Introduction
Bladder cancer (BCa) is the most common tumor in the
urinary system (1). In recent years,
the number of deaths caused by BCa has increased year by year,
ranking 13th among all tumors, which poses a huge impact on human
health (2). At present, therapeutic
strategies, including surgery, chemotherapy and radiotherapy are
applied in the treatment of BCa. Nevertheless, the 5-year survival
of BCa is still low owing to the high recurrent rate and rapid
progression (3). Previous studies
have found that the microenvironment of tumor immunity is closely
related to the progression of BCa (4). Some novel treatments are applied for
BCa, such as the targeted drug Balversa, neoadjuvant chemotherapy
and radiotherapy (5–7). It is of significance to clearly uncover
the pathogenesis of BCa, thus improving the diagnostic and
therapeutic efficacies.
Development of high-throughput sequencing technology
deepens gene research (8). circRNA
is newly discovered and is considered to have a huge role in tumor
progression (9). Previous studies
have suggested that circRNA may become a potential target for tumor
prediction and treatment (10). The
circRNA has a cyclic structure composed of covalent bonds,
characterized as high stability, high abundance, and high
conservation compared with other non-coding RNAs. Functionally,
circRNA is involved in rearrangement of gene information,
prevention of gene degradation, and RNA folding (11). circRNAs have been reported to exert a
crucial role in many types of tumors, serving as oncogenes or tumor
suppressors (12–14).
A previous study demonstrated that circ-FNTA
(circ_0084171) is abnormally upregulated in BCa (15). circ-FNTA locates on chr8:
42914234-42932507 with the cleavage sequence length of 582 bp. In
the circbase database (http://www.circbase.org/cgi-bin/listsearch.cgi), the
annotated gene of circ-FNTA is FNTA (farnesyltransferase, CAAX box,
alpha, NCBI Gene 3782, transcript NM_002027) (16). Farnesyltransferase inhibitors (FTIs)
are proved to inhibit the activation of multiple tumor muteins and
delay tumor progression (17). It is
speculated that circ-FNTA may be important in the progression of
BCa. This study mainly explored the expression pattern and
biological function of circ-FNTA in BCa, and the potential
mechanism.
Patients and methods
Sample collection
BCa tissues (n=40) and matched normal tissues (n=40)
were surgically resected, immediately placed in liquid nitrogen and
preserved at −80°C. None of enrolled BCa patients received
preoperative anti-tumor therapies. Patients and their families in
this study have been fully informed. This study was approved by
Ethics Committee of Linyi Cancer Hospital (Linyi, China). All the
patients provided written informed consent. This study was
conducted in accordance with the Declaration of Helsinki.
Cell culture
Human bladder immortalized epithelium cells
(SV-HUC-1) and BCa cells (5637, T24, RT4 and UM-UC-3) were provided
by the American Type Culture Collection (ATCC). Cells were cultured
in Roswell Park Memorial Institute 1640 (RPMI-1640) containing 10%
fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 µg/ml streptomycin. Cells were maintained at
37°C, in 5% CO2 incubator. Medium was replaced every 2–3
days.
Transfection
Transfection plasmids were provided by Sangon
Biotech. Cells were pre-seeded in the 6-well plates and transfected
using Lipofactamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) at 50–70% confluence. At 24–48 h, cells were harvested for
subsequent experiments.
RNA extraction and quantitative
real-time polymerase chain reaction (qRT-PCR)
RNA extraction from cells was performed using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was
reverse transcribed into complementary deoxyribose nucleic acid
(cDNA) using Primescript RT Reagent (TaKaRa). The obtained cDNA was
subjected to qRT-PCR using SYBR®Premix Ex Taq™ (TaKaRa).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used
as internal references. Each sample was performed in triplicate,
and relative level was calculated by 2−ΔΔCt. Primer
sequences are listed in Table I.
 | Table I.Sequences of transfection primers. |
Table I.
