Introduction
Breast cancer is one of the most common gynecologic
tumors, mainly occurring in the elderly (1). A previous review reported the most
common genes involved in epigenetic modifications in patients with
breast cancer (2). Most breast
cancer-associated mortalities are caused by local breast cancer
cell migration and distant metastasis (3). A review reported genetic analyses and
inherited gene mutations in patients with breast cancer (4,5).
Although advances in molecular diagnosis and medical treatments,
including surgical techniques, radiation, chemotherapy and gene
target therapy, have improved the 5-year survival rate of patients
with breast cancer, the overall clinical outcomes remain poor
(6–9). It is therefore essential to determine
potential target proteins to inhibit breast cancer growth and
metastasis.
Neurensin-2 (NRSN2) is a small neuronal membrane
protein that is localized in small vesicles of neural cells
(10). A previous study revealed
that NRSN2 can promote non-small cell lung cancer cell
proliferation via the PI3K/AKT/mTOR pathway (11). Tang et al (12) reported that NRSN2 overexpression was
associated with malignant phenotypes in ovarian cancer, suggesting
that it could be considered as a target for ovarian cancer
treatment. However, Wang et al (13) demonstrated that NRSN2 upregulation
inhibited cell proliferation and survival via the PI3K/AKT/mTOR
pathway in hepatocellular carcinoma. These findings encouraged the
present study to further investigate the role of NRSN2 in breast
cancer cells.
The PI3K/AKT/mTOR, p65 and NF-κB signaling pathways
contribute to breast cancer progression. Therefore, the present
study hypothesized that NRSN2 may regulate breast cancer cell
proliferation via the PI3K/AKT/mTOR and NF-κB pathways (14). The results from the present study
demonstrated that NRSN2 overexpression significantly increased the
proliferation, invasion and metastasis of breast cancer cells,
suggesting that NRSN2 may be considered as a potential therapeutic
target for breast cancer treatment via downregulation of the
PI3K/AKT/mTOR and NF-κB signaling pathways.
Materials and methods
Ethical statements
This study was conducted in strict accordance with
the recommendations of the Guide for the Care and Use of Laboratory
Animals of the Tianjin Medical University. The protocol was
approved by the Chinese Association for Laboratory Animal
Operations. All surgery and euthanasia were performed under sodium
pentobarbital anesthesia (intravenous, 35 mg/kg). Mice were
sacrificed via cervical decapitation.
Patients and tissues
A total of 24 patients with breast cancer were
recruited in Peking University between May 2015 and October 2016.
Their average age was 54.5±24.5 years (range, 30–79 years). Breast
cancer and adjacent noncancerous tissues were obtained from
patients who underwent tumor resection and stored at −80°C prior to
immunohistochemistry (IHC) and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analyses. Patients who had
previously undergone radiotherapy, chemotherapy or administration
of any other drug were excluded from this study. All patients
provided written informed consent prior to any procedures of this
study. The patient study was approved by the Ethics Committee of
Peking University (approval no. PEK20150524).
Cell line, chemicals and reagents
The breast tumor cell lines MDA-MB-231 and BT549,
and the normal breast cell line MCF-10A were purchased from the
American Type Culture Collection. All cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (Invitrogen;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.) and placed at 37°C in
a humidified incubator containing 5% CO2. Cells were
treated with the NF-κB inhibitor caffeic acid phenethyl ester
(CAPE; 70 mM; Apex Biotechnology Corp.), PI3K inhibitor (70 mM;
Sigma-Aldrich; Merck KGaA) or PBS as control for 12 h at 37°C for
further experiments (15).
Small interfering RNA (siR)-NRSN2
transfection
All siRs (siR-NRSN2, 5′-CAATCTTCTGTGCAGACTATC-3′;
siR-NC, 5′-CGAGGACAGGCTGATCTTCC-3′) were synthesized by Invitrogen;
Thermo Fisher Scientific, Inc. MDA-MB-231 cells (1×106
cells/well) were cultured in six-well plates and transfected with
150 pM siR-NRSN2 or si-control using a Cell Line Nucleofector kit
(cat. no. VCA-1003; Lonza Group, Ltd.) according to manufacturer's
protocol. The efficiency of siR-NRSN2 transfection was verified via
western blotting at 72 h following transfection, prior to
subsequent experiments.
NRSN2 overexpression
An expression plasmid (pRK5- hNRSN2) with a Flag tag
at the C-terminus was constructed by Invitrogen (Thermo Fisher
Scientific, Inc.). MDA-MB-231 cells (1×104) were seeded
in 6-well plates (Corning Inc.) and transiently transfected with
pRK5-hNRSN2 (2 µg) or pRK5-control (pControl) (2 µg) using
Lipofectamine® 2000 (cat. no. 11668-027; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The efficiency of NRSN2 overexpression was verified by
western blotting at 72 h following transfection, prior to
subsequent experiments.
RT-qPCR
Total RNA was isolated from tissues or cells by
using an RNAeasy Mini kit (Qiagen, Inc.). The expression of NRSN2
in tissues and cells was measured using a Hairpin-it™ RT-qPCR kit
(Invitrogen; Thermo Fisher Scientific, Inc.). NRSN2 expression
levels were measured in an iCycler thermal cycler (Bio-Rad
Laboratories, Inc.) using iQ SYBR Green Supermix (Bio-Rad
Laboratories, Inc.). The thermocycling conditions were: 95°C for
120 sec; followed by 45 cycles at 95°C for 30 sec, 56°C for 20 sec
and 65°C for 30 sec. The primers were designed as follows: NRSN2,
forward 5′-CGGAGACGCAGGTCCAGAGGGAT-3′, reverse
5′-TATGCATCAACTGTTTATTGAAAGG-3′; and β-actin, forward
5′-GTGGGCGCCCAGGCACCA-3′ and reverse 5′-CTCCTTAATGTCACGCACGATTT-3′.
