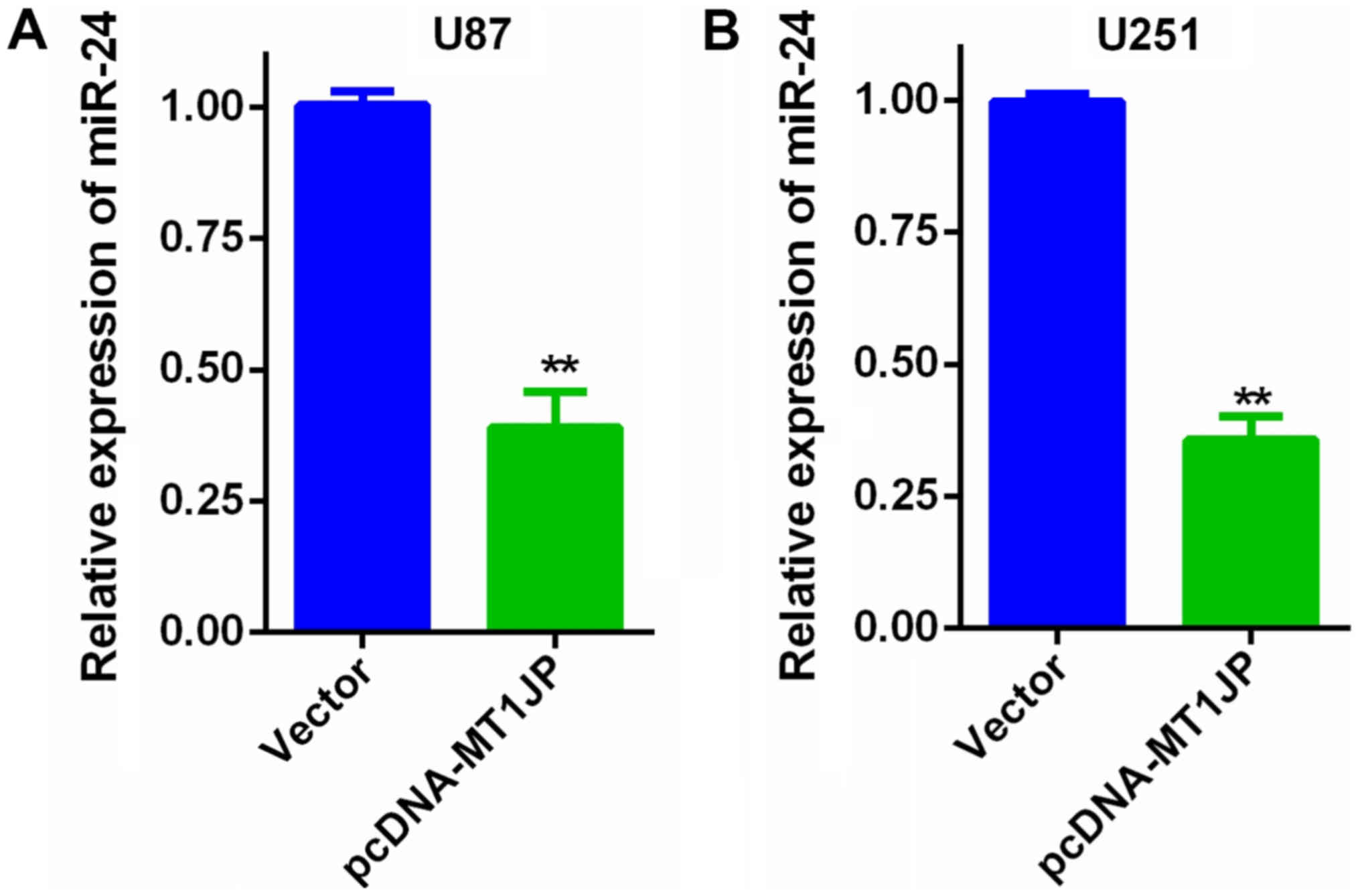
miR-24 expression in glioma
Overexpression of MT1JP suppressed miR-24 expression
in U87 cells (Fig. 4A) and U251
cells (Fig. 4B). Additionally, the
expression level of miR-24 in glioma and paired adjacent normal
tissues was evaluated by RT-qPCR. The miR-24 expression level was
significantly upregulated in glioma tissues compared with that in
the control normal tissues (Fig.
