Introduction
Long non-coding RNAs (lncRNAs) are transcripts
longer than 200 nucleotides that lack protein-coding potential. Due
to the simultaneous and synergistic availability of genome and
transcriptome sequences, lncRNAs were discovered by analyzing the
mouse transcriptome based on the functional annotation of 60,770
full-length cDNAs in 2002 (1).
Djebali et al (2) have also
used sequencing technology to reveal that <2% of the human
genome encodes proteins, and three-quarters of the human genome is
actively transcribed into non-coding RNAs. LncRNAs are highly
conserved (>95%) in mammals (3),
indicating their biological functionality. However, for a long
time, a large number of ncRNAs have been regard as dark matter or
transcriptional noise.
In recent years, the general application of
genome-wide analysis, which includes microarrays and
high-throughput sequencing technologies, have revealed various
important biological functions of lncRNAs in cell differentiation,
development and numerous diseases, especially cancer (4). In cancer, lncRNAs exhibit
tissue-specific expression and are transcriptionally regulated by
key tumor suppressors or oncogenes, which can influence cell cycle
regulation, survival, immune response and pluripotency (5). For example, lncRNA taurine upregulated
gene 1 (TUG1) is a direct transcriptional target of p53 and affects
cell proliferation in human non-small cell lung and laryngeal
cancer (6). Likewise, lncRNA
plasmacytoma variant translocation 1 (PVT1) is adjacent to
proto-oncogene Myc in the same genomic region of 8q24, and its
amplification is associated with the Myc gene copy number gain,
thus increasing Myc protein levels in cancer (7).
Although the above examples represent only a small
proportion of lncRNA effects, they indicate the large diversity in
the functions of lncRNAs in cancer. Based on these mechanisms,
lncRNAs are attractive potential therapeutic targets and biomarkers
of cancer, and genome-wide analysis in combination with novel
computational strategies may further advance the clinical
application of lncRNAs. In this review, the emerging roles of
lncRNAs in cancer based on genome-wide analysis and the potential
clinical applications of lncRNAs were summarized.
Identification of lncRNAs in cancer using
genome analysis
Aberrant expression of lncRNAs in
cancer
LncRNAs exhibit tissue-specific expression, and
their expression level consistently changes in cancer (8). Considering their limitations in
large-sample tests and tedious operation protocols, traditional
molecular methods such as reverse transcription-PCR and