Sequences of transfection primers.
| Genes | Primer sequence |
|---|
| miRNA cDNA |
|
miRNA-451a | Primer
5′-AAAAAAACCGTTACCATTACTGAGTT-3′ |
| U6 | Primer
5′-GCAAATTCGTGAAGCGTTCCATA-3′ |
| qRT-PCR primer |
|
circ-FNTA | Forward
5′-GCCCAAAAACTATCAAGTTTGGCAT-3′ |
|
| Reverse
5′-ATAACCCATTGTCGATGCTGCC-3′ |
|
S1PR3 | Forward
5′-TCTCCGAAGGTCAAGGAAGA-3′ |
|
| Reverse
5′-TCAGTTGCAGAAGATCCCATTC-3′ |
|
GAPDH | Forward
5′-TCCTCTGACTTCAACAGCGACAC-3′ |
|
| Reverse
5′-GAGCAACACAGATGAACCGC-3′ |
|
miRNA-451a | Forward
5′-GGCCCTCGAGCTTTTGACCACCCCTTAACC-3′ |
|
| Reverse
5′-CCCGGGGCGGCCGCACAATGAATTATAATACAAT-3′ |
| U6 | Forward
5′-AGAAGGCTGGGGCTCATTTG-3′ |
|
| Reverse
5′-AGGGGCCATCCACAGTCTTC-3′ |
Cell Counting Kit (CCK-8)
Cells were seeded in the 96-well plate with
5×103 cells per well. At the appointed time points,
absorbance value at 450 nm of each sample was recorded using the
CCK-8 kit (Dojindo Laboratories) for depicting the viability
curve.
5-Ethynyl-2′-deoxyuridine (EdU)
proliferation assay
Cells were inoculated into 96-well plates with
1×105 cells per well, and labeled with 100 µl of EdU
reagent (50 µM) per well for 2 h. After washing with phosphate
buffered saline (PBS), the cells were fixed in 50 µl of fixation
buffer, decolored with 2 mg/ml glycine and permeated with 100 µl of
penetrant. After washing with PBS once, cells were stained with
AdoLo and 4′,6-diamidino-2-phenylindole DAPI) in the dark for 30
min. EdU-positive ratio was determined under a fluorescent
microscope.
Transwell invasion assay
Cell density was adjusted to 3×104
cells/ml. Suspension (100 µl) was applied to the upper Transwell
chamber (Corning Inc.). Into the lower chamber, 600 µl of medium
containing 20% FBS was applied. After 24 h of incubation, cells
migrated to the lower chamber were fixed in methanol for 15 min,
stained with crystal violet for 20 min and counted using a
microscope. The number of migratory cells was counted in 5 randomly
selected fields per sample (×200).
Target gene prediction
Target genes of circ-FNTA and miRNA-451a were
predicted on Starbase (http://starbase.sysu.edu.cn/) (18) and TargetScan (http://www.Targetscan.org) (19). Predicted miRNAs on both websites were
depicted by Venn diagram. The network of target genes of miRNA-451a
was depicted using Cytoscape software v.3.5.1.
Dual-luciferase reporter gene
assay
Based on the predicted binding sites, we constructed
pmirGLO-circ-FNTA-mut, pmirGLO-circ-FNTA-wt, pmirGLO-S1PR3-mut and
pmirGLO-S1PR3-wt. Cells were co-transfected with miRNA-451a
mimics/NC and wild-type/mutant-type vectors using Lipofectamine
2000. After 48 h, co-transfected cells were collected for
determining luciferase activity using a dual-luciferase reporter
assay system (Promega Cooperation).
Statistical analysis
GraphPad Prism 6 (La Jolla) was used for data
analyses. Data were expressed as mean ± standard deviation.
Intergroup differences were analyzed by the t-test. Kaplan-Meier
method was introduced for survival analysis. Two-tailed P<0.05
was considered as statistically significant.