5A). Furthermore, the expression of miR-24 was higher in the
higher WHO graded samples compared with that in the lower WHO
graded samples (Fig. 5B).
Furthermore, miR-24 expression was significantly
upregulated in the glioma cell lines SHG-44, U87 and U251 compared
with HA cells (Fig. 5C). Notably,
miR-24 expression was negatively correlated with MT1JP expression
in glioma tissues (Fig. 5D).
Finally, Kaplan-Meier survival analysis suggested that the overall
survival time of patients with glioma was significantly improved in
patients with lower miR-24 expression compared with that in the
higher expression group (Fig.
5E).
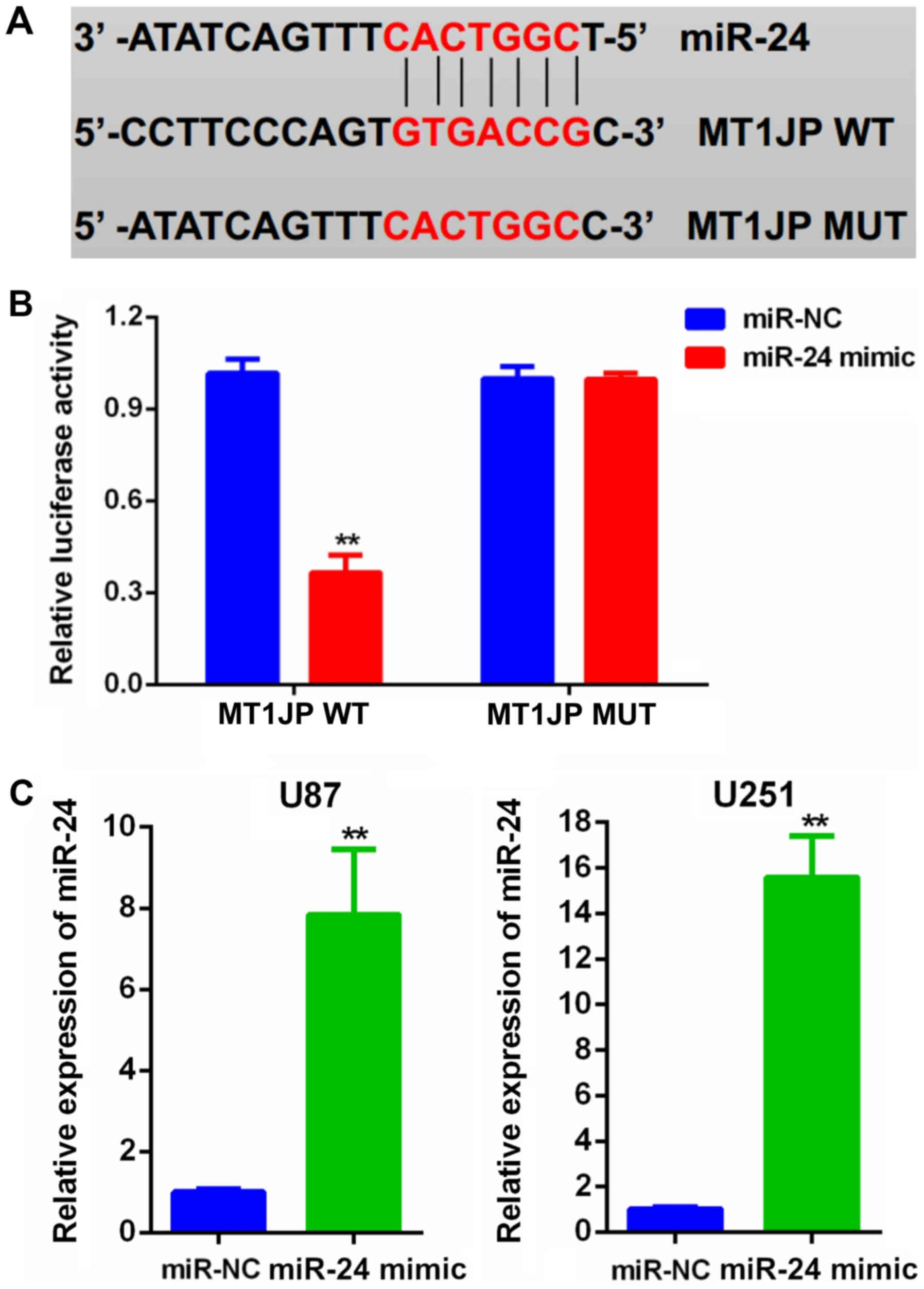
MT1JP is a direct target of
miR-24
Bioinformatics analysis of the predicted
lncRNA-miRNA interactions revealed potential binding sequences
between MT1JP and miR-24 (Fig. 6A)
(10). Subsequently, a luciferase
reporter assay confirmed that miR-24 overexpression significantly
decreased the luciferase activity of the MT1JP-WT reporter compared
with a negative control oligo, whereas miR-24 did not affect the
luciferase activity of the MT1JP-MUT reporter in U87 cells
(Fig. 6B). In addition, following
transfection with miR-24 mimics, the mRNA expression level of
miR-24 in both U87 and U251 cells was significantly increased
(Fig. 6C).
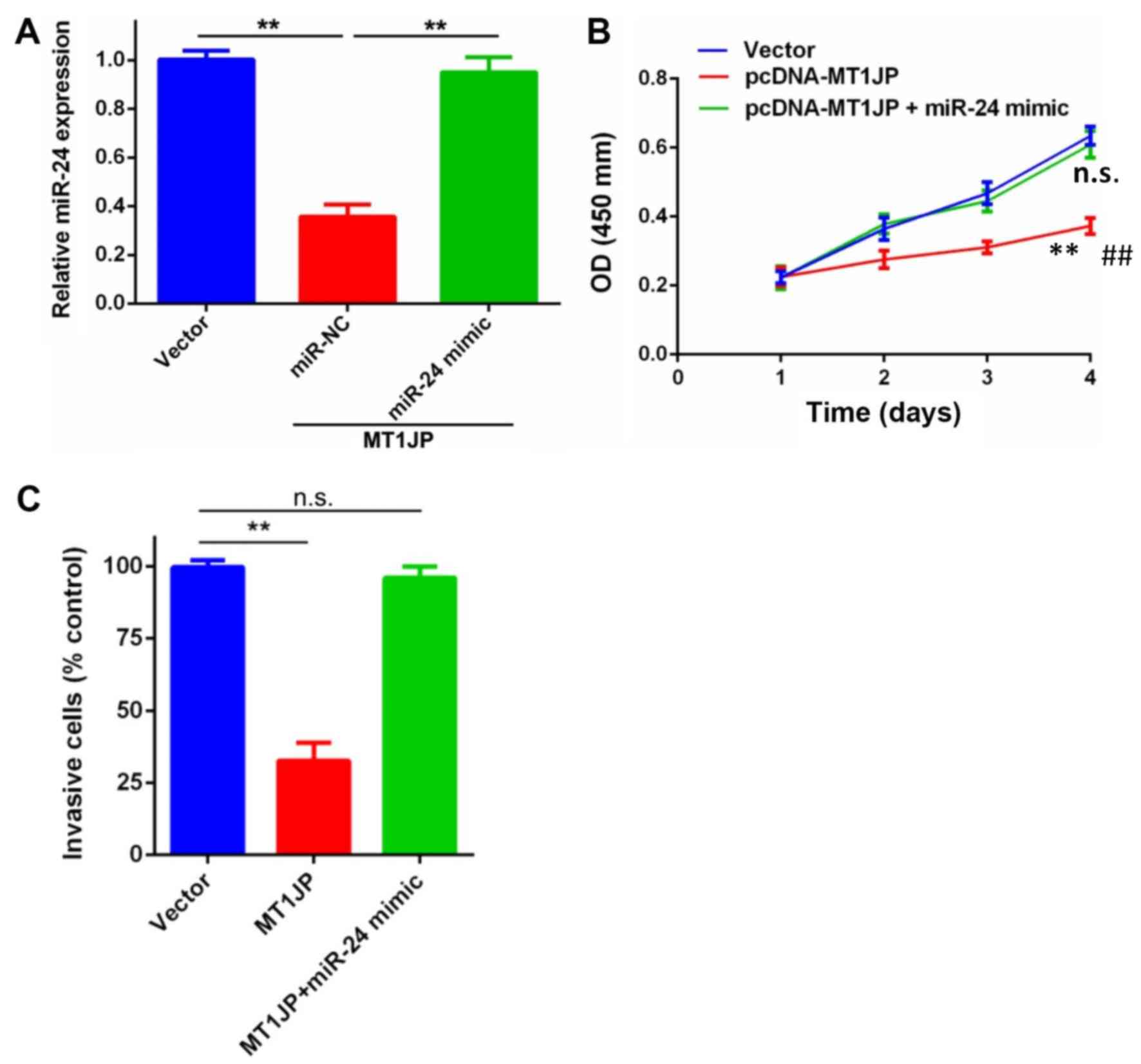