fluorescence in situ hybridization do not suffice for the
study of lncRNAs (9). The general
implementation of microarrays (Table
I) and high-throughput sequencing (Table II) technology overcomes these
difficulties and makes great progress in cancer. To delineate
genome-wide lncRNA expression patterns, polyA+ RNA sequencing
(RNA-seq) data from 6,503 samples, including tumors, normal samples
and cancer cell lines, were applied (10). Based on these data, a non-parametric
method termed Sample Set Enrichment Analysis was developed to
assess differential expression. Using this approach, known lncRNAs
in breast and prostate cancer, including upregulated oncogenic
lncRNA hox transcript antisense intergenic RNA (HOTAIR) and
prostate cancer antigen-3 (PCA3), downregulated tumor suppressor
lncRNA maternally expressed gene 3 (MEG3) were identified, and a
myriad of unannotated lncRNAs exhibiting potential tissue- and
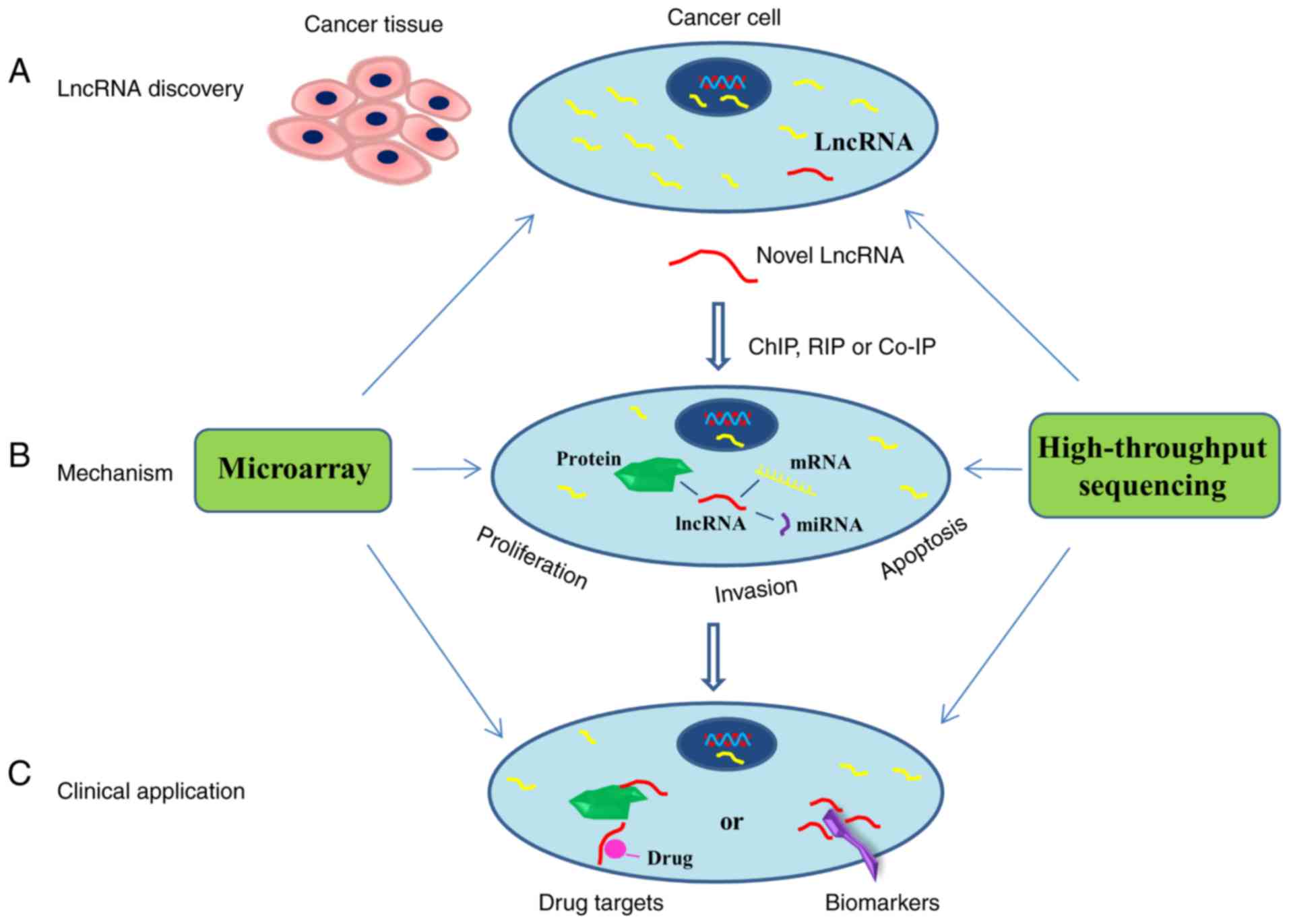
cancer-specific gene signatures were also identified (Fig. 1A) (10).
 | Table I.Typical cancer-associated lncRNAs
identified by microarray. |
Table I.
Typical cancer-associated lncRNAs
identified by microarray.
| Author, year | LncRNA | Cancer type | Biological
function | Mechanism | (Refs.) |
|---|
| Rinn et al,
2007; Botti et al, 2017 | HOTAIR | Breast, gastric,
colorectal and cervical | Promotes
metastasis | Acts as scaffold
for the chromatin repressors PRC2 and LSD1. Silences HoxD and other
gene loci | (11,12) |
| He et al,
2017 | MEG3 | Hepatocellular | Tumor suppressor;
inhibits cell growth and induces apoptosis | miR-29 regulates
expression of MEG3 in methylation-dependent pattern | (13) |
| Tee et al,
2016 | MALAT1 | Neuroblastoma,
bladder, lung and gastric | Promotes cell
proliferation and metastasis | Promotes
tumor-driven angiogenesis by upregulating pro-angiogenic gene
expression | (31) |
| Ji et al,
2015; Zhang et al, 2017 | LINC00152 | Hepatocellular and
lung | Promotes
proliferation, invasion, metastasis and apoptosis | Affects mechanistic
targets of mTOR, Akt and EGFR pathways | (33,35) |
| Zhu et al,
2016 | LOC572558 | Bladder | Tumor suppressor;
inhibits proliferation and induces apoptosis | Regulates the p53
signaling pathway | (37) |
| Prensner et
al, 2014; Smolle et al, 2017 | SCHLAP1 | Prostate | Biomarker for
metastatic | Unknown
progression | (54,55) |
| Li et al,
2016 | GAS5 | Breast | Prognostic marker;
suppresses cancer proliferation | Acts as a molecular
sponge for miR-21, leading to the de-repression of phosphatase and
tensin homologs | (57) |
 | Table II.Typical cancer-associated lncRNAs
identified by high-throughput sequencing. |
Table II.