Relative mRNA expression changes were calculated using the
2−ΔΔCq method (16). The
results are expressed compared to β-actin expression.
Cell proliferation assay
Cell proliferation was determined using a Cell
Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc.)
according to the manufacturer's instructions. Briefly, pRK5-hNRSN2
or SiR-NRSN2-transfected MDA-MB-231 cells and their controls were
seeded in 96-well plates at a density of 1×103/well and
cultured at 37°C in a 5% CO2 atmosphere for 48 h. CCK-8
solution (10 µl) was added to each well for 2 h. The cell
proliferation was monitored by measuring the absorbance at 450 nm
using a microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
For the colony formation assay, pRK5-hNRSN2 or
SiR-NRSN2-transfected MDA-MB-231 cells were seeded in 6-well plates
at a density of 1×103/well and cultured at 37°C in a 5%
CO2 atmosphere for 7 days until visible colonies were
formed. Cells were stained with 5% crystal violet for 10 min at
room temperature. The numbers of colonies were then counted using a
light microscope at ×20 magnification.
Cell migration and invasion assay
MDA-MB-231 cells were transfected with pRK5-hNRSN2
or siR-NRSN2. Matrigel-uncoated and -coated migration inserts (8-µm
pore size; Corning Inc.) were used for migration and invasion
assays, respectively.
For the migration assay, MDA-MB-231 cells
(1×104) in DMEM were plated into the upper chamber with
the non-coated membrane. For the invasion assay, pRK5-hNRSN2 or
siR-NRSN2-transfected cells were prepared at a density of
1×104 cells in 500 µl serum-free DMEM in the upper
chamber and the lower chamber contained DMEM with 5% FBS. Cells
were seeded in the upper chamber of a BD BioCoat Matrigel Invasion
Chamber (BD Biosciences) according to the manufacturer's
instructions. Following incubation for 48 h, cells were fixed with
4% paraformaldehyde for 1 h at room temperature and stained with
0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 10 min at 37°C.
The membranes were mounted onto a glass slide with antifade
mounting medium (cat. no. P0126; Beyotime Institute of
Biotechnology), and the number of migrating and invading tumor
cells was counted in at least three randomly selected fields under
a light microscope (Olympus Corporation) at ×200 magnification.
Western blotting
MDA-MB-231 cells were transfected with pRK5-hNRSN2
or siR-NRSN2. Following transfection, cells were lysed using
radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck KGaA)
containing protease-inhibitor (Sigma-Aldrich; Merck KGaA) and were
centrifuged at 12,000 × g, at 4°C for 10 min. Breast cancer tissues
(10 mg) were lysed using radioimmunoprecipitation assay buffer
containing protease inhibitor and were centrifuged at 1,000 × g at
4°C for 10 min. Supernatant was collected for protein analysis. The
concentrations of protein were measured using a BCA Protein
Concentration Assay kit (Beyotime Institute of Biotechnology).
Protein samples (50 µg per lane) were separated by sodium dodecyl
sulfate-polyacrylamide electrophoresis (SDS-PAGE) on a 10% gel and
transferred to polyvinylidene difluoride (PVDF; Bio-Rad
Laboratories, Inc.) membrane. Membranes were incubated with rabbit
anti-human primary antibodies against NRSN2 (1:1,000; cat. no.
ab237739; Abcam), PI3K (1:1,000; cat. no. ab32089; Abcam),
phosphorylated (p)-PI3K (1:1,000; cat. no. ab154598; Abcam), p65
(1:1,000; cat. no. ab16502; Abcam), p-p65 (1:1,000; cat. no.
ab86299; Abcam), IκBα (1:1,000; cat. no. ab7217; Abcam) and p-IκBα
(1:1,000; cat. no. ab133462; Abcam), β-actin (1:5,000; cat. no.
20536-1-AP; ProteinTech Group, Inc.), AKT (1:1,000; cat. no.
51077-1-AP; ProteinTech Group, Inc.), p-mTOR (1:1,000; cat. no.
ab2731; Abcam), and mTOR (1:1,000; cat. no. ab109268; Abcam.),
overnight at 4°C. The membranes were subsequently incubated with
HRP-conjugated anti-rabbit IgG secondary antibodies (diluted
1:5,000; cat. no. A9169; Sigma-Aldrich; Merck KGaA) for 24 h at
4°C. Enhanced chemiluminescence reagent (Millipore; Merck KGaA) was
used to visualize the bands. Quantitation of the signal intensities
were analyzed using the Quantity One software package (version 2.0;
Bio-Rad Laboratories, Inc.).
Animal study
Pathogen-free female Balb/c (8-week-old; 20–25 g
body weight) nude mice were purchased from Slack Co., Ltd. All mice
were treated in accordance with the China Legislation on the
Protection of Animals and the Guide for the Care and Use of
Laboratory Animals. The study was approved by the ethics committees
of Shanxi Traditional Chinese Medical University. Animals were
housed in a temperature-controlled facility at 23±1°C with 50±5%
humidity under a 12-h light/dark cycle. All mice had free access to
food and water. The siR-NRSN2-transfected MDA-MB-231, pRK5-control
vector-transfected MDA-MB-231 cells or MDA-MB-231 cells
(1×107) were subcutaneously injected into the flanks of
female Balb/c mice. The tumor volume was calculated every 3 days
and calculated as Volume = (D × d2)/2 (D represents the
maximal diameter, d represents the minimal one). The mice were
sacrificed on day 30 following anesthesia.
