Introduction
Cervical cancer (CC) is one of the most common
gynecological carcinomas worldwide with an incidence rate of ~3.2%
in 2018 (1). Although advancements
in radiotherapy and immunotherapy have been applied (2), ~50% of patients with late FIGO stage
CC, a clinical stage system based on the location and metastasis of
Tumor (3), remain to suffer from
recurrence and metastasis. A previous study demonstrated that the
overactivation of oncogene pathways such as Wnt/β-catenin was
associated with the progression of CC (4). A number of oncogenes have been reported
to be engaged in the overactivation of the Wnt/β-catenin pathway,
including long non-coding RNA (lncRNA) CALML3-AS1, which was
demonstrated to be upregulated and promote cell proliferation and
metastasis in CC (5). It is believed
that inhibiting the Wnt/β-catenin pathway could partially restore
the biocharacter of the oncogene induced to CC (6). Furthermore, numerous biomarkers
predicting the prognosis of CC exert their function through the
Wnt/β-catenin pathway (7). However,
the present study intended to establish novel biomarkers predicting
prognosis and therapeutic targets for clinical application.
lncRNAs are members of non-coding RNAs (ncRNAs) that
participate in a number of physiological and pathological
processes, including cell division and differentiation (8). LncRNAs have been reported to be
involved in the progression of several different types of
carcinoma, including gastric, breast and lung cancer (9,10).
Several studies have demonstrated that lncRNAs contribute to
multiple-processes involved in CC, such as cell proliferation,
angiogenesis and tissue invasion (11–13).
The present study scanned The Cancer Genome Atlas
(https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
database and identified RP11-480I12.5 as significantly upregulated
in CC. The differences in the expression of RP11-480I12.5 between
normal and CC tissues were detected. Subsequently, the biological
function of RP11-480I12.5 was determined through a series of
experiments and it was demonstrated that RP11-480I12.5 promotes the
cervical cancer cell proliferation, migration and invasion of CC
through the epithelial-to-mesenchymal transition (EMT) and
Wnt/β-catenin signaling pathways.
Materials and methods
Tissue samples
All human tissues were obtained from the Department
of Gynaecology and Obstetrics, Jinan Women and Children Health
Hospital (Jinan, China) after confirmation by a pathologist from
July 2014 to January 2015. The inclusion criteria was as follows:
Diagnosed as CC by pathologists between July 2014 and January 2015
in the Department of Gynaecology and Obstetrics, Jinan Women and
Children Health Hospital. Patients with metastasis were excluded. A
total of 50 patients were enrolled, (range, 45–71 years) and the
mean age was 60.1±3.5 years old. A total of 50 cases were divided
into four groups, according to their clinical features and the TNM
stage system (14): Stage I, 7
cases; stage II, 19 cases; stage III, 9 cases and stage IV, 15
cases. Non-cancerous tissues were taken >5 cm from the tumor
site and confirmed by a pathologist as normal tissue. All patients
provided written informed consent and the present study was
approved by the institution's Institutional Review Board.
Cell culture
The normal cervical epithelial ECT cell line
PCS-480-011, and the human CC cell lines were obtained from the
American Type Culture Collection. SiHa (HTB-35), HeLa229 (CCL-2.1)
and MS751 (HTB-34) cells were cultured in DMEM (Sigma-Aldrich;
Merck KGaA), supplemented with 10% fetal bovine serum (FBS)
(Invitrogen; Thermo Fisher Scientific, Inc.) and
penicillin-streptomycin (PS), whereas the C33A (HTB-31), HeLa
(CCL-2) and Caski (CRM-CRL-1550) cell lines were cultured in DMEM
F-12 medium (cat. no., 11330057; Thermo Fisher Scientific, Inc.)
containing 1 0% FBS, at 37°C under 5% CO2. SiHa cells
are squamous cell (epidermoid) carcinoma grade II; HeLa 229,
adenocarcinoma; MS751, epidermoid carcinoma and ECT cells are
normal epithelium cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cancer cell lines and
cancer tissues and normal tissues using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. qPCR was performed using the SYBRGreen
detection RT-PCR system (Takara Bio, Inc.) with RT mix (cat. no.,
RR036B; Takara Bio, Inc.) and SYBRGreen (cat. no., 740703; Takara
Bio, Inc.). Thermocycling conditions were as follows: Initial
denaturation: 95°C for 5 sec; 40 cycles, 95°C for 5 sec of
denaturation, 95°C for 35 sec and 60°C for 30 sec for annealing and
elongation and 60°C for 30 sec for final extension Actin was
applied for normalization. The following primer sequences were used
for the qPCR: E-cadherin: Forward, 5′-AACTCCACCTCCTGAAGCTG-3′ and
reverse, 5′-TTGCTTGACCTACTGCCAGA-3′; N-cadherin: Forward,
5′-TCCACCTACCTCCTGAAGCTG-3′ and reverse, 5′-TTGACCACCAGCTGTGAC-3′;
Snail: Forward, 5′-ACCAACTACCAACACCAAG-3′ and reverse,
5′-TACCCACCAAGCTGTAG-3′; Slug: Forward, 5′-GGTATCATGGTCGGTATGGGT-3′
and reverse, 5′-TCTTTCAGCAGTGGTGGAGA-3′; Vimentin: Forward,
5′-TCCTACGTTGGTGATGAAGCT-3′ and reverse,
5′-TTCTCTTTCAGCAGTGGTGG-3′; Actin: Forward,
5′-TCATGGTCGGTATGGGTCAA-3′ and reverse, 5′-TCAGCAGTGGTGGAGAAAGA-3′;
and RP11-480I12.5: Forward, 5′-ACGTTGGTGATGAAGCTCAA-3′ and reverse,
5′-GCAGTGGTGGAGAAAGAGTA-3′. All experiments were performed in
triplicate. The 2−ΔΔCq method was used to calculate
relative RNA expression (15–17).