Typical cancer-associated lncRNAs
identified by high-throughput sequencing.
| Author, year | LncRNA | Cancer type | Biological
function | Mechanism | (Refs.) |
|---|
| Tseng et al,
2014 | PVT1 | Breast, ovarian and
hepatocellular | Promotes
proliferation | Controls Myc levels
by regulating protein stability | (7) |
| Russell et
al, 2015 | CASC15 | Neuroblastoma and
metastatic melanoma | Tumor suppressor;
inhibits proliferation and migratory | As a mediator of
neural growth and differentiation to impact initiation and
progression | (17) |
| Pan et al,
2016 | GAS8-AS1 | Papillary
thyroid | Tumor suppressor;
inhibits cancer growth | Unknown | (22) |
| Liu et al,
2015 | SIRT1-AS | Hepatocellular | Promotes
proliferation | Binds to SIRT1 mRNA
at 3′UTR, masks the microRNA-29c binding site and stabilizes SIRT1
mRNA | (23) |
| Sánchez et
al, 2014 | PR-lncRNA-1 | Colorectal | Tumor suppressor;
inhibits cell survival and proliferation | Modulates gene
expression response to DNA damage downstream of p53 | (41) |
| Hünten et
al, 2015; Kaller et al, 2017 | LINC01021 | Colorectal | Inhibits
proliferation | A novel direct p53
target gene | (42,43) |
| Merry et al,
2015; Somasundaram et al, 2018 | DACOR1 | Colon | Growth
suppressor | Changes DNA
methylation patterns without affecting DNMT1 protein levels | (47,48) |
| Kondo et al,
2017 | JHDM1D-AS1 | Pancreatic | Promotes
tumorigenesis | Regulates
angiogenesis in response to nutrient starvation | (49) |
| Hu et al,
2019 | LINK-A | Breast | Downregulates
cancer antigenicity | Inactivation of
protein kinase A pathways | (50) |
Rinn et al (11) have reported that lncRNA HOTAIR is a
spliced and polyadenylated RNA with 2,158 nucleotides and 6 exons,
according to a DNA microarray. Patients with high HOTAIR expression
levels exhibit an increased incidence of malignancy and lymph node
metastasis (12). Another lncRNA,
MEG3, which is expressed in normal tissues, has been demonstrated
to be either lost or decreased in a number of human tumors and
tumor-derived cell lines; MEG3 is a tumor suppressor involved in
the etiology, progression and chemosensitivity of cancers (13). In addition to well-known lncRNAs, a
growing number of novel lncRNAs have been identified, such as
adenocarcinoma- and immune-associated lncRNAs. A lncRNA expression
profile in lung adenocarcinoma and matched adjacent normal lung
tissues measured by microarray demonstrated that 1,048 lncRNAs were
upregulated and 1,997 lncRNAs were downregulated in lung
adenocarcinoma (14). Among these
lncRNAs, nine lncRNAs were validated; the expression of
NONHSAT077036 was associated with N classification and clinical
stage (14). Wang et al
(15) also used the Chinese Glioma
Genome Atlas microarray to identify 344 immune-associated lncRNAs.
Further validation indicated that the nine immune-associated lncRNA
signature, including SNHG8, ST20-AS1, PGM5-AS1, LINC00937,
MIR155HG, AGAP2-AS1, MAPKAPK5-AS1, HCG18 and TUG1, exhibited
prognostic value for anaplastic gliomas.
Genetic alteration of lncRNAs in
cancer
Microarrays and sequencing technologies, especially
whole genome sequencing, have been prominently applied to identify
the genetic alterations of lncRNAs in cancer, including single
nucleotide polymorphisms (SNPs), copy number alterations (CNAs) and
point mutations (16). For example,
a common disease genome-wide association study identified a SNP
within a tumor suppressor lncRNA cancer-associated susceptibility
candidate 15 (CASC15) at 6p22, which was associated with CASC15-S
differential expression (17).
CBioPortal (http://www.cbioportal.org/) is an open-access resource
for the interactive examination of multidimensional cancer genomics
data sets that include information regarding somatic mutations,
CNAs, RNA expression, DNA methylation and protein and
phosphoprotein abundance (18,19).