IHC
Human breast tissues or mouse cancer tissues were
analyzed for NRSN2 expression using IHC, as previously described
(17). Briefly, tumor tissue samples
were fixed in 10% formalin for 12 h at room temperature and
embedded in paraffin blocks for 8 h at room temperature. Thin
tissue sections (4-µm thick) were de-waxed and rehydrated. Tissue
sections were immersed for 15 min at room temperature in 0.3%
H2O2 (diluted with 100% methanol) to block
endogenous peroxidase activity. The tumor sections were incubated
with specific primary antibodies: Rabbit anti-human or mouse
antibodies against rabbit anti-human/mouse NRSN2 (1:1,000; cat. no.
ab237739; Abcam) overnight at 4°C. Tumor tissues were then
incubated with horseradish peroxidase-conjugated goat anti-rabbit
immunoglobulin G secondary antibody (1:5,000; cat. no. PV-6001;
OriGene Technologies, Inc.) for 2 h at room temperature. Finally,
tissue sections were stained with 3–3′-diaminobenzidine
tetrahydrochloride (Sigma-Aldrich; Merck KGaA) for 1 h at room
temperature and lightly counter-stained by hematoxylin for 30 min
at room temperature. A Ventana Benchmark automated staining system
was used to detect NRSN2 protein expression in tumor tissues
(Olympus BX51; Olympus Corporation) at ×200 magnification. The
quantification of NRSN2 density was analyzed using Image J software
(version 4.6; National Institutes of Health).
Statistical analysis
All data were expressed as the mean ± standard
deviation. Experiments were repeated at least three times.
Statistical analyses were performed using Student's t-test or
one-way ANOVA followed by Tukey's honestly significant difference
post hoc test. Data were analyzed using SPSS Statistics 19.0
software (IBM Corp.) and GraphPad Prism 5.0 (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
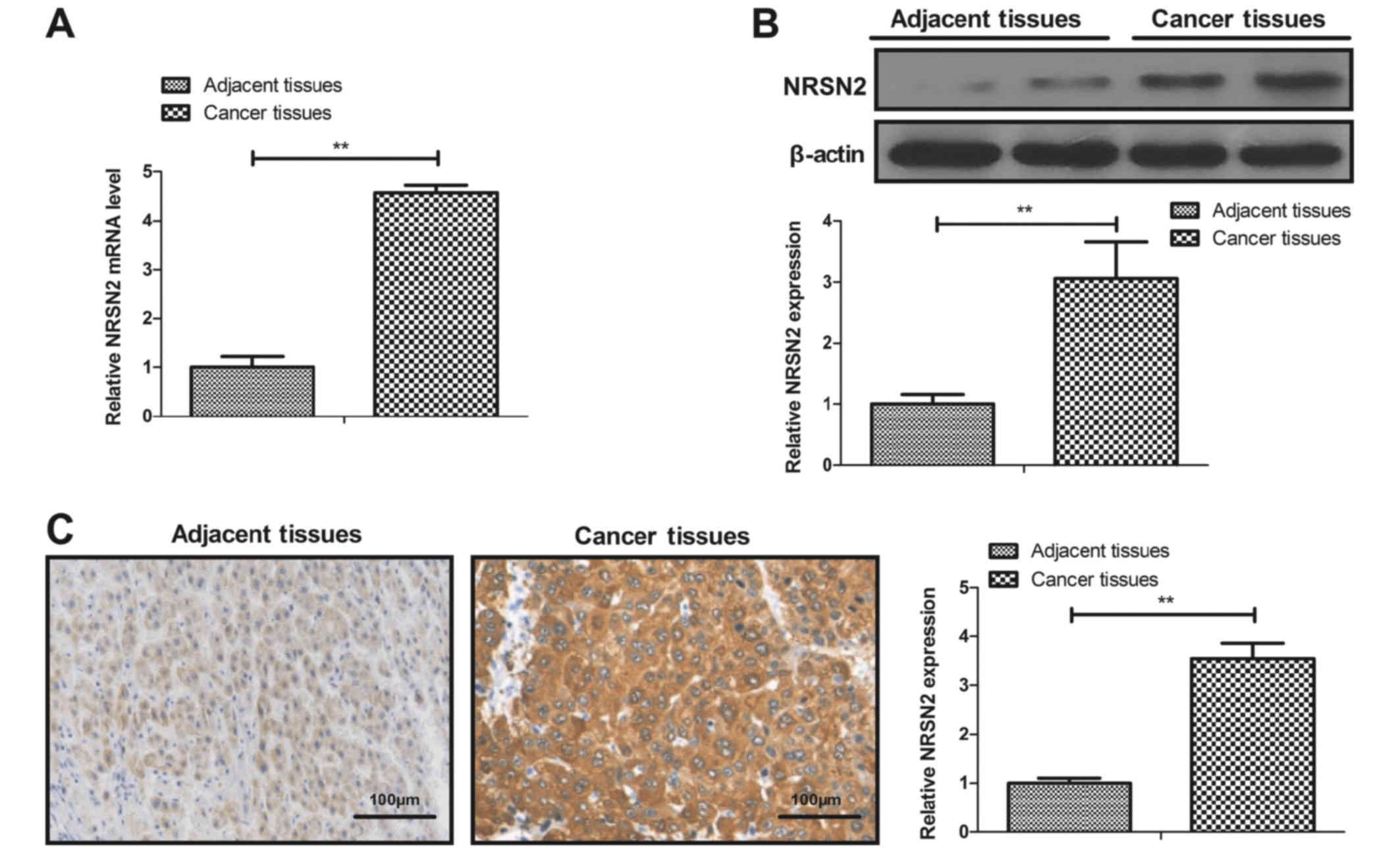
NRS2N expression is elevated in breast
cancer tissues and cell lines
The mRNA and protein levels of NRSN2 in breast
cancer tissues and cell lines were determined. As presented in
Fig. 1A and B, mRNA and protein
levels of NRSN2 were significantly increased in breast cancer
tissues compared with adjacent tissues. Furthermore, the results
from IHC demonstrated that NRSN2 was highly expressed in breast
cancer tissues compared with adjacent tissues (Fig. 1C). In addition, as presented in
Fig. 1D and E, the mRNA and protein
levels of NRSN2 were significantly increased in the breast cancer
cell lines BT549 and MDA-MB-231 compared with the MCF-10A cell
line. These results demonstrated that NRSN2 was highly expressed in
breast cancer tissues and cell lines, which suggested that NRSN2
may have a role in breast cancer progression.