Colony formation assay
Transfected CC cells were seeded in 6-well plates at
a density of 500 cells/well. Following a 2-week culture period,
cell colonies were fixed for 20 min with 4% paraformaldehyde and
stained with 1% crystal violet at room temperature. The colonies
were examined and counted under a light microscope (Olympus FV100;
magnification, ×4). The colony index was the ratio of the colony
number and the cells seeded onto the plates.
Transwell and Matrigel assays
Matrigel chambers were used in order to determine
the invasion ability of the cervical cancer cells described above,
according to the manufacturer's protocol (https://www.thermofisher.com/order/catalog/product/140644?SID=srch-srp-140644).
A total of 5×104 cells/well were resuspended in 200 µl
plain medium (DMEM or DMEM F-12 described above) with no FBS in the
upper chamber of a Transwell system, whereas the lower chamber was
filled with 0.5 ml of medium supplemented with 10% FBS. BML284 (5
µg/ml) was added to the complete medium when necessary. Following
incubation at 37°C for 24 h, the invasive cells were fixed with
100% methanol at room temperature for 20 min and stained with 0.5%
crystal violet for 10 min. The migratory cells in the lower chamber
were observed using an inverted microscope (magnification, ×200).
For the migration assay, the indicated cells (5×104)
were plated onto uncoated upper chambers. Cells were fixed with
100% methanol at room temperature and stained with 0.5% crystal
violet. The number of transmembrane cells was observed under a
light microscope (Nikon Corporation) at ×200 magnification.
Wound healing assay
A total of 1×105 cells/well were seeded
in 6-well plates. Cells were incubated with medium (1% FBS and 1%
PS with the corresponding medium as aforementioned) at 37°C for 48
h. Cells were not scratched and were not cultured in gapped inserts
with barriers. Once the cells had reached 100% confluency, equal
wounds were made with the 1 ml-pipette and the medium was replaced
with total plain medium with no FBS. Following incubation for 24 h,
the relative width was measured by the ratio of area and length.
The area was calculated through ImageJ 1.49 (National Institutes of
Health), according to the manufacturer's protocol, images were
obtained with light microscope Olympus FV100 at 200×
magnification.
Lentiviral production and stable cell
line construction
Lentiviral vectors expressing a short hairpin RNA
(shRNA, shRNA-1, shRNA-2 4 µg), empty vector (4 µg) as control and
RP11-480I12.5 (4 µg) were co-transfected with the packaging
vectors, psPAX2 (1 µg) and pMD2G (3 µg) (Addgene) into 293FT cells
(cat. no., ACS-4500; ATCC) for lentiviral production using
Lipofectamine® 3,000, according to the manufacturer's
protocol. In order to establish stable cell lines, cells were
transduced by using the aforementioned lentiviruses with polybrene
(8 mg/ml, Sigma-Aldrich; Merck KGaA). HeLa cell line was infected
with the shRNA lentiviruses to knock down HeLa, while C33A was
infected with overexpressed lentivirus and control lentivirus.
Following incubation for 72 h at 37°C, cervical cancer cells
described above were supplemented with 2 mg/ml puromycin for 3
days. The details of the shRNA and plasmids are presented in
Table SI.
CCK-8 assay
Stable CC cells were seeded in 96-well plates at a
density of 200 cells/well. Following cell culture with complete
medium (10% FBS added) for 24, 48 and 72 h, respectively, CCK-8
(Beyotime Institute of Biotechnology) was added and incubated for 2
h at 37°C. The absorbance was measured at 450 nm and the viability
was normalized to the 0 h.
Edu assay
Transfected CC cells were seeded in 96-well plates
at a density of 500 cells/well. Following incubation for 24 h at
37°C, Edu was added according to the manufacturer's protocol
(RayBiotech). Five fields of view were randomly selected and images
were captured using an Olympus FV100 microscope with red light
fluorescence (magnification, ×200). The percentage of
Edu+ cells was calculated by the confocal
automatically.