Inhibitory effect of MT1JP on glioma
cell proliferation and invasion is dependent on miR-24
Rescue experiments were performed to determine
whether MT1JP-mediated inhibition of glioma cell proliferation and
invasion was miR-24-dependent. miR-NC or miR-24 mimic were
co-transfected into U87 cells transfected with pcDNA-MT1JP. These
results confirmed that miR-NC or miR-24 mimic was stably
transfected into U87 cells that were already transfected with
pcDNA-MT1JP (Fig. 7A). CCK-8 and
Matrigel invasion assays suggested that the inhibition of
proliferation and invasion of U87 cells that was induced by MT1JP
overexpression was partially abolished in the presence of the
miR-24-3p mimic (Fig. 7B and C).
These data indicated that the tumor suppressive function of MT1JP
in glioma cells involved the negative regulation of miR-24.
Discussion
Glioma cells were reported to carry heterogeneous
genetic molecular aberrations and the molecular mechanism of glioma
development is highly complex (13,14). An
increasing number of studies have revealed that lncRNAs have
pivotal roles in the development of glioma (15–18).
lncRNA small nucleolar RNA host gene 12 promotes glioma progression
via an miR-101-3p/forkhead box P1 axis (19). Long intergenic non-coding RNA 152
facilitates glioma cell proliferation and invasion by interacting
with miR-16 (20).
The present study demonstrated that MT1JP expression
was significantly decreased in glioma tissues and cell lines. The
decreased expression of MT1JP was significantly associated with
poor prognosis. Furthermore, overexpression of MT1JP inhibited the
proliferation and invasion of glioma cells. These data suggest that
MT1JP has pivotal roles in glioma development.
lncRNAs can regulate the development of various
human malignancies, including glioma, via several different
mechanisms, including transcriptional and post-transcriptional
regulations (21,22). Accumulating evidence has revealed
that lncRNAs function as a competing endogenous RNA or a molecular
sponge to regulate the expression and biological functions of
miRNAs (23,24). For example, lncRNA retinal non-coding
RNA 3 promotes prostate cancer development via regulation of
miR-185 (25). LncRNA X inactive
specific transcript sponges miR-34a, facilitating colon cancer
development (26). A previous study
demonstrated that MT1JP inhibits proliferation, migration and
invasion, and promotes apoptosis of gastric cancer cells (10). Bioinformatics analysis suggested that
miR-24 was a potential target of MT1JP, and was previously reported
as a oncogenic miRNA in glioma (27). Thus, it was hypothesized that MT1JP
may inhibit glioma development by acting as a competing endogenous
RNA to modulate the function of miR-24.