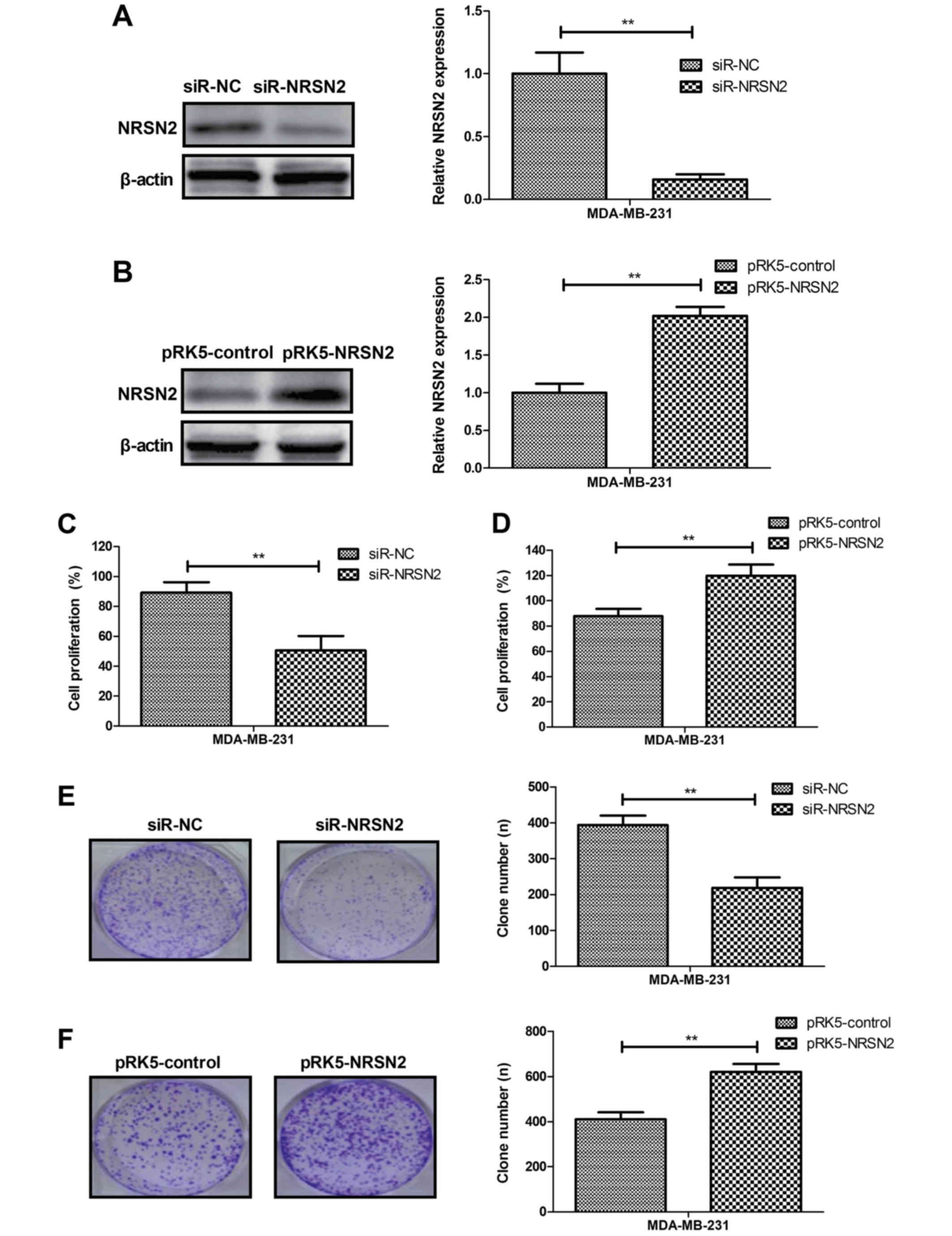
NRSN2 knockdown inhibits the
proliferation of breast cancer cells in vitro
The role of NRSN2 in the breast cancer cell line
MDA-MB-231 was further investigated. As presented in Fig. 2A and B, NRSN2 knockdown (siR-NRSN2)
and overexpression (pRK5-NRSN2) significantly decreased and
increased, respectively, NRSN2 expression in MDA-MB-231 cells.
Furthermore, NRSN2 knockdown inhibited MDA-MB-231 cell
proliferation (Fig. 2C), whereas
NRSN2 overexpression promoted MDA-MB-231 cell proliferation
(Fig. 2D). Colony formation assays
demonstrated that NRSN2 overexpression increased the numbers of
MDA-MB-231 cell colonies, whereas NRSN2 knockdown decreased the
numbers of MDA-MB-231 cells colonies formed (Fig. 2E and F). These results demonstrated
that NRSN2 knockdown could inhibit breast cancer cell proliferation
in vitro.