Western blotting
The cells (HeLa-NC, HeLa-sh, HeLa-sh2, C33A-NC and
C33A-OV) were collected and lysed with RIPA buffer (Beyotime
Institute of Biotechnology) and cell protein was obtained. The
protein was quantified using the BCA kit (Beyotime Institute of
Biotechnology), according to the manufacturer's protocol. Proteins
(20 µg/lane) were separated via SDS-PAGE (12% gel). The protein was
transferred onto a polyvinylidene fluoride membrane and blocked
with 5% milk for 1 h at room temperature. The membranes were
labeled with the corresponding primary antibodies; N-cad (cat. no.
13116), E-cad (cat. no. 14472), Vimentin (cat. no. 5741), Snail
(cat. no. 3879), Slug (cat. no. 9585), β-catenin (cat. no. 8480),
PCNA (cat. no., 13110), MMP7 (cat. no., 3807), MMP9 (cat. no.,
13667) and β-actin (cat. no. 3700) (all 1:1,000; all from Cell
Signaling Technology, Inc.), and incubated overnight at 4°C.
Following the primary incubation, membranes were incubated with the
goat anti-rabbit and goat anti-mouse secondary antibody (1:10,000;
cat. no. 5724, KPL, Inc.) for 1 h at room temperature. Protein
bands were visualized using ECL (cat. no., 32209; Thermo Fisher
Scientific, Inc.), the relative level of protein was normalized to
β-actin.
TCGA database analysis
The datasets of dysregulated genes of cervical
cancers was downloaded from TCGA database, the cervical carcinoma
dataset (18), and a total of 306
cancers and 13 normal tissues were enrolled. The relative
expression was analyzed by two-tailed paired Student's t-test.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 18; SPSS, Inc.) Data are presented as the mean ±
standard deviation. All experiments were repeated in triplicate.
The χ2 test was used to analyze clinicopathological
characteristics. The survival Kaplan-Meier analysis was applied by
log-rank analysis. For patients enrolled, RP11-480I12.5 high was
defined as those exhibiting higher than the average RNA level of
the whole cohort of all the patients, while the remaining were
defined as RP11-480I12.5 low. Comparisons between the two groups
(normal tissue and cancer tissue) were evaluated using two-tailed
paired Student's t-test and a similar variance was confirmed.
One-way ANOVA was applied when making comparisons in datasets,
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
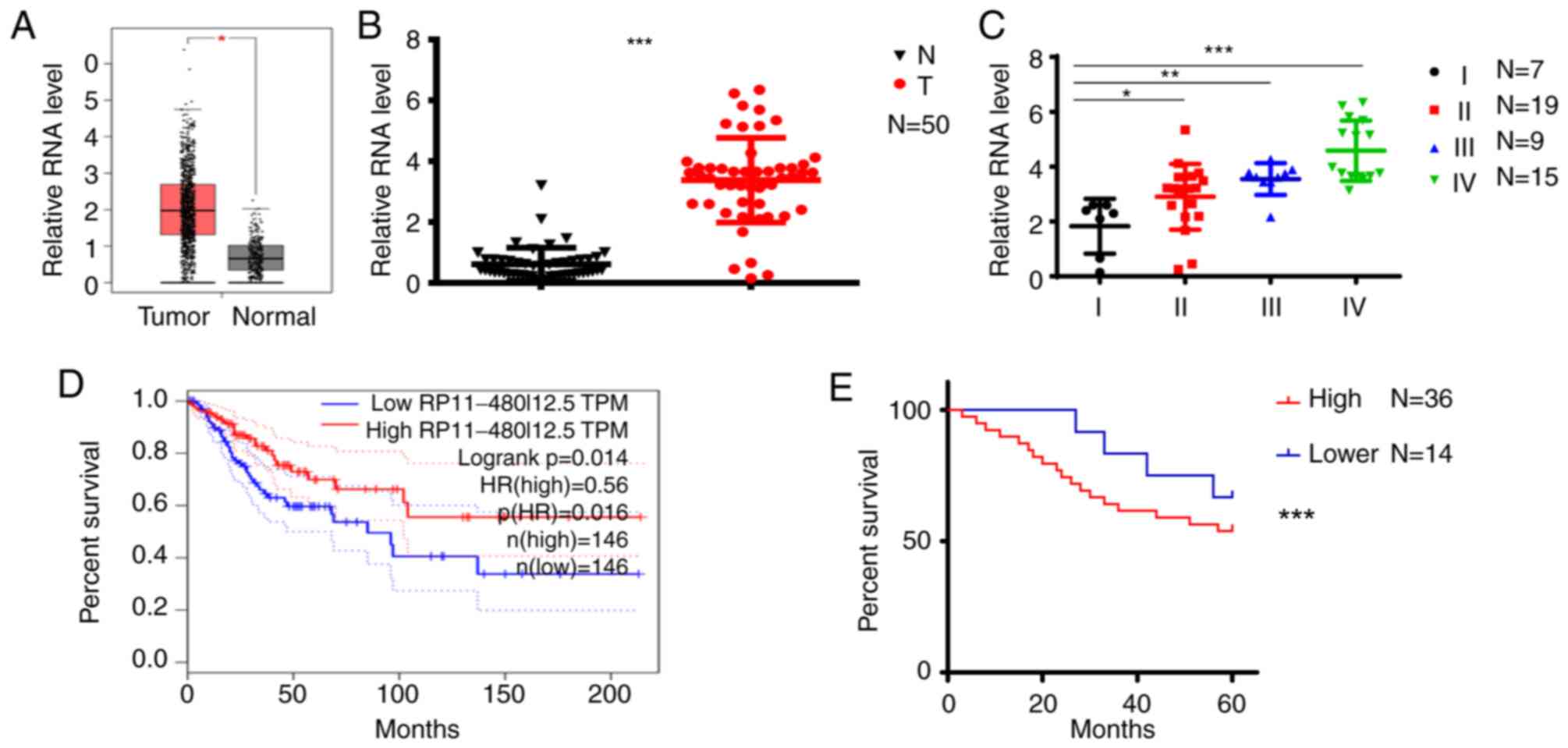
RP11-480I12.5 is upregulated in CC
tissues and associated with a poor prognosis
The present study scanned TCGA database and
identified RP11-480I12.5 as one of the lncRNAs contributing to the
progression of CC (Fig. 1A). To
further verify this hypothesis, the present study analyzed the
expression differences in 50 paired normal and tumor tissues.