CBioPortal provides access to data from >5,000 tumor samples
from 105 cancer studies in The Cancer Genome Atlas (TCGA) (20). A previous study assessed 1,000 cases
of breast invasive carcinoma in this database; the results
demonstrated that 577 lncRNAs exhibited alterations among the 2,730
analyzed lncRNAs (21). The
deregulation of 11 lncRNAs, primarily due to CNA, was associated
with poor overall survival rate; the results also suggested that
greater distance from the oncogene Myc was associated with smaller
alterations of CNA, which indicated that 8q24.21 was the center of
CAN (21). Another team also
supports this conclusion; Tseng et al (7) analyzed genome-wide data from two large
cancer databases, including Progenetix and TCGA, and consistently
observed in 98% of cases a co-gain of Myc and PVT1 across a wide
variety of cancer types harboring the amplified 8q24 region, such
as ovarian, oesophageal and breast cancer. According to their
study, PVT1 RNA and Myc protein expression were associated in
primary human tumors, and the copy number of PVT1 was co-increased
in >98% of cancer cases with the increase of Myc copy
numbers.
Pan et al (22) identified somatic mutations for
papillary thyroid carcinoma (PTC) in the Chinese population using
402 pairs of tumor and normal tissues, of which 91 were
characterized by exome sequencing and 311 by Sanger sequencing; the
results demonstrated that the lncRNA GAS8-AS1 was the secondary
most frequently altered gene and acted as a novel tumor suppressor
in PTC. Liu et al (23)
identified a single-nucleotide mutation (622U>C) in the SIRT1-AS
sequence using gene-sequencing technology; bioinformatics analysis
revealed that this mutation led to an alteration in the secondary
structure of lncRNA SIRT1-AS and prevented it from binding to SIRT1
mRNA.
Mechanisms of lncRNAs identified by
genome-wide analyses in cancer
The mechanisms of lncRNAs likely depend on their
secondary or tertiary structures (24). Nuclear lncRNAs may act as molecular
scaffolds and aid in alternative splicing or chromatin structure
modification (25). Cytoplasmic
lncRNAs may modulate translation and promote or inhibit mRNA
degradation, acting as miRNA sponges (25,26). In
addition to identifying novel lncRNAs, genome-wide analysis also
serves important roles in lncRNA mechanistic studies, such as to
determine its functional importance or to probe its mechanistic
properties (27). Recently, these
techniques were combined with approaches used to study gene-gene,
gene-protein and protein-protein interactions, including chromatin
immunoprecipitation (ChIP), RNA binding protein immunoprecipitation
(RIP) and co-immunoprecipitation (Co-IP), to further assess the
mechanistic role of lncRNAs in cancer (Fig. 1B) (4).
The mechanisms of lncRNAs in cancer
identified by microarrays
Microarrays involve hybridizing a modified
complementary DNA (cDNA) or a complementary RNA template of target
transcripts to a set of short oligonucleotide probes immobilized on
a solid surface (28). When bound to
the probes, these modifications enable either direct or indirect
florescence detection, and a digital signal is used to quantify the
level of gene expression (28).
Therefore, microarray offers a highly sensitive, highly selective
and high-throughput method for the analysis of mRNAs and lncRNAs in
biology and disease (29).
The intergenic lncRNA MALAT1, was originally
identified as a prognostic marker of lung cancer metastasis, and
was associated with metastasis in other cancer types, such as
breast, prostate, and pancreatic cancer (30). Microarray-based differential gene
expression analysis has revealed that FGF2 is significantly
downregulated by MALAT1, and that MALAT1-mediated FGF2 protein
secretion from neuroblastoma cells induces vasculature formation to
promote tumor angiogenesis (31). In
addition, LINC00152 is involved in the pathogenesis of cancer; the
detailed mechanism was unknown until microarray-based analysis was
performed (32). LINC00152 can
affect the mammalian target of rapamycin, phosphatidylinositol
3-kinase/AKT and the epidermal growth factor receptor signaling
pathways as a molecular indicator that assists in the regulation of
the associated target gene expression (33–36).
Furthermore, using a high-throughput phospho-proteome microarray,
Zhu et al (37) have
demonstrated that Akt, mouse double minute 2 homolog and p53
phosphorylation are reduced when lncRNA LOC572558 is stably
upregulated, suggesting that LOC572558 may be a tumor suppressor
and regulate the p53 signaling pathway in bladder cancer.
The mechanisms of lncRNAs in cancer
identified by sequencing
Sequencing involves the generation of
cDNA-containing primers by PCR for library construction and
analysis (38). The library is
subsequently sequenced and quantified for each oligonucleotide base
(38). The advantages of sequencing
include a direct measure of the genome and its ability to discover
novel transcripts, alternate transcript starts and stops, splicing
variants and single nucleotide polymorphisms (39). Compared with microarray, sequencing
offers more information when combined with functional experiments
in mechanistic studies of lncRNAs (9).