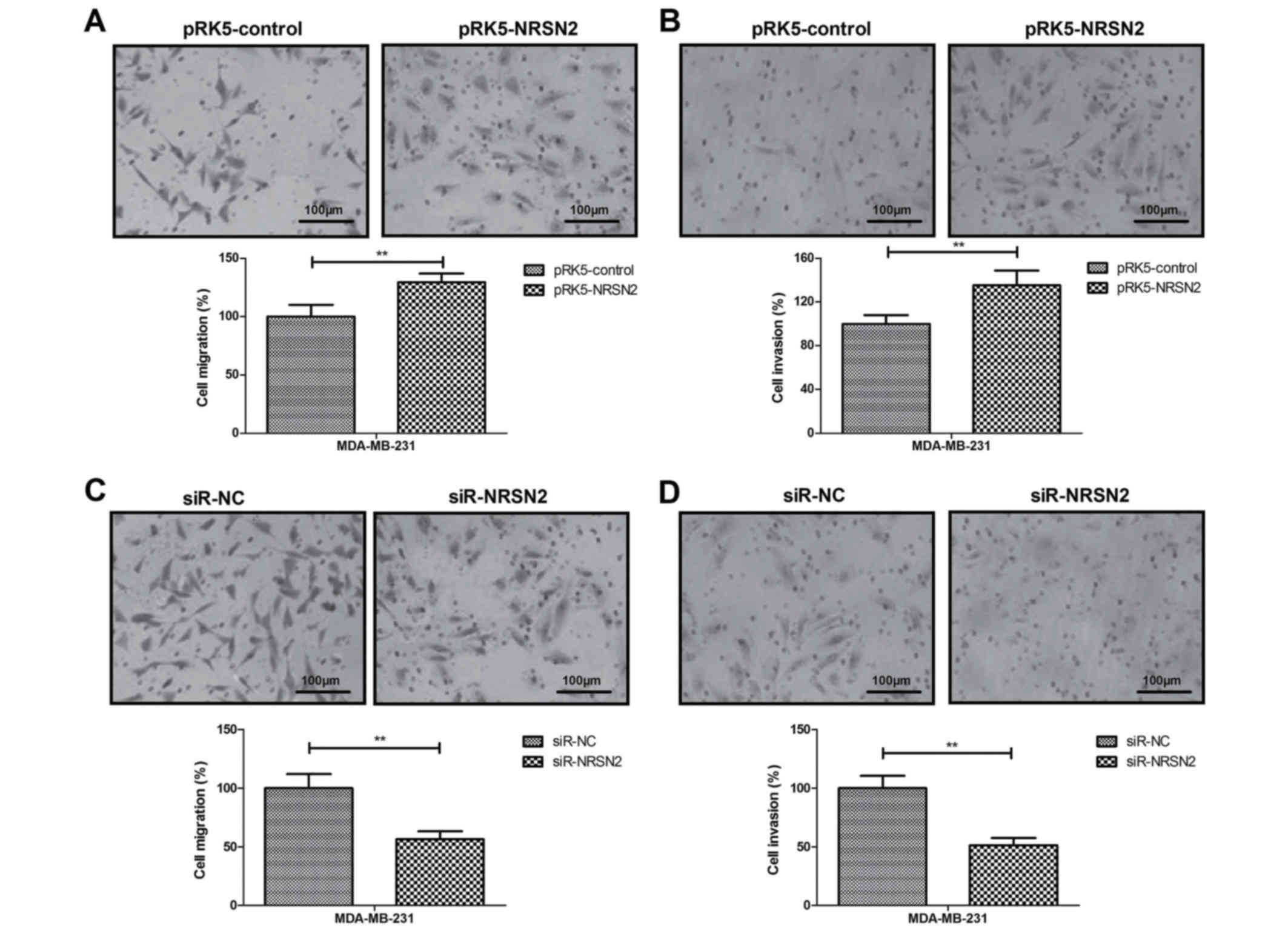
NRSN2 promotes tumor cell migration
and invasion in vitro
The effects of NRSN2 on cell motility were analyzed
using migration and invasion assays. The results demonstrated that
NRSN2 overexpression promoted the migration and invasion of
MDA-MB-231 cells (Fig. 3A and B),
whereas NRSN2 knockdown inhibited the migration and invasion of
MDA-MB-231 cells (Fig. 3C and
D).
NRSN2 promotes proliferation,
migration and invasion of breast cancer cells by activating
PI3K/AKT/mTOR and NF-κB signaling pathways
The potential molecular mechanisms mediated by NRSN2
were investigated in MDA-MB-231 cells. The results demonstrated
that NRSN2 knockdown significantly inhibited the phosphorylation of
PI3K, AKT and mTOR in MDA-MB-231 cells (Fig. 4A). Furthermore, NRSN2 knockdown
significantly decreased IκBα and P65 phosphorylation in MDA-MB-231
cells (Fig. 4B). NRSN2
overexpression induced opposing effects (Fig. 4C and D). In addition, the NF-κB
inhibitor CAPE (NF-κBIR) inhibited the pro-proliferation effects of
NRSN2 (NF-κBIR) in MDA-MB-231 cells (Fig. 4E). Furthermore, NF-κBIR suppressed
the NRSN2 overexpression-induced (NF-κBIR-NRSN2) migration and
invasion of MDA-MB-231 cells (Fig. 4F
and G). Treatment of NRSN2 overexpressing cells with PI3K
inhibitor (PI3KIR-NRSN2) inhibited NRSN2 overexpression-induced
proliferation, migration and invasion of MDA-MB-231 cells (Fig. 4H-J). However, PI3K inhibitor or NF-κB
inhibitor induced no effects on the proliferation, migration or
invasion of MDA-MB-231 cells. These results suggested that NRSN2
may regulate the proliferation and aggressiveness of MDA-MB-231
cells through PI3K/AKT/MTOR and NF-κB signaling pathways.