RP11-480I12.5 was significantly upregulated in CC tissues compared
with normal tissues (P<0.001; Fig.
1B). Subsequently, the. The level of RP11-480I12.5 was
significantly higher in the later stages of CC compared with stage
I tumors (P<0.001; Fig. 1C).
Furthermore, the present study analyzed the association between
RP11-480I12.5 and the overall survival of patients. Both TCGA
database (P<0.001; Fig. 1D) and
the in-house database containing the aforementioned 50 cases used
in the present study (P<0.001; Fig.
1E) demonstrated that RP11-480I12.5 was negatively associated
with prognosis.
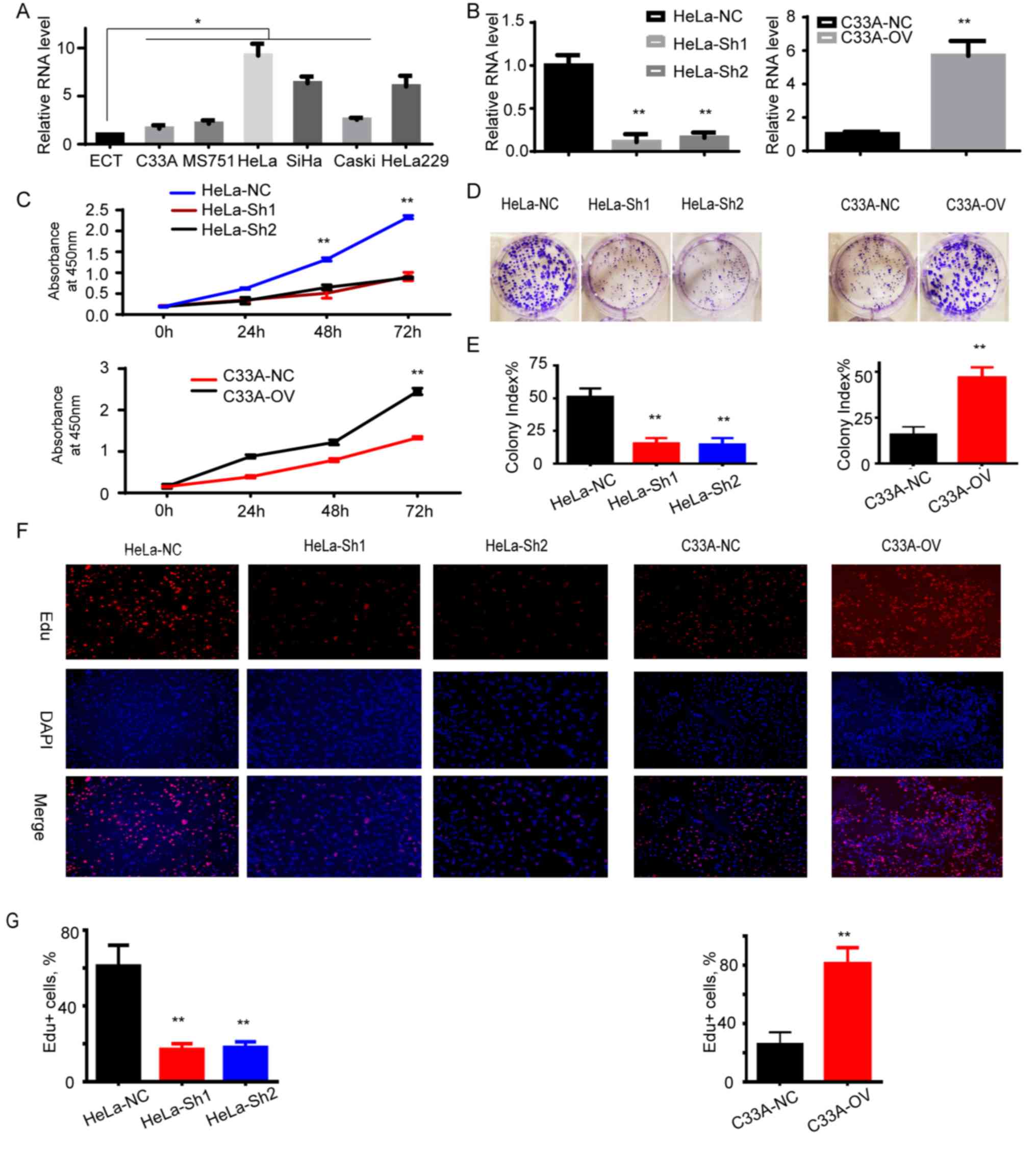
RP11-480I12.5 promotes the
proliferation of CC
The present study demonstrated that RP11-480I12.5 is
upregulated in CC and negatively associated with prognosis. In
order to further determine the biological function of
RP11-480I12.5, the present study examined its expression pattern in
HeLa and C33A CC cell lines described above. The CC cell line was
demonstrated to express a significantly higher level of
RP11-480I12.5 compared with the normal control tissue, ECT