Results
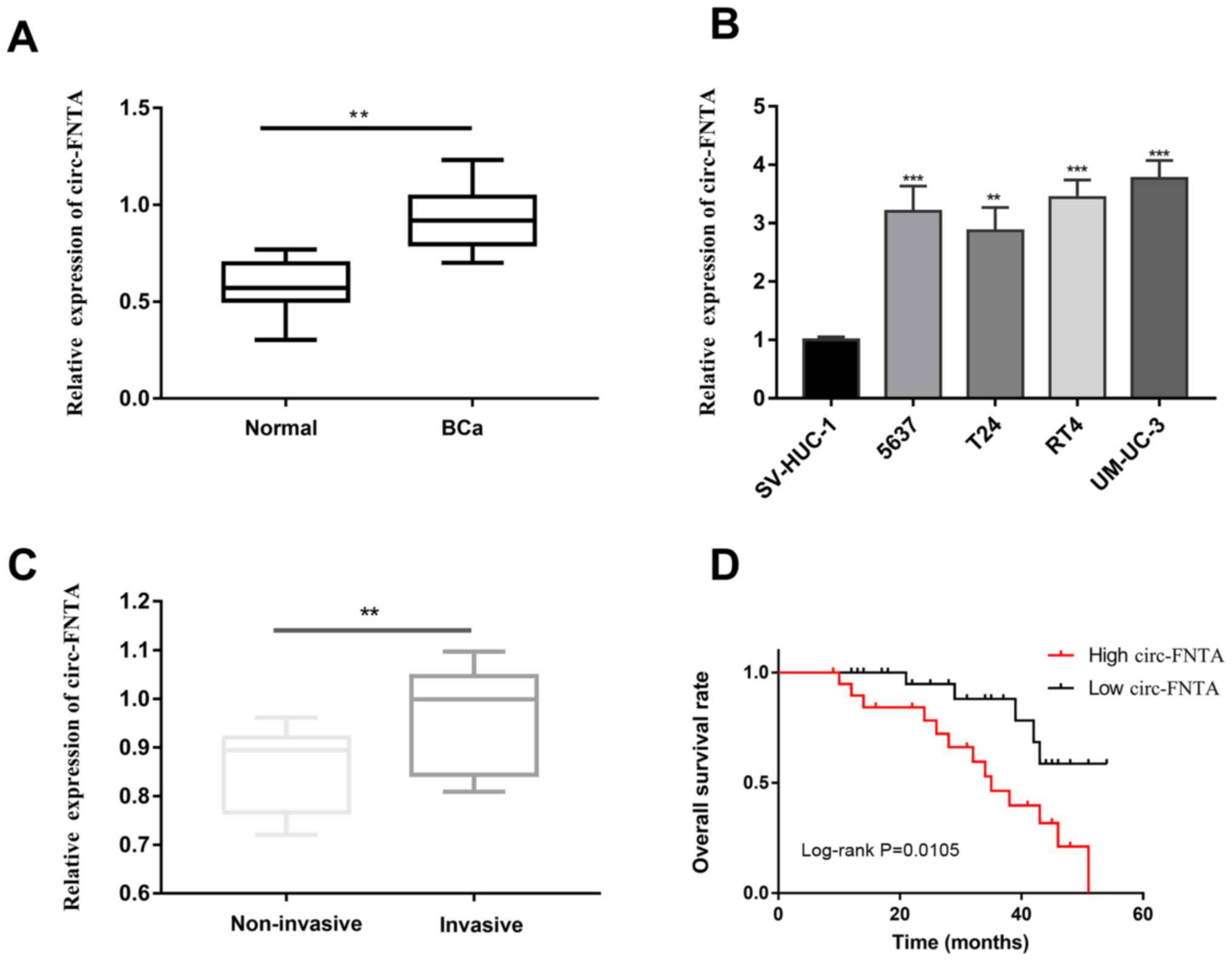
circ-FNTA is upregulated in BCa
QRT-PCR showed higher abundance of circ-FNTA in BCa
tissues relative to normal ones (Fig.
1A). Similarly, its level was higher in BCa cells than that of
bladder epithelial cells (Fig. 1B).
According to the invasion status of the enrolled BCa patients, they
were classified into non-invasive group and invasive group.
circ-FNTA was upregulated in the invasive group compared with that
of the non-invasive group (Fig. 1C).
Through analyzing the follow-up data of BCa patients, it is found
that high level of circ-FNTA predicted worse prognosis of BCa
(Fig. 1D). It is suggested that
circ-FNTA may exert a carcinogenic role in the progression of
BCa.