To test this hypothesis, a series of experiments
were conducted in the present study, and the present results
suggested that miR-24 expression was upregulated in glioma tissues
and was negatively correlated with MT1JP expression. Additionally,
overexpression of MT1JP significantly decreased miR-24 expression.
The luciferase reporter assay further confirmed that miR-24 was a
direct target of MT1JP. Notably, rescue experiments suggested that
the tumor suppressive function of MT1JP in glioma cells may be
dependent on miR-24. Xu et al (27) reported that miR-24 promotes glioma
cell proliferation by targeting MAX interactor 1, dimerization
protein. Collectively, the present results supported this
regulatory mechanism, with MT1JP promoting the progression of
glioma by acting as a competing endogenous RNA to inhibit the
function of miR-24.
In conclusion, the present findings suggested that
MT1JP expression was downregulated in glioma tissues compared with
that in control normal tissues. Overexpression of MT1JP inhibited
glioma cell proliferation and invasion by acting as a competing
endogenous RNA and directly sponging miR-24. The present results
suggested the important function of MT1JP in glioma development and
provided a further understanding of the regulatory mechanisms
mediated by competing endogenous RNA underlying glioma development.
MT1JP may be useful as an important diagnostic biomarker and
therapeutic target in glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on request.
Authors' contributions
JC, JYL and SY performed the majority of the
experiments and were the major contributors in writing the
manuscript. JC, JL, WL and RZ collected and analysed data. JC, CQ
and GD made substantial contributions to the design of the study,
drafted the manuscript and revised it critically for important
intellectual content. GD gave final approval of the version to be
published.
Ethics approval and consent to
participate
The present study was conducted with the approval of
the Ethics and Research Committees of The First Affiliated Hospital
of Gannan Medical University (Jiangxi, China) and was performed in
accordance with the Declaration of Helsinki. All patients provided
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sareddy GR, Viswanadhapalli S, Surapaneni
P, Suzuki T, Brenner A and Vadlamudi RK: Novel KDM1A inhibitors
induce differentiation and apoptosis of glioma stem cells via
unfolded protein response pathway. Oncogene. 36:2423–2434. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Venkatesh HS, Tam LT, Woo PJ, Lennon J,
Nagaraja S, Gillespie SM, Ni J, Duveau DY, Morris PJ, et al:
Targeting neuronal activity-regulated neuroligin-3 dependency in
high-grade glioma. Nature. 549:533–537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Almiron Bonnin DA, Havrda MC, Lee MC, Liu
H, Zhang Z, Nguyen LN, Harrington LX, Hassanpour S, Cheng C and
Israel MA: Secretion-mediated STAT3 activation promotes
self-renewal of glioma stem-like cells during hypoxia. Oncogene.
37:1107–1118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Battistelli C, Cicchini C, Santangelo L,
Tramontano A, Grassi L, Gonzalez FJ, de Nonno V, Grassi G, Amicone
L and Tripodi M: The Snail repressor recruits EZH2 to specific
genomic sites through the enrollment of the lncRNA HOTAIR in
epithelial-to-mesenchymal transition. Oncogene. 36:942–955. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Tao Y and Liao Q: Long noncoding
RNA: A crosslink in biological regulatory network. Brief Bioinform.
2017.
|
|
7
|
Wang X, Vukovic L, Koh HR, Schulten K and
Myong S: Dynamic profiling of double-stranded RNA binding proteins.
Nucleic Acids Res. 43:7566–7576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S,
Jiang Z, Xu J, Liu Q and Cao X: The STAT3-binding long noncoding
RNA lnc-DC controls human dendritic cell differentiation. Science.