(P<0.05; Fig. 2A). Subsequently,
the present study established the RP11-480I12.5 stable knockdown
cell line in HeLa cells and the RP11-480I12.5 overexpression cell
line in C33A cells. The relative RNA level is presented in Fig. 2B (P<0.001), which indicated that
it increased in RP11-480I12.5 overexpression cell lines, while
decreased in knocked down cell lines, indicating that stable cell
lines were established. Those cells with higher levels of
RP11-480I12.5 demonstrated a higher absorbance at 450 nm
(P<0.001; Fig. 2C), a higher
colony index (P<0.001; Fig. 2D and
E) and a higher percentage of Edu+ cells
(P<0.001; Fig. 2F and G). The
results of the present study indicate that RP11-480I12.5 promotes
cellular proliferation of CC.
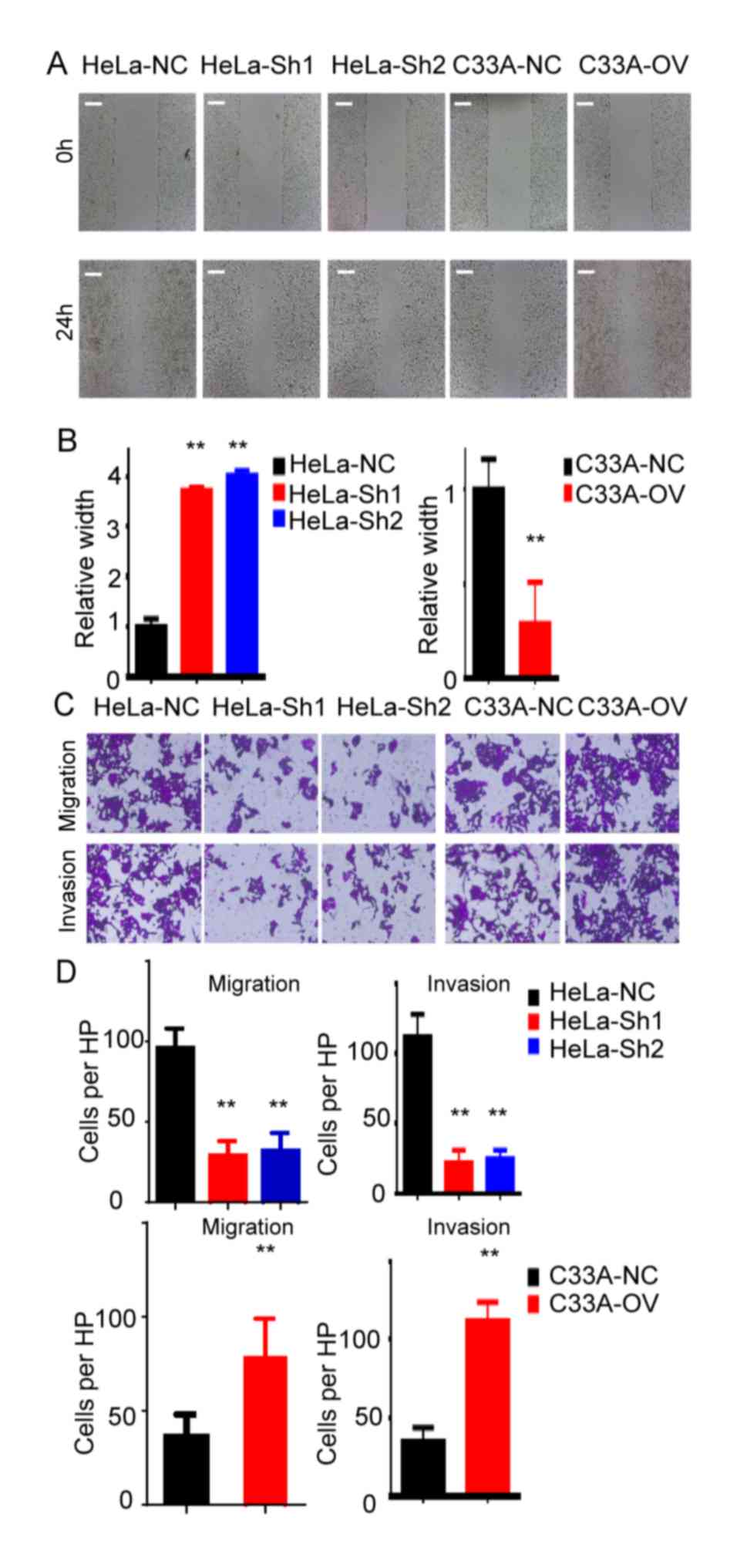
RP11-480I12.5 promotes the migration
and invasion of CC
Recurrence and metastasis are the most common
reasons contributing to the mortality of patients with cancer
(19). The present study analyzed
the migration and invasion abilities of the indicated cell lines.