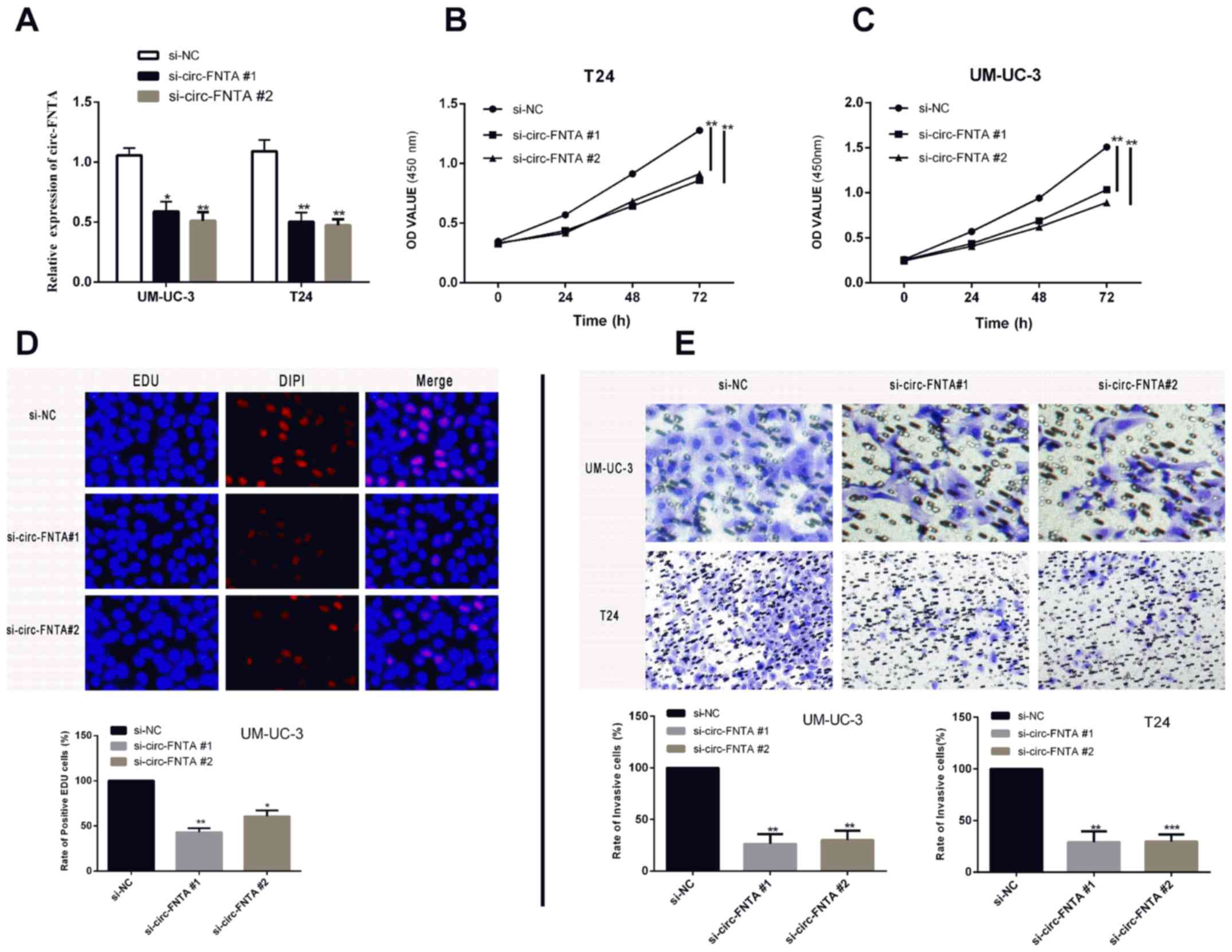
Knockdown of circ-FNTA suppresses
proliferative and invasive abilities of BCa
T24 and UM-UC-3 cell lines were selected for the
following in vitro experiments. We constructed two circ-FNTA
siRNAs (si-circ-FNTA #1 and si-circ-FNTA #2). Transfection of
si-circ-FNTA #1 or si-circ-FNTA #2 markedly downregulated circ-FNTA
level in BCa cells (Fig. 2A). CCK-8
assay showed reduced viability in T24 and UM-UC-3 cells transfected
with si-circ-FNTA #1 or si-circ-FNTA #2 (Fig. 2B, 2C). Knockdown of circ-FNTA
markedly decreased the ratio of EdU-positive cells, suggesting
inhibited proliferative ability of BCa cells (Fig. 2D). Transwell assay showed that
knockdown of circ-FNTA in T24 and UM-UC-3 cells markedly decreased
the ratio of invasive cells, indicating an attenuated invasive
ability (Fig. 2E). Hence, silence of
circ-FNTA was proved to attenuate proliferative and invasive
abilities of BCa cells.