A number of studies have demonstrated the
association between endogenous p53 activity and the expression of
lncRNAs in cancer progression (40–43). By
integrating RNA-seq with p53 ChIP-seq analyses of HCT116 cells
under DNA damage, two lncRNAs, PR-lncRNA-1 and PR-lncRNA-10, were
identified (41). These lncRNAs are
required for the efficient binding of p53 to its target genes,
modulating p53 activity at the transcriptional, but not the
post-transcriptional level, and contributing to apoptotic induction
in response to DNA damage in cancer (41). The effect of p53 activation and
genome-wide DNA binding of p53 was also determined by RNA-seq in
combination with ChIP-seq analysis in SW480 cells (42). A total of 393 lncRNAs, including 270
upregulated and 123 downregulated lncRNAs, were identified as
differentially regulated by p53. In addition, 18 lncRNAs
demonstrating p53 binding near the corresponding gene TSS and 17
lncRNAs displaying p53 chromatin occupancy in a 20-kbp region
surrounding the corresponding gene TSS were identified (42). LINC01021 has been identified as a
novel direct p53 target gene, the ectopic expression of which
inhibits colorectal cancer cell proliferation and chemosensitivity
(42,43).
A number of lncRNAs affect gene expression though
interacting with epigenetic processes (25). Enhancer of zeste homolog 2 (EZH2),
the catalytic component of polycomb repressive complex 2 (PRC2),
possesses histone-lysine N-methyltransferase activity and plays an
essential role in cancer (44).
RIP-seq has demonstrated that the lncRNA MALAT1 binds with EZH2 to
suppresses the tumor suppressor PCDH10 and promote gastric cellular
migration and invasion (45). In
addition to histone methylation, DNA methylation is another crucial
epigenetic marker typically associated with repressed genes in
human cells (46). Merry et
al (47) identified a subset of
lncRNAs that interacted with DNMT1 in HCT116 cells though RIP-seq.
DNMT1-associated colon cancer-repressed lncRNA 1 (DACOR1) exhibited
high tissue-specific expression in the normal colon tissue, but was
repressed in a panel of colon tumors and patient-derived colon
cancer cell lines (47). The genomic
occupancy sites of DACOR1 significantly overlap with known
differentially methylated regions, and dysregulation of DACOR1
contributes to aberrant DNA methylation patterns in colon tumors
(47,48).
LncRNAs also serve pivotal roles in tumor
progression and malignant transformation by regulating the
nutrient-starved tumor microenvironment or immune checkpoints.
Kondo et al (49) identified
the nutrient starvation-responsive lncRNA JHDM1D antisense 1 using
ChIP-seq and formaldehyde-assisted isolation of regulatory element
sequencing, which was demonstrated to be upregulated and to promote
tumorigenesis by regulating angiogenesis in response to nutrient
starvation. In addition, Hu et al (50) reported that the oncogenic lncRNA long
intergenic non-coding RNA for kinase activation (LINK-A)
inactivated tumor suppressor pathways and downregulated antigen
presentation through inactivation of protein kinase A pathways,
thus contributing to decreased immunosurveillance and escaping from
immune checkpoints by regulating immune cell gene expression
programs. These results provided the basis for developing
combination immunotherapy treatment regimens.
Cancer diagnostics and therapies based on
genome-wide analysis of lncRNAs
The emerging role of lncRNAs, especially their
highly tissue- and developmental stage-specific expression,
provides a new range of possibilities for the diagnosis and
treatment of cancer. The lncRNAs may serve as diagnostic biomarkers
detected in the plasma, urine or tissue or contained in exosomes,
microvesicles or apoptotic bodies and as treatment targets for
drugs or radiotherapy (Fig. 1C)
(51). Based on genome-wide
analysis, the prognostic values of various lncRNAs for patients
with cancer were evaluated, and the reported number of lncRNAs with
clinical implications has increased (52).
The first lncRNA to be approved by the Food and Drug
Administration was PCA3, which was used as a urine biomarker in
patients with prostate cancer (53).