344:310–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G,
Chen X, Lu Y, Guo M, Luo J and Chen R: LncRNA MT1JP functions as a
tumor suppressor by interacting with TIAR to modulate the p53
pathway. Oncotarget. 7:15787–15800. 2016.PubMed/NCBI
|
|
10
|
Xu Y, Zhang G, Zou C, Zhang H, Gong Z,
Wang W, Ma G, Jiang P and Zhang W: LncRNA MT1JP suppresses gastric
cancer cell proliferation and migration through
MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 46:2445–2459.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boussiotis VA and Charest A:
Immunotherapies for malignant glioma. Oncogene. 37:1121–1141. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stepanenko AA and Kavsan VM:
Karyotypically distinct U251, U373, and SNB19 glioma cell lines are
of the same origin but have different drug treatment sensitivities.
Gene. 540:263–265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao S, Wang Y, Li J, Lv M, Niu H and Tian
Y: Tumor-suppressive function of long noncoding RNA MALAT1 in
glioma cells by suppressing miR-155 expression and activating FBXW7
function. Am J Cancer Res. 6:2561–2574. 2016.PubMed/NCBI
|
|
16
|
Meng L, Ma P, Cai R, Guan Q, Wang M and
Jin B: Long noncodingRNA ZEB1-AS1 promotes the tumorigenesis of
glioma cancer cells by modulating the miR-200c/141-ZEB1 axis. Am J
Transl Res. 10:3395–3412. 2018.PubMed/NCBI
|
|
17
|
Zong Z, Song Y, Xue Y, Ruan X, Liu X, Yang
C, Zheng J, Cao S, Li Z and Liu Y: Knockdown of LncRNA SCAMP1
suppressed malignant biological behaviours of glioma cells via
modulating miR-499a-5p/LMX1A/NLRC5 pathway. J Cell Mol Med.
8:5048–5062. 2019. View Article : Google Scholar
|
|
18
|
Liu H, Li C, Yang J, Sun Y, Zhang S, Yang
J, Yang L, Wang Y and Jiao B: Long noncoding RNA
CASC9/miR-519d/STAT3 positive feedback loop facilitate the
gliomatumourigenesis. J Cell Mol Med. 12:6338–6344. 2018.
View Article : Google Scholar
|
|
19
|
Sun Y, Liu J, Chu L, Yang W, Liu H, Li C
and Yang J: Long noncoding RNA SNHG12 facilitates the tumorigenesis
of glioma through miR-101-3p/FOXP1 axis. Gene. Nov 15–2018.(Epub
ahead of print). View Article : Google Scholar
|
|
20
|
Chen X, Li D, Gao Y, Tang W, Iw L, Cao Y
and Hao B: Long intergenic noncoding RNA 00152 promotes glioma cell
proliferation and invasion by interacting with MiR-16. Cell Physiol
Biochem. 46:1055–1064. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engreitz JM, Haines JE, Perez EM, Munson
G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES: Local
regulation of gene expression by lncRNA promoters, transcription
and splicing. Nature. 539:452–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Song Z, Feng C, Lu Y, Zhou Y, Lin Y
and Dong C: The long non-coding RNA SUMO1P3 facilitates breast
cancer progression by negatively regulating miR-320a. Am J Transl
Res. 9:5594–5602. 2017.PubMed/NCBI
|
|
24
|
Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou
Z, Chang C, Qi J and Yeh S: Estrogen receptor β promotes renal cell
carcinoma progression via regulating LncRNA
HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene.
37:5037–5053. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian C, Deng Y, Jin Y, Shi S and Bi H:
Long non-coding RNA RNCR3 promotes prostate cancer progression
through targeting miR-185-5p. Am J Transl Res. 10:1562–1570.
2018.PubMed/NCBI
|
|
26
|
Sun N, Zhang G and Liu Y: Long non-coding
RNA XIST sponges miR-34a to promotes colon cancer progression via
Wnt/β-catenin signaling pathway. Gene. 665:141–148. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu W, Liu M, Peng X, Zhou P, Zhou J, Xu K,
Xu H and Jiang S: miR-24-3p and miR-27a-3p promote cell
proliferation in glioma cells via cooperative regulation of MXI1.
Int J Oncol. 42:757–766. 2013. View Article : Google Scholar : PubMed/NCBI
|