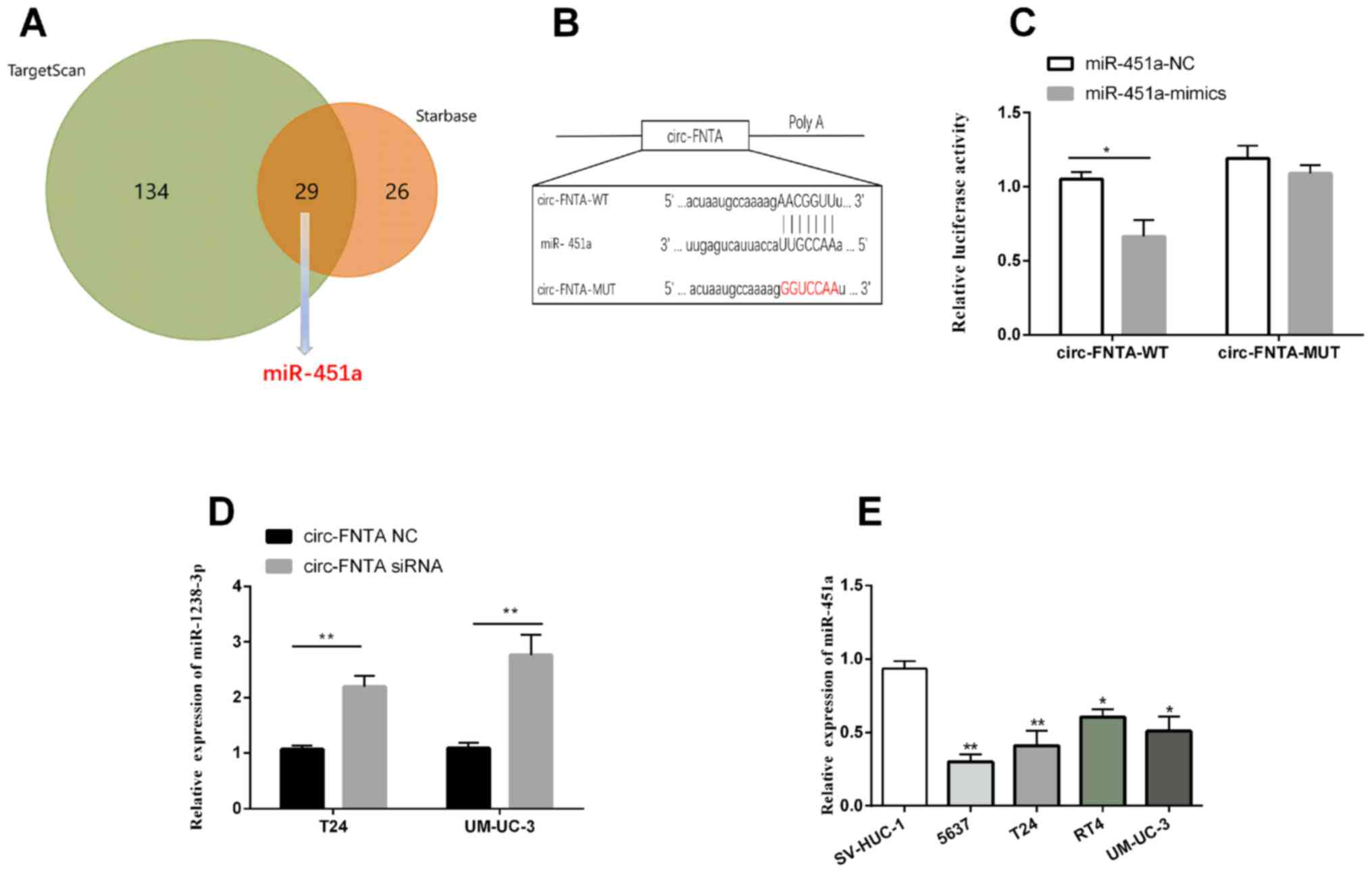
circ-FNTA targets miRNA-451a
According to the prediction on Starbase and
TargetScan, a total of 29 overlapped target miRNAs of circ-FNTA
were discovered (Fig. 3A).
miRNA-451a is previously reported to be downregulated in BCa
(20). It is predicted to be the
downstream target of circ-FNTA among the 29 overlapped ones. Hence,
we focused on the potential role of miRNA-451a in the progression
of BCa. Through bioinformatics analysis, potential binding sites
between circ-FNTA and miRNA-451a were identified (Fig. 3B). A remarkable decline in luciferase
activity was observed after co-transfection of miRNA-451a mimics
and pmirGLO-circ-FNTA-wt, confirming the binding relationship
between circ-FNTA and miRNA-451a (Fig.