A large-scale high-throughput study was performed using a
microarray of prostate tumors from patients who experienced
metastasis (n=212) and those who did not (n=333), and the lncRNA
SCHLAP1 was discovered as one of the optimal predictive genes for
metastasis (54). This was further
validated using a clinical-grade urine test in a Clinical
Laboratory Improvement Amendments-certified laboratory; SCHLAP1 has
been suggested to be a promising biomarker of the aggressive
clinical course of prostate cancer (55). Therapeutic resistance to trastuzumab
caused by dysregulation of lncRNAs often poses a major obstacle in
the clinical management of HER2-positive breast cancer (56). In addition, microarray results
indicated that the expression of the lncRNA GAS5 was decreased in
SKBR-3/Tr cells and breast cancer tissues from trastuzumab-treated
patients, suggesting that GAS5 may serve as a novel prognostic
marker and candidate drug target for HER2-positive breast cancer
(57). Collectively, the prognostic
values of these lncRNAs for patients with cancer were assessed by
meta-analysis, which may facilitate improvements in the diagnosis
and prognosis of cancer treatment (58–60).
Although the majority of pioneering studies remain in the early
stage, they still encourage further examination of the diagnostics
and therapeutic opportunities of lncRNAs in cancer.
Conclusion and future perspective
The development of genome-wide analysis has resulted
in a tremendous advance in the understanding of the underlying
mechanisms of lncRNAs. The extensive functions and mechanisms of
lncRNAs in cancers are a hot area of research. This review provides
an overview of lncRNAs that were identified by microarray and
sequencing in cancer, including their functions and mechanisms, as
well as their potential use in cancer diagnosis and therapies.
However, this review also provides information that remains far
from the realistic clinical application of lncRNAs in the treatment
of cancer. The techniques that detect lncRNA interactions with key
genes and proteins, the use of dependable animal models for
cancer-associated lncRNAs and the implementation of reliable
computational calculation methods of genome-wide analysis should be
developed for future functional and mechanistic studies of lncRNAs.
In addition, large clinical trials should be performed to confirm
these potential lncRNA biomarkers. Furthermore, genome-wide
analyses, which reduce analytic time and cost, will be
indispensable in future studies and in clinical applications of
lncRNAs in cancer.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
XXR conceived the review, researched the literature
and wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that they have no competing
interests.
References
|
1
|
Okazaki Y, Furuno M, Kasukawa T, Adachi J,
Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al:
Analysis of the mouse transcriptome based on functional annotation
of 60,770 full-length cDNAs. Nature. 420:563–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jathar S, Kumar V, Srivastava J and
Tripathi V: Technological developments in lncRNA biology. Adv Exp
Med Biol. 1008:283–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng GX, Do BT, Webster DE, Khavari PA
and Chang HY: Dicer-microRNA-Myc circuit promotes transcription of
hundreds of long noncoding RNAs. Nat Struct Mol Biol. 21:585–590.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Z, Wang X, Cao S, Han X, Wang Z,
Zhao X, Liu X, Li G, Pan X and Lei D: The long noncoding RNA TUG1
promotes laryngeal cancer proliferation and migration. Cell Physiol
Biochem. 49:2511–2520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sánchez Y and Huarte M: Long non-coding
RNAs: Challenges for diagnosis and therapies. Nucleic Acid Ther.
23:15–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Veneziano D, Di Bella S, Nigita G, Laganà
A, Ferro A and Croce CM: Noncoding RNA: Current deep sequencing
data analysis approaches and challenges. Hum Mutat. 37:1283–1298.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Botti G, Marra L, Malzone MG, Anniciello
A, Botti C, Franco R and Cantile M: LncRNA HOTAIR as prognostic
circulating marker and potential therapeutic target in patients
with tumor diseases. Curr Drug Targets. 18:27–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He Y, Luo Y, Liang B, Ye L, Lu G and He W:
Potential applications of MEG3 in cancer diagnosis and prognosis.
Oncotarget. 8:73282–73295. 2017.PubMed/NCBI
|
|
14
|
Peng Z, Wang J, Shan B, Yuan F, Li B, Dong
Y, Peng W, Shi W, Cheng Y, Gao Y, et al: Genome-wide analyses of
long noncoding RNA expression profiles in lung adenocarcinoma. Sci
Rep. 7:153312017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang W, Zhao Z, Yang F, Wang H, Wu F,
Liang T, Yan X, Li J, Lan Q, Wang J and Zhao J: An immune-related
lncRNA signature for patients with anaplastic gliomas. J
Neurooncol. 136:263–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Russell MR, Penikis A, Oldridge DA,
Alvarez-Dominguez JR, McDaniel L, Diamond M, Padovan O, Raman P, Li
Y, Wei JS, et al: CASC15-S is a tumor suppressor lncRNA at the 6p22
neuroblastoma susceptibility locus. Cancer Res. 75:3155–3166. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin C, Yuan G, Hu Z, Zeng Y, Qiu X, Yu H
and He S: Bioinformatics analysis of the interactions among lncRNA,
miRNA and mRNA expression, genetic mutations and epigenetic
modifications in hepatocellular carcinoma. Mol Med Rep.