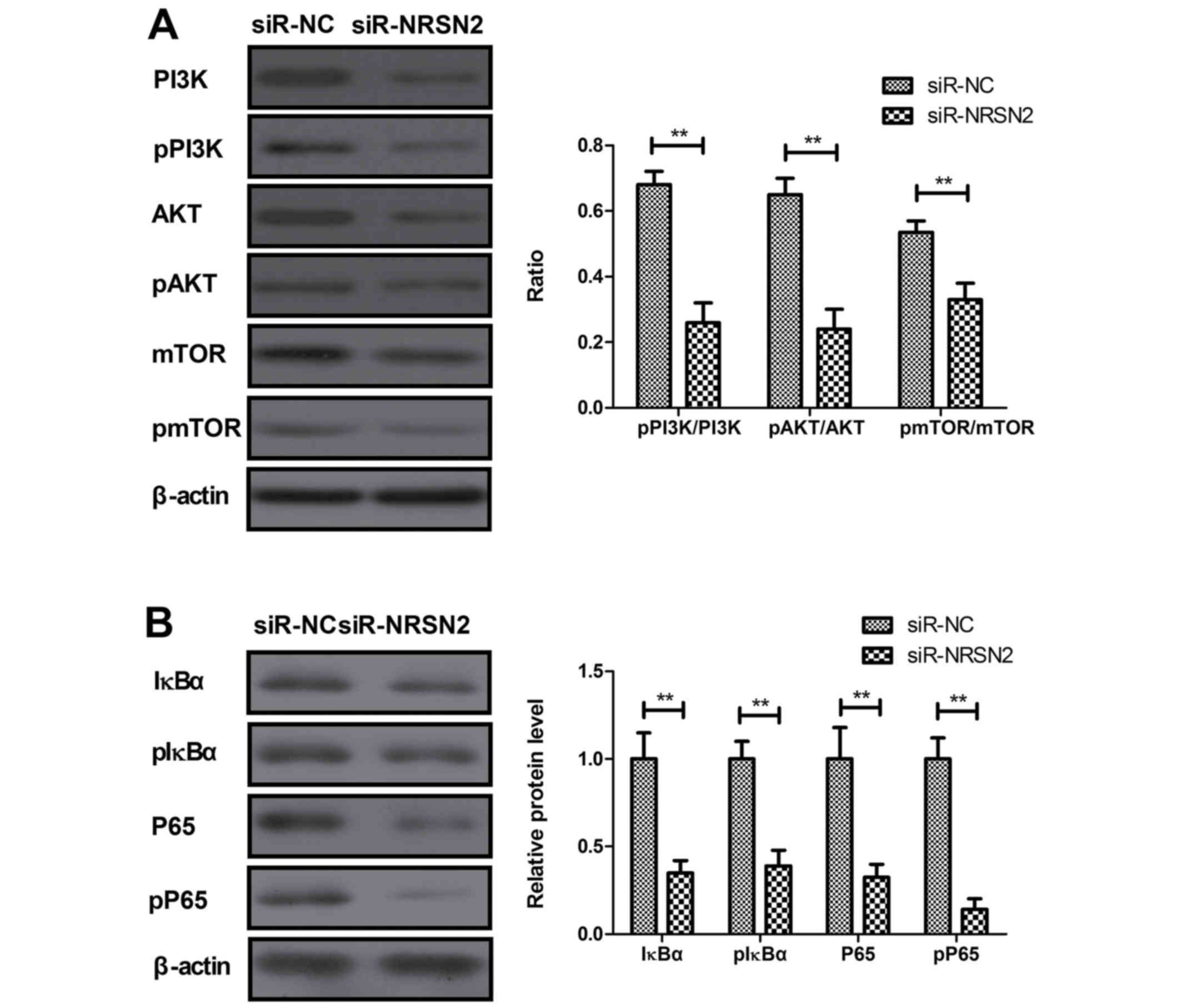 | Figure 4.NRSN2 promotes proliferation,
migration and invasion of breast cancer cells by activating
PI3K/AKT/mTOR and NF-κB signaling pathways. (A) NRSN2 knockdown
significantly inhibited the phosphorylation of PI3K, AKT and mTOR
in MDA-MB-231 cells. (B) NRSN2 knockdown significantly decreased
the levels of IκBα and P65 phosphorylation in MDA-MB-231 cells.
Control, PBS-treated cells. NC, negative control; NRSN2,
neurensin-2; ns, not significant; p-, phosphorylated; si, small
interfering (RNA); si-RNRSN2, siRNA against NRSN2; pRK5-NRSN2,
NRSN2 overexpression vector; NF-κBIR, NF-κB inhibitor; PI3KIR, PI3K
inhibitor. **P<0.01. NRSN2 promotes proliferation, migration and
invasion of breast cancer cells by activating PI3K/AKT/mTOR and
NF-κB signaling pathways. (C) NRSN2 overexpression promoted the
phosphorylation of PI3K, AKT and mTOR in MDA-MB-231 cells. (D)
NRSN2 overexpression promoted the levels of p-IκBα and p-P65 in
MDA-MB-231 cells. Control, PBS-treated cells. NC, negative control;
NRSN2, neurensin-2; ns, not significant; p-, phosphorylated; si,
small interfering (RNA); si-RNRSN2, siRNA against NRSN2;
pRK5-NRSN2, NRSN2 overexpression vector; NF-κBIR, NF-κB inhibitor;
PI3KIR, PI3K inhibitor. **P<0.01. NRSN2 promotes proliferation,
migration and invasion of breast cancer cells by activating
PI3K/AKT/mTOR and NF-κB signaling pathways. (E) NF-κBIR inhibited
the pro-proliferation effects of NRSN2 in MDA-MB-231 cells. NF-κBIR
suppressed the migration (F) and invasion (G) of MDA-MB-231 cells.
PI3KIR suppressed NRSN2-promoted (H) proliferation, (I) migration
and (J) invasion of MDA-MB-231 cells. Control, PBS-treated cells.