3C). Expression level of miRNA-451a was markedly upregulated by
transfection of si-circ-FNTA #1 in T24 and UM-UC-3 cells (Fig. 3D). It was found that miRNA-451a was
downregulated in BCa cells relative to bladder epithelial cells
(Fig. 3E). The above data
demonstrated that circ-FNTA targeted miRNA-451a and negatively
regulated its level in BCa.
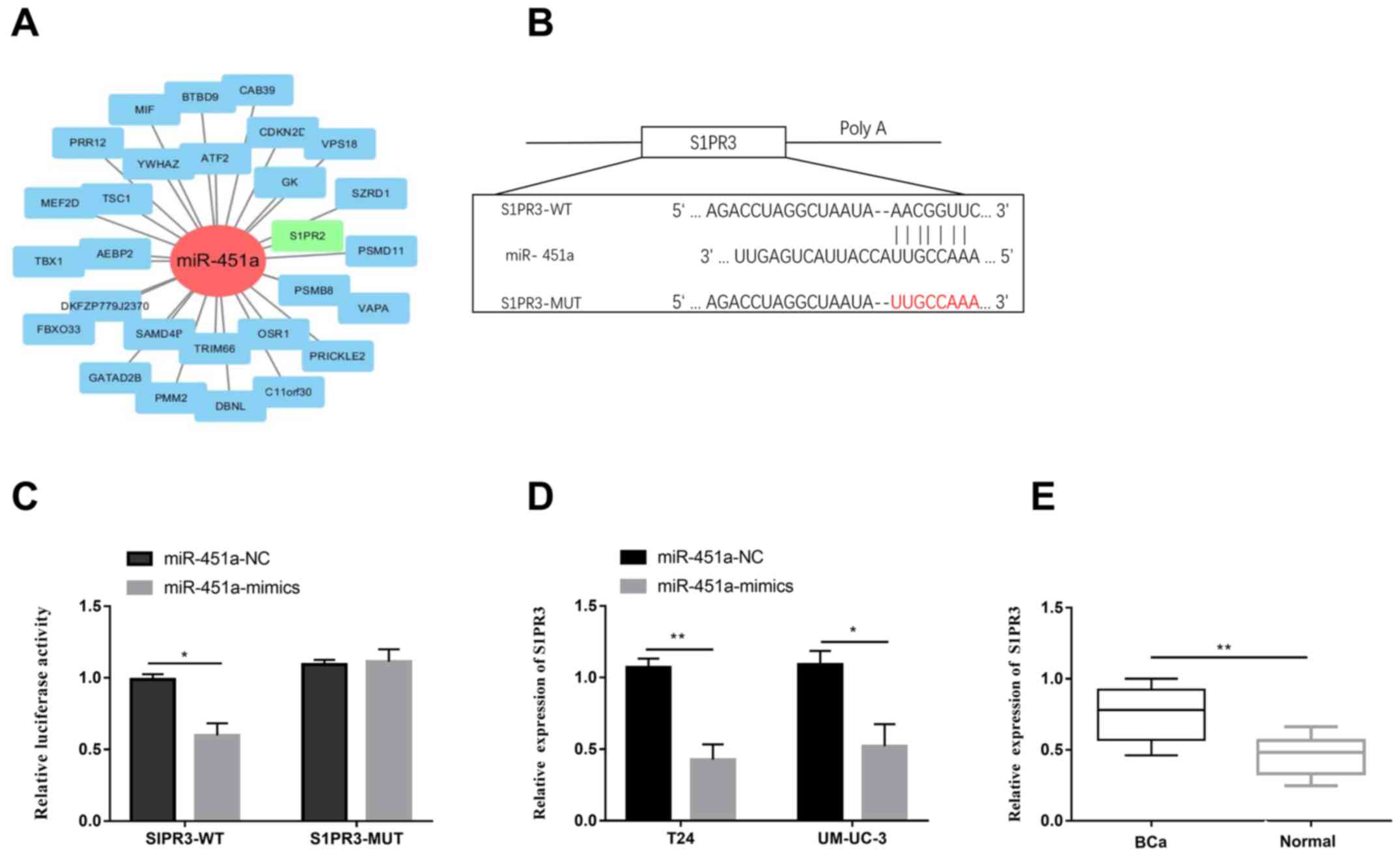
miRNA-451a regulates its target gene
S1PR3
Through analyzing the database, S1PR3 was predicted
to be the target gene of miRNA-451a (Fig. 4A). A relevant study reported the
involvement of S1PR3 in the progression of BCa (21). Potential binding sites between
miRNA-451a and S1PR3 were identified (Fig. 4B). Subsequently, dual-luciferase
reporter gene assay verified the binding relationship between
miRNA-451a and S1PR3 (Fig. 4C).