19:1356–1364. 2019.PubMed/NCBI
|
|
21
|
Liu H, Li J, Koirala P, Ding X, Chen B,
Wang Y, Wang Z, Wang C, Zhang X and Mo YY: Long non-coding RNAs as
prognostic markers in human breast cancer. Oncotarget.
7:20584–20596. 2016.PubMed/NCBI
|
|
22
|
Pan W, Zhou L, Ge M, Zhang B, Yang X,
Xiong X, Fu G, Zhang J, Nie X, Li H, et al: Whole exome sequencing
identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid
carcinoma driver alternations. Hum Mol Genet. 25:1875–1884. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Wu W and Jin J: A novel mutation in
SIRT1-AS leading to a decreased risk of HCC. Oncol Rep.
34:2343–2350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zampetaki A, Albrecht A and Steinhofel K:
Long non-coding RNA structure and function: Is there a link? Front
Physiol. 9:12012018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao RW, Wang Y and Chen LL: Cellular
functions of long noncoding RNAs. Nat Cell Biol. 21:542–551. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu XS, Wang F, Li HF, Hu YP, Jiang L,
Zhang F, Li ML, Wang XA, Jin YP, Zhang YJ, et al: LncRNA-PAGBC acts
as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO
Rep. 18:1837–1853. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Signal B, Gloss BS and Dinger ME:
Computational approaches for functional prediction and
characterisation of long noncoding RNAs. Trends Genet. 32:620–637.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jaksik R, Iwanaszko M, Rzeszowska-Wolny J
and Kimmel M: Microarray experiments and factors which affect their
reliability. Biol Direct. 10:462015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi Y and Shang J: Long noncoding RNA
expression profiling using arraystar LncRNA microarrays. Methods
Mol Biol. 1402:43–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun Y and Ma L: New insights into long
non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel).
11:2019. View Article : Google Scholar
|
|
31
|
Tee AE, Bing L, Song R, Li J, Pasquier E,
Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, et al: The
long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by
up-regulating pro-angiogenic gene expression. Oncotarget.
7:8663–8675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G
and Sun B: LINC00152 promotes proliferation in hepatocellular
carcinoma by targeting EpCAM via the mTOR signaling pathway.
Oncotarget. 6:42813–42824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Li M, Dong C, Ma Y, Xiao L, Zuo S,
Gong Y, Ren T and Sun B: Linc00152 knockdown inactivates the
Akt/mTOR and Notch1 pathways to exert its anti-hemangioma effect.
Life Sci. 223:22–28. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Xiang C, Wang Y, Duan Y, Liu C,
Jin Y and Zhang Y: lncRNA LINC00152 knockdown had effects to
suppress biological activity of lung cancer via EGFR/PI3K/AKT
pathway. Biomed Pharmacother. 94:644–651. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu J, Guo J, Jiang Y, Liu Y, Liao K, Fu Z
and Xiong Z: Improved characterization of the relationship between
long intergenic non-coding RNA Linc00152 and the occurrence and
development of malignancies. Cancer Med. 8:4722–4731. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu Y, Dai B, Zhang H, Shi G, Shen Y and
Ye D: Long non-coding RNA LOC572558 inhibits bladder cancer cell
proliferation and tumor growth by regulating the AKT-MDM2-p53
signaling axis. Cancer Lett. 380:369–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Podnar J, Deiderick H, Huerta G and
Hunicke-Smith S: Next-generation sequencing RNA-Seq library
construction. Curr Protoc Mol Biol. 106:4.21.1–19. 2014. View Article : Google Scholar
|
|
39
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin T, Hou PF, Meng S, Chen F, Jiang T, Li
ML, Shi ML, Liu JJ, Zheng JN and Bai J: Emerging roles of p53
related lncRNAs in cancer progression: A systematic review. Int J
Biol Sci. 15:1287–1298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sánchez Y, Segura V, Marín-Béjar O, Athie
A, Marchese FP, González J, Bujanda L, Guo S, Matheu A and Huarte
M: Genome-wide analysis of the human p53 transcriptional network
unveils a lncRNA tumour suppressor signature. Nat Commun.