NC, negative control; NRSN2, neurensin-2; ns, not significant; p-,
phosphorylated; si, small interfering (RNA); si-RNRSN2, siRNA
against NRSN2; pRK5-NRSN2, NRSN2 overexpression vector; NF-κBIR,
NF-κB inhibitor; PI3KIR, PI3K inhibitor. **P<0.01. |
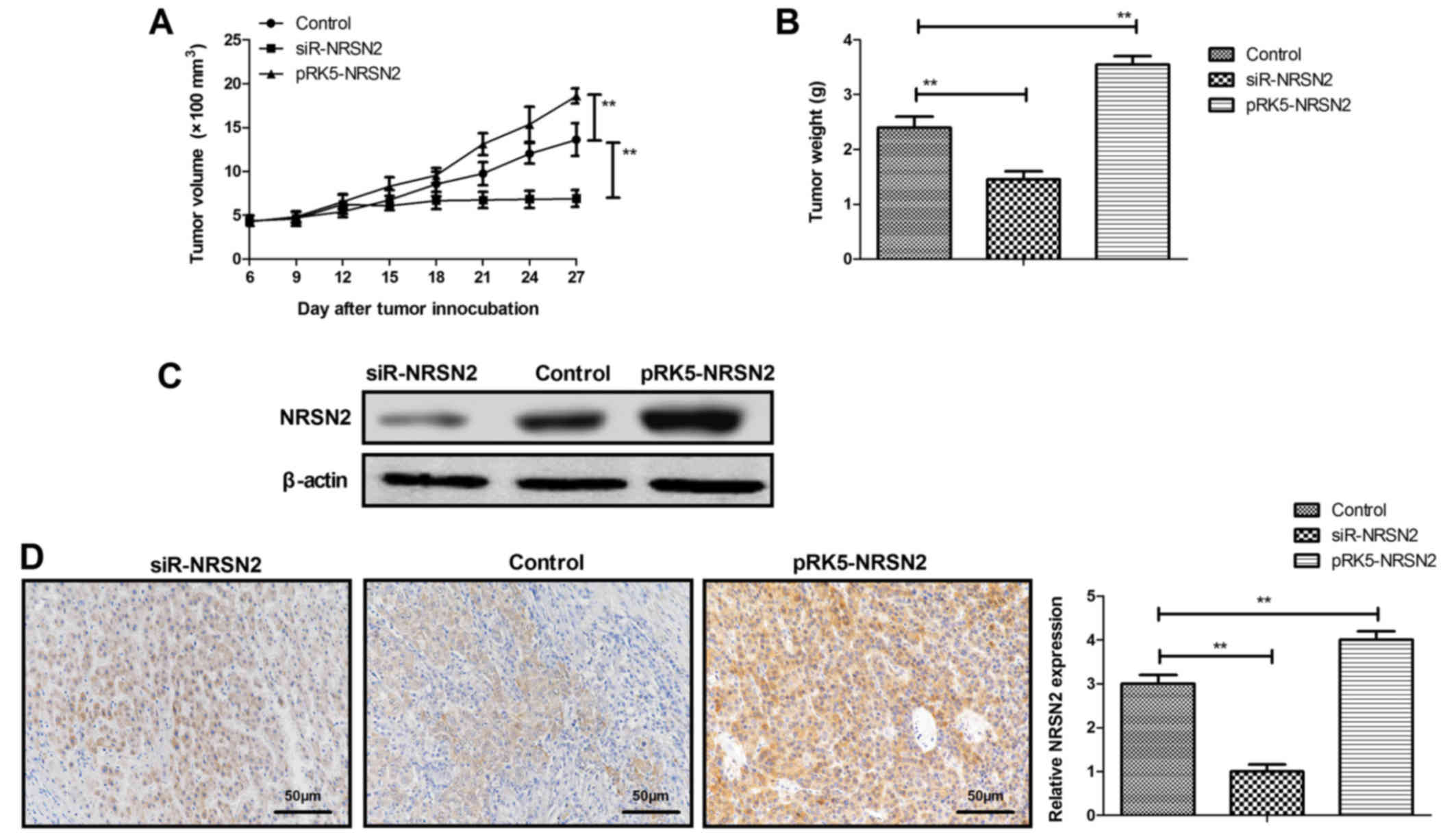
NRSN2 overexpression promotes tumor
growth in vivo
The role of NRSN2 in tumor growth was further
investigated in vivo in subcutaneous breast cancer xenograft
nude mice. The results demonstrated that tumor growth was faster in
nude mice injected with pRK5-NRSN2 plasmid-transfected MDA-MB-231
cells than in nude mice injected with pRK5-control vector (control;
Fig. 5A and B). However, injection
of siR-NRSN2 plasmid-transfected MDA-MB-231 cells induced smaller
tumor volume and weight compared with in nude mice injected with
cells transfected pRK5-control vector (Fig. 5A and B). The results from western
blotting and IHC demonstrated that NRSN2 protein expression was
increased in pRK5-NRSN2 plasmid-transfected tumor tissues compared
with in siR-NRSN2 plasmid-transfected tumor tissues (Fig. 5C and D). These results suggested that
NRSN2 may promote tumor growth in vivo.
Discussion
Breast cancer is the most common female cancer
worldwide. It is commonly diagnosed at advanced stages and exhibits
rising incidence and mortality rates (18). Previous studies have indicated that
NRSN2 is highly expressed in numerous human cancer cells, including
lung cancer and hepatocellular carcinoma, and may therefore be
considered as a potential target for human cancer treatment
(10,11,13). The
results from the present study suggested that NRSN2 may serve an
important role in the carcinogenesis and progression of breast
cancer in vitro and in vivo. The results demonstrated
that NRSN2 was highly expressed in breast cancer tissues and cells,
and may therefore stimulate the progression of breast cancer and
promote the proliferation of breast cancer cells. These findings
also indicated that NRSN2 knockdown may inhibit the migration and
invasion of breast cancer cells in vitro. Notably, NRSN2
knockdown potentially regulated human breast cancer proliferation
via PI3K/AKT/mTOR and NF-κB signaling pathway inactivation.
Tumor markers have been widely used for the
diagnosis of early stage breast cancer in patients (19–21). The
present study demonstrated that NRSN2 was significantly upregulated
in breast cancer tissues compared with adjacent noncancerous
tissues. Although a previous study reported that NRSN2
downregulation promoted cell proliferation and survival via the
PI3K/AKT/mTOR pathway in hepatocellular carcinoma (13), the results from the present study
demonstrated that NRSN2 downregulation significantly inhibited
breast cancer cell proliferation and aggressiveness. Notably, NRSN2
controlled human breast cancer proliferation via the regulation of
PI3K/AKT/mTOR and NF-κB signaling pathways; however, NF-κB
inhibitor or PI3K inhibitor had no effects on the proliferation,
migration and invasion of MDA-MB-231 cells. These results suggested
that NRSN2 may regulate the proliferation and aggressiveness of
breast cancer cells via PI3K/AKT/mTOR and NF-κB signaling
pathways.