Transfection of miRNA-451a mimics remarkably downregulated S1PR3
level in T24 and UM-UC-3 cells (Fig.
4D). In addition, S1PR3 was downregulated in BCa tissues
compared with those of controls (Fig.
4E). As a result, S1PR3 was demonstrated to be the target gene
of miRNA-451a. It is suggested that circ-FNTA/miRNA-451a/S1PR3 axis
exerted carcinogenic role in BCa.
Discussion
The role of circRNAs in urinary tumors has been well
studied (22). Plenty of circRNAs
have been discovered participating in the progression of BCa. For
example, circRNA-cTFRC absorbs miRNA-107 to regulate target gene
expression, and thereafter aggravates the progression of BCa
(23). CircGprc5a is upregulated in
BCa. It induces the upregulation of Gprc5a through a polypeptide,
and further stimulates the progression of BCa (24). circRNA-PRMT5 accelerates EMT of BCa
through sponging miRNA-30c (25).
This study mainly explored the role of circ-FNTA, a newly
discovered circRNA, in regulating the progression of BCa.
The reference gene for circ-FNTA is the FNTA gene
located on chromosome 8. FNTA is considered to be a key gene for
tumor progression through activating the Ras-MAPK pathway. FTI
alleviates tumor progression through blocking the activation of
Ras-MAPK pathway (26). Abnormal
copy numbers of FNTA are believed to cause pathological changes of
breast cancer, which are key targets for developing drugs (27). CeRNA theory proposes that circRNA
sponges miRNA to influence the target gene expression, thus
influencing the pathological progression (28).
S1PR3 (sphingosine-1 phosphate receptor 3) is a key
receptor gene for tumor progression. For example, in lung
adenocarcinoma, S1PR3 expression is upregulated and closely related
to the activated TGF-β/SMAD pathway. S1PR3 activation can promote
malignant progression of lung cancer (29). In addition, S1PR3 induces expansion
of cancer stem cells by activating Notch signaling pathway, and
S1PR3 may be a potential target for tumor therapy (30). S1PR3 is a molecular marker for tumor
progression of BCa, which exerts prognostic potential (21). It is suggested that S1PR3 has a
carcinogenic role in aggravating the malignant progression of
tumors.
In this study, circ-FNTA was upregulated in BCa
tissues and cell lines. Through analyzing the clinical data of BCa
patients, circ-FNTA was found to be highly expressed in invasive
BCa patients relative to the non-invasive ones. In vitro
experiments demonstrated that silence of circ-FNTA attenuated
proliferative and invasive abilities of BCa. Subsequently, through
online prediction and dual-luciferase reporter gene assay
verification, miRNA-451a was confirmed to be the target of
circ-FNTA and S1PR3 was found to be the target gene of miRNA-451a.
Our study identified the role of circ-FNTA/miRNA-451a/S1PR3 axis in
aggravating the progression of BCa.
In conclusion, circ-FNTA accelerates the
proliferative and invasive abilities of BCa through absorbing
miRNA-451a to regulate the S1PR3 level, and indicates a poor
prognosis of BCa patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JT and LZ designed the study and performed the
experiments, JT and JF established the animal models, LZ and JX
collected the data, TR and HG analyzed the data, JT and LZ prepared
the manuscript. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Linyi Cancer Hospital (Linyi, China). Signed informed consents were
obtained from the patients and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declared no conflict of interest.