The results of the present study demonstrated that cells with
higher levels of RP11-480I12.5 indicated an improved migration
ability. The cells harbored improved migration ability in the
wounds healing assay in HeLa-NC and C33A-OV cells compared with the
control cells (P<0.01; Fig. 3A and
B). The Transwell and matrigel assays demonstrated that cells
with higher levels of RP11-480I12.5 had enhanced migration and
invasion abilities (P<0.01; Fig. 3C
and D).
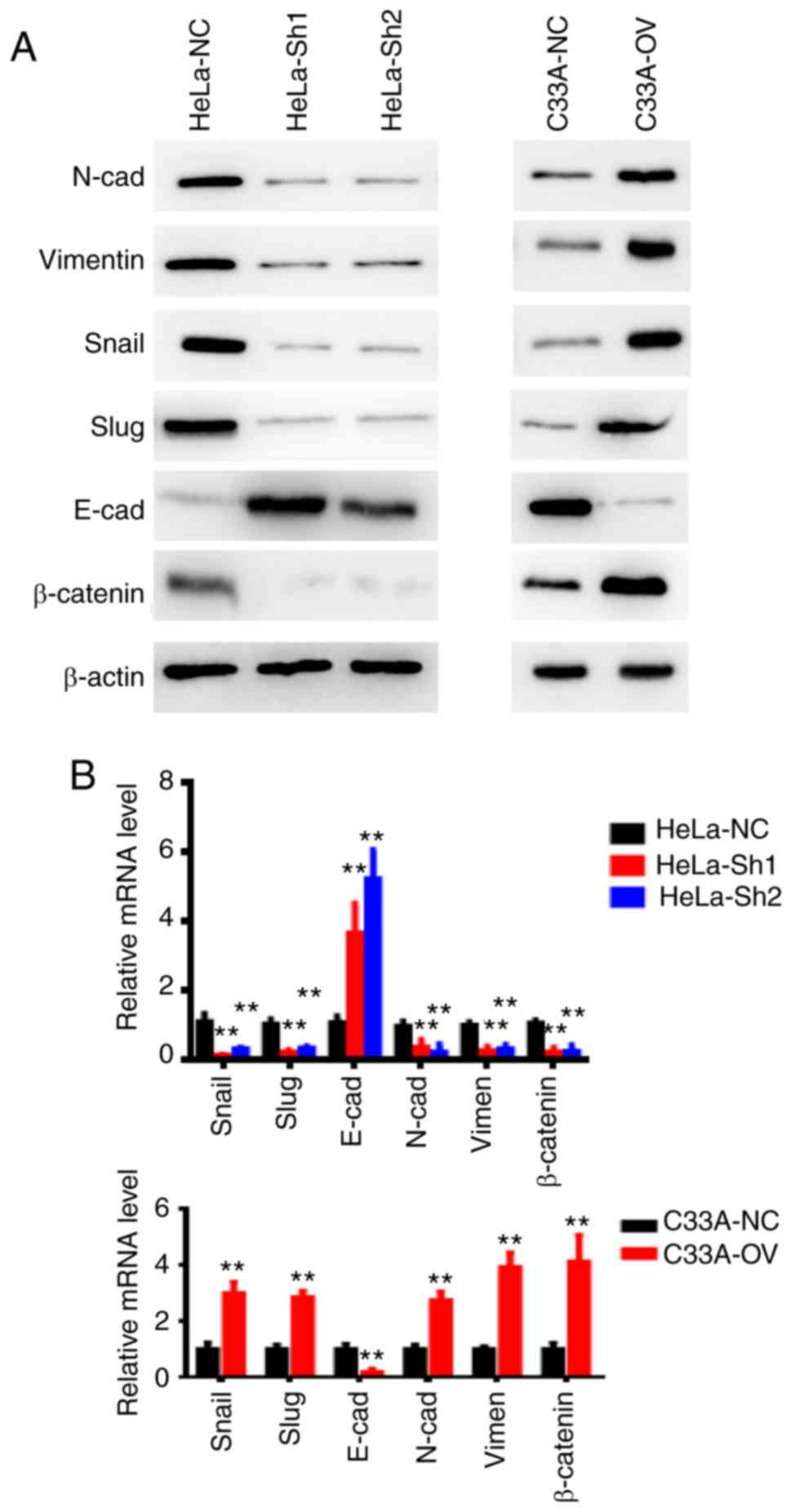
RP11-480I12.5 induces EMT by promoting
the Wnt/β-catenin pathway
The present study previously demonstrated that
RP11-480I12.5 promotes cell proliferation, migration and invasion.