5:58122014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hünten S, Kaller M, Drepper F, Oeljeklaus
S, Bonfert T, Erhard F, Dueck A, Eichner N, Friedel CC, Meister G,
et al: p53-regulated networks of protein, mRNA, miRNA and lncRNA
expression revealed by integrated pulsed stable isotope labeling
with amino acids in cell culture (pSILAC) and next generation
sequencing (NGS) analyses. Mol Cell Proteomics. 14:2609–2629. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kaller M, Götz U and Hermeking H: Loss of
p53-inducible long non-coding RNA LINC01021 increases
chemosensitivity. Oncotarget. 8:102783–102800. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamaguchi H and Hung MC: Regulation and
role of EZH2 in cancer. Cancer Res Treat. 46:209–222. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan
S, Yu Q, Li YY, Kang Y, Li H, et al: MALAT1 long ncRNA promotes
gastric cancer metastasis by suppressing PCDH10. Oncotarget.
7:12693–12703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Michalak EM, Burr ML, Bannister AJ and
Dawson MA: The roles of DNA, RNA and histone methylation in ageing
and cancer. Nat Rev Mol Cell Biol. 20:573–589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Merry CR, Forrest ME, Sabers JN, Beard L,
Gao XH, Hatzoglou M, Jackson MW, Wang Z, Markowitz SD and Khalil
AM: DNMT1-associated long non-coding RNAs regulate global gene
expression and DNA methylation in colon cancer. Hum Mol Genet.
24:6240–6253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Somasundaram S, Forrest ME, Moinova H,
Cohen A, Varadan V, LaFramboise T, Markowitz S and Khalil AM: The
DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA
methylation in colon cancer. Clin Epigenetics. 10:1272018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kondo A, Nonaka A, Shimamura T, Yamamoto
S, Yoshida T, Kodama T, Aburatani H and Osawa T: Long noncoding RNA
JHDM1D-AS1 promotes tumor growth by regulating angiogenesis in
response to nutrient starvation. Mol Cell Biol. 37:2017. View Article : Google Scholar
|
|
50
|
Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A,
Egranov SD, Zhang Y, Xia W, Gong J, et al: Oncogenic lncRNA
downregulates cancer cell antigen presentation and intrinsic tumor
suppression. Nat Immunol. 20:835–851. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
de Kok JB, Verhaegh GW, Roelofs RW,
Hessels D, Kiemeney LA, Aalders TW, Swinkels DW and Schalken JA:
DD3(PCA3), a very sensitive and specific marker to detect prostate
tumors. Cancer Res. 62:2695–2698. 2002.PubMed/NCBI
|
|
54
|
Prensner JR, Zhao S, Erho N, Schipper M,
Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A,
Siddiqui J, et al: RNA biomarkers associated with metastatic
progression in prostate cancer: A multi-institutional
high-throughput analysis of SChLAP1. Lancet Oncol. 15:1469–1480.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Smolle MA, Bauernhofer T, Pummer K, Calin
GA and Pichler M: Current insights into long non-coding RNAs
(LncRNAs) in prostate cancer. Int J Mol Sci. 18:2017. View Article : Google Scholar
|
|
56
|
Merry CR, Mcmahon S, Thompson CL, Miskimen
KL, Harris LN and Khalil AM: Integrative transcriptome-wide
analyses reveal critical HER2-regulated mRNAs and lincRNAs in HER2+
breast cancer. Breast Cancer Res Treat. 150:321–334. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen
W and Wei Q: Downregulation of LncRNA GAS5 causes trastuzumab
resistance in breast cancer. Oncotarget. 7:27778–27786.
2016.PubMed/NCBI
|
|
58
|
Fradet V, Toren P, Nguile-Makao M, Lodde
M, Lévesque J, Léger C, Caron A, Bergeron A, Ben-Zvi T, Lacombe L,
et al: Prognostic value of urinary prostate cancer antigen 3 (PCA3)
during active surveillance of patients with low-risk prostate
cancer receiving 5α-reductase inhibitors. BJU Int. 121:399–404.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ma W, Chen X, Ding L, Ma J, Jing W, Lan T,
Sattar H, Wei Y, Zhou F and Yuan Y: The prognostic value of long
noncoding RNAs in prostate cancer: A systematic review and
meta-analysis. Oncotarget. 8:57755–57765. 2017.PubMed/NCBI
|
|
60
|
Liu L, Meng T, Yang XH, Sayim P, Lei C,
Jin B, Ge L and Wang HJ: Prognostic and predictive value of long
non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their
effects on colorectal cancer cell proliferation, migration and
invasion. Cancer Biomark. 22:283–299. 2018. View Article : Google Scholar : PubMed/NCBI
|