References
|
1
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soerjomataram I, Lortet-Tieulent J, Parkin
DM, Ferlay J, Mathers C, Forman D and Bray F: Global burden of
cancer in 2008: A systematic analysis of disability-adjusted
life-years in 12 world regions. Lancet. 380:1840–1850. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen D, Li SG, Chen JY and Xiao M: miR-183
maintains canonical Wnt signaling activity and regulates growth and
apoptosis in bladder cancer via targeting AXIN2. Eur Rev Med
Pharmacol Sci. 22:4828–4836. 2018.PubMed/NCBI
|
|
4
|
Stone L: Bladder cancer: Mastering the
immune microenvironment. Nat Rev Urol. 14:6392017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sargos P, Baumann BC, Eapen L,
Christodouleas J, Bahl A, Murthy V, Efstathiou J, Fonteyne V,
Ballas L, Zaghloul M, et al: Risk factors for loco-regional
recurrence after radical cystectomy of muscle-invasive bladder
cancer: A systematic-review and framework for adjuvant
radiotherapy. Cancer Treat Rev. 70:88–97. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grossman HB: Bladder cancer: Neoadjuvant
is new again. Lancet Oncol. 12:830–831. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morales-Barrera R, Suárez C, de Castro AM,
Racca F, Valverde C, Maldonado X, Bastaros JM, Morote J and Carles
J: Targeting fibroblast growth factor receptors and immune
checkpoint inhibitors for the treatment of advanced bladder cancer:
New direction and New Hope. Cancer Treat Rev. 50:208–216. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic circular RNA functions as a miR-21 sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang
Y, Bu Y, Zhao M, Zhang S and Zhang X: Long non-coding RNA HULC
activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in
HBV-related hepatocellular carcinoma. Cancer Lett. 454:158–170.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Chen M, Jiang N, Shi K and Qian
R: A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the
proliferation of lung adenocarcinoma. Cancer Biol Ther.
20:1127–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He J, Chen J, Ma B, Jiang L and Zhao G:
CircLMTK2 acts as a novel tumor suppressor in gastric cancer.
Biosci Rep. 39:392019. View Article : Google Scholar
|
|
15
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sebti SM and Adjei AA: Farnesyltransferase
inhibitors. Semin Oncol. 31 (Suppl 1):28–39. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42D:D92–D97. 2014. View Article : Google Scholar
|
|
19
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
20
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Go H, Kim PJ, Jeon YK, Cho YM, Kim K, Park
BH and Ku JY: Sphingosine-1-phosphate receptor 1 (S1PR1) expression
in non-muscle invasive urothelial carcinoma: Association with poor
clinical outcome and potential therapeutic target. Eur J Cancer.
51:1937–1945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang
X, Chen W, Han Y, Shao L, Gao Y, et al: Genome-wide identification
of cancer-specific alternative splicing in circRNA. Mol Cancer.
18:352019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Su H, Tao T, Yang Z, Kang X, Zhang X, Kang
D, Wu S and Li C: Circular RNA cTFRC acts as the sponge of
microRNA-107 to promote bladder carcinoma progression. Mol Cancer.
18:272019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S,
Feng Y, Chen L, Yang J and Tian F: circGprc5a promoted bladder
oncogenesis and metastasis through Gprc5a-targeting Peptide. Mol
Ther Nucleic Acids. 13:633–641. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dangle PP, Zaharieva B, Jia H and Pohar
KS: Ras-MAPK pathway as a therapeutic target in cancer - emphasis
on bladder cancer. Recent Patents Anticancer Drug Discov.
4:125–136. 2009. View Article : Google Scholar
|
|
27
|
Chin K, DeVries S, Fridlyand J, Spellman
PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et
al: Genomic and transcriptional aberrations linked to breast cancer
pathophysiologies. Cancer Cell. 10:529–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qi X, Lin Y, Chen J and Shen B: Decoding
competing endogenous RNA networks for cancer biomarker discovery.
Brief Bioinform. Jan 30–2019.(Epub ahead of print).
doi.org/10.1093/bib/bbz006. View Article : Google Scholar
|
|
29
|
Zhao J, Liu J, Lee JF, Zhang W, Kandouz M,
VanHecke GC, Chen S, Ahn YH, Lonardo F and Lee MJ: TGF-β/SMAD3
pathway stimulates sphingosine-1 phosphate receptor 3 expression:
Implication of sphingosine-1 phosphate receptor 3 in lung
adenocarcinoma progression. J Biol Chem. 291:27343–27353. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hirata N, Yamada S, Shoda T, Kurihara M,
Sekino Y and Kanda Y: Sphingosine-1-phosphate promotes expansion of
cancer stem cells via S1PR3 by a ligand-independent Notch
activation. Nat Commun. 5:48062014. View Article : Google Scholar : PubMed/NCBI
|