To further investigate the potential underlying molecular mechanism
involved, the present study examined EMT markers and β-catenin in
the indicated cells. The mesenchymal markers, including N-cadherin,
Vimentin, Snail and Slug were decreased in cervical cancer cells
with higher levels of RP11-480I12.5, while the epithelial markers
such as E-cadherin were increased, as expected. These results are
consistent with the aforementioned proliferation and migration and
invasion functions. The relative mRNA levels of the EMT markers in
the HeLa and C33A cell lines are presented in Fig. 4. The corresponding changes
demonstrated in the proliferation marker and the matrix
metalloproteinase protein are presented in Fig. S1. The Wnt pathway activator, BML284
was subsequently added to the C33A cell lines. The results of the
present study demonstrated that RP11-480I12.5 activates the
Wnt/β-catenin pathway in a synergistic manner with BML 284
(Fig. S2), this may be due to the
fact that they exert their function in different ways. Overall, the
results of the present study indicate that RP11-480I12.5 induces
the EMT of CC through the Wnt/β-catenin pathway.
Discussion
Human CC is one of the most common malignancies
worldwide. Patients with late-stage CC still suffer from recurrence
and metastasis; however, the exact molecular mechanisms of
tumorigenesis and progression remain unknown (19). Therefore, further studies are
required in order to determine additional biomarkers and
therapeutic targets.
LncRNAs are ncRNAs that play an important role in a
number of physiological and pathological processes, including cell
differentiation and division (20,21). An
increasing number of studies have demonstrated that lncRNAs are
involved in the tumorigenesis and progression of multiple types of
cancer, including gastric and colon cancer (22,23).
LncRNAs are known to exert their function on CC via multiple steps
(24). For example, the lncRNA
LOC105374902 promotes the malignancy of CC cells by acting as a
sponge of miR-1285-3p, a process enhanced by TNF-α (25). Furthermore, the lncRNA SBF2-AS1
promotes the progression of CC by regulating the miR-361-5p/FOXM1
axis (26), while the lncRNA
BDNF-Anti-Sense is downregulated in CC and possesses anti-tumor
functions by negatively associating with brain-derived neurotrophic
factor (27). RP11-480I12.5 is a
newly identified lncRNA and, to the best of our knowledge, has not
yet been extensively studied. The potential function and molecular
mechanism underlying RP11-480I12.5 in CC remain unknown. Thus, the
present study examined the level of RP11-480I12.5 in CC and its
association with prognosis. Patients with higher levels of
RP11-480I12.5 experienced shorter overall survival and disease-free
survival. In order to determine the biological function of
RP11-480I12.5, the present study detected the RNA level of
RP11-480I12.5 in the CC cell line and established stable knockdown
and overexpression cell lines in HeLa and C33A cells. Cells with
higher levels of RP11-480I12.5 indicated improved proliferation,
migration and invasion ability than cells with lower levels of
RP11-480I12.5. The present study subsequently detected mesenchymal
and epithelial markers in the indicated cells. RP11-480I12.5 was
demonstrated to promote the transition from an epithelial status to
a mesenchymal status.
Overall, the results of the present study
demonstrate that RP11-480I12.5 promotes the cell proliferation,
migration and invasion ability of human CC by inducing EMT through
the Wnt/β-catenin pathway.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LS designed the present study. LS, YL and LZ were
engaged into the performance of the research, analysis of the data
and editing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Jinan Women
and Children Health Hospital Institutional Review Board. The
Bioethics Commission number is 140707-003. Written consent was
obtained from all patients prior to tissue collection. The
Bioethics Commission number was 140707-003.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Langabeer SE: ‘JAK2 V617F mutation in
cervical cancer related to HPV & STIs’-letter. J Cancer Prev.
24:59–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nevin PE, Garcia PJ, Blas MM, Rao D and
Molina Y: Inequities in cervical cancer care in indigenous Peruvian
women. Lancet Glob Health. 7:e556–e557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Gregorio A, Widschwendter P, Ebner F,
Friedl T, Huober J, Janni W and de Gregorio N: Influence of the new
FIGO classification for cervical cancer on patient survival: A
retrospective analysis of 265 histologically confirmed cases with
FIGO stages IA to IIB. Oncology. 1–7. Oct 8–2019.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song T, Hou X and Lin B: MicroRNA-758
inhibits cervical cancer cell proliferation and metastasis by
targeting HMGB3 through the WNT/β-catenin signaling pathway. Oncol
Lett. 18:1786–1792. 2019.PubMed/NCBI
|
|
5
|
Belt EJ, Brosens RP, Delis-van Diemen PM,
Bril H, Tijssen M, van Essen DF, Heymans MW, Beliën JA, Stockmann
HB, Meijer S and Meijer GA: Cell cycle proteins predict recurrence
in stage II and III colon cancer. Ann Surg Oncol. 19 (Suppl
3):S682–S692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W, Su X, Li S, Wang Y, Wang Q and
Zeng H: Inhibiting MNK selectively targets cervical cancer via
suppressing eIF4E-Mediated β-catenin activation. Am J Med Sci.
358:227–234. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lan Y, Xiao X, Luo Y, He Z and Song X:
FEZF1 is an independent predictive factor for recurrence and
promotes cell proliferation and migration in cervical cancer. J
Cancer. 9:3929–3938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang T, Wang J, Zhou Y, Zhao Y, Hang D
and Cao Y: LncRNA CASC2 is up-regulated in osteoarthritis and
participates in the regulation of IL-17 expression and chondrocyte
proliferation and apoptosis. Biosci Rep. 39(pii): BSR201824542019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Z and Yang H: Upregulation of the
long noncoding RNA ADPGK-AS1 promotes carcinogenesis and predicts
poor prognosis in gastric cancer. Biochem Biophys Res Commun.
513:127–134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan BS, Yang MC, Singh S, Chou YC, Chen
HY, Wang MY, Wang YC and Chen RH: LncRNA NORAD is repressed by the
YAP pathway and suppresses lung and breast cancer metastasis by
sequestering S100P. Oncogene. 38:5612–5626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding X, Jia X, Wang C, Xu J, Gao SJ and Lu
C: Correction to: A DHX9-lncRNA-MDM2 interaction regulates cell
invasion and angiogenesis of cervical cancer. Cell Death Differ.
Feb 25–2019.(Epub ahead of print). View Article : Google Scholar
|
|
12
|
Shan S, Li HF, Yang XY, Guo S, Guo Y, Chu
L, Xu MJ and Xin DM: Higher lncRNA CASC15 expression predicts poor
prognosis and associates with tumor growth in cervical cancer. Eur
Rev Med Pharmacol Sci. 23:507–512. 2019.PubMed/NCBI
|
|
13
|
Hsu W, Liu L, Chen X, Zhang Y and Zhu W:
LncRNA CASC11 promotes the cervical cancer progression by
activating Wnt/beta-catenin signaling pathway. Biol Res. 52:332019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu YM, Ni LQ, Wang SS, Lv QL, Chen WJ and
Ying SP: Outcome and prognostic factors in cervical cancer patients
treated with surgery and concurrent chemoradiotherapy: A
retrospective study. World J Surg Oncol. 16:182018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin Y, Zhai H, Ouyang Y, Lu Z, Chu C, He Q
and Cao X: Knockdown of PKM2 enhances radiosensitivity of cervical
cancer cells. Cancer Cell Int. 19:1292019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blum A, Wang P and Zenklusen JC: SnapShot:
TCGA-analyzed tumors. Cell. 173:5302018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Zhou C, Qin Q, Liu Z and Li P:
LncRNA LEF1-AS1 regulates the migration and proliferation of
vascular smooth muscle cells by targeting miR-544a/PTEN axis. J
Cell Biochem. 120:14670–14678. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo ZH, Walid AA, Xie Y, Long H, Xiao W,
Xu L, Fu Y, Feng L and Xiao B: Construction and analysis of a
dysregulated lncRNA-associated ceRNA network in a rat model of
temporal lobe epilepsy. Seizure. 69:105–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Yang G, Liu N, Zhou Z, Cao B, Zhou
P and Yang B: Effect of lncRNA BNC2-AS1 on the proliferation,
migration and invasion of gastric cancer cells. Clin Lab. 64:2018.
View Article : Google Scholar
|
|
23
|
Liu A and Liu L: Long non-coding RNA
ZEB2-AS1 promotes proliferation and inhibits apoptosis of colon
cancer cells via miR-143/bcl-2 axis. Am J Transl Res. 11:5240–5248.
2019.PubMed/NCBI
|
|
24
|
Tian Y, Wang YR and Jia SH: Knockdown of
long noncoding RNA DLX6-AS1 inhibits cell proliferation and
invasion of cervical cancer cells by downregulating FUS. Eur Rev
Med Pharmacol Sci. 23:7307–7313. 2019.PubMed/NCBI
|
|
25
|
Feng Y, Ma J, Fan H, Liu M, Zhu Y, Li Y
and Tang H: TNF-α-induced lncRNA LOC105374902 promotes the
malignant behavior of cervical cancer cells by acting as a sponge
of miR-1285-3p. Biochem Biophys Res Commun. 513:56–63. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao F, Feng J, Yao H, Li Y, Xi J and Yang
J: LncRNA SBF2-AS1 promotes the progression of cervical cancer by
regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol.
47:776–782. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang H, Liu C, Yan T, Wang J and Liang W:
Long noncoding RNA BDNF-AS is downregulated in cervical cancer and
has anti-cancer functions by negatively associating with BDNF. Arch
Biochem Biophys. 646:113–119. 2018. View Article : Google Scholar : PubMed/NCBI
|