Currently, the PI3K/AKT/mTOR signaling pathway is
considered as a potential target for breast cancer therapy
(22). A recent study reported that
intermittent hypoxia induces the overexpression of prometastatic
genes in breast cancer cells via NF-κB, such as tenascin-C (an
essential factor of the metastatic niche) and matrix
metalloproteinase 9, and induces pro-inflammatory processes, via
cyclooxygenase-2 (COX-2) for example (23). According to previous studies
(24–26), alterations of the PI3K/AKT/mTOR and
NF-κB signaling pathways were investigated in breast cancer cell
lines following NRSN2 overexpression or knockdown.
Shen et al (27) reported that the tumor volume in
6-week-old female NOD/SCID mice injected with wild-type MDA-MB-231
cells (1×106 cells in 30 µl PBS) was ~200 mm3
at day 22. In the present study, specific pathogen-free female
Balb/c nude mice were injected with MDA-MB-231 cells transfected
with pRK5-hNRSN2- or pRK5-vector (control; 1×107 cells)
or MDA-MB-231 cells to analyze the role of NRSN2 in breast cancer
growth. The tumor volume of wild-type MDA-MB-231-injected mice at
day 27 was ~2,100 mm3. The difference in the tumor
volumes between the present study and the study by Shen et
al (27) may be due to the types
of mice used and the number of cells injected. The results from the
present study demonstrated that NRSN2 stimulated PI3K/AKT/mTOR and
NF-κB phosphorylation in MDA-MB-231 cells, which further promoted
breast cancer cells growth both in vitro and in vivo.
However, only 8 mice were used in this study to explore the
inhibitory effects of NRSN2. Further investigation using a larger
sample size would therefore be needed to determine other signaling
pathways associated with NRSN2. In addition, the present study did
not use ideal controls when investigating the effects of NF-κB
inhibitor or PI3K inhibitor on the proliferation, migration and
invasion of breast cancer cells, as an overexpression-only group
was not included. The effects of NF-κB inhibitor or PI3K inhibitor
on NRSN2 expression will also be further analyzed in the
future.
In conclusion, the results from the present study
demonstrated that NRSN2 was overexpressed in breast cancer tissues
and cells, and that it significantly promoted the proliferation and
aggressiveness of breast cancer by activating PI3K/AKT/mTOR and
NF-κB signaling pathways. These findings suggested that NRSN2 may
be considered as a potential therapeutic target for breast
cancer.
Acknowledgement
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FR and WZ performed all experiments. SL and HR
prepared for the experiments, analyzed and collected data. YG
designed the study and wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The animal study was approved by the Chinese
Association for Laboratory Animal Operations. The patient study was
approved by the Ethic Committee of Peking University (approval no.
PEK20150524). All patients provided written informed consent prior
to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeSantis CE, Fedewa SA, Goding Sauer A,
Kramer JL, Smith RA and Jemal A: Breast cancer statistics, 2015:
Convergence of incidence rates between black and white women. CA
Cancer J Clin. 66:31–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iranshahi N, Zafari P, Yari KH and
Alizadeh E: The most common genes involved in epigenetics
modifications among Iranian patients with breast cancer: A
systematic review. Cell Mol Biol (Noisy-le-grand). 62:116–122.
2016.PubMed/NCBI
|
|
3
|
Araki K and Ito Y: A review multigene
assays for clinical utility in breast cancer. Gan To Kagaku Ryoho.
43:1332–1340. 2016.(In Japanese). PubMed/NCBI
|
|
4
|
Lucius K and Trukova K: Integrative
therapies and cardiovascular disease in the breast cancer
population: A review, part 2. Integr Med (Encinitas). 14:33–40.
2015.PubMed/NCBI
|
|
5
|
Lucius K and Trukova K: Integrative
therapies and cardiovascular disease in the breast cancer
population: A review, part 1. Integr Med (Encinitas). 14:22–29.
2015.PubMed/NCBI
|
|
6
|
Leuteritz K, Weißflog G, Barthel Y,
Brähler E, Zwerenz R, Wiltink J and Beutel ME: Therapeutic alliance
and treatment outcome in psychodynamic psychotherapy of depressed
breast cancer patients: The same old story or different from other
populations? Breast Cancer. 24:765–773. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moreno Ayala MA, Gottardo MF, Asad AS,
Zuccato C, Nicola A, Seilicovich A and Candolfi M: Immunotherapy
for the treatment of breast cancer. Expert Opin Biol Ther.
17:797–812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allaire BT, Ekwueme DU, Poehler D, Thomas
CC, Guy GP Jr, Subramanian S and Trogdon JG: Breast cancer
treatment costs in younger, privately insured women. Breast Cancer
Res Treat. 164:429–436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Holzel D, Eckel R, Bauerfeind I, Baier B,
Beck T, Braun M, Ettl J, Hamann U, Kiechle M, Mahner S, et al:
Improved systemic treatment for early breast cancer improves cure
rates, modifies metastatic pattern and shortens post-metastatic
survival: 35-year results from the munich cancer registry. J Cancer
Res Clin Oncol. 143:1701–1712. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
An Y, Amr SS, Torres A, Weissman L,
Raffalli P, Cox G, Sheng X, Lip V, Bi W, Patel A, et al: SOX12 and
NRSN2 are candidate genes for 20p13 subtelomeric deletions
associated with developmental delay. Am J Med Genet B
Neuropsychiatr Genet. 162B:832–840. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang XY, Kuang JL, Yan CS, Tu XY, Zhao
JH, Cheng XS and Ye XQ: NRSN2 promotes non-small cell lung cancer
cell growth through PI3K/Akt/mTOR pathway. Int J Clin Exp Pathol.
8:2574–2581. 2015.PubMed/NCBI
|
|
12
|
Tang W, Ren A, Xiao H, Sun H and Li B:
Highly expressed NRSN2 is related to malignant phenotype in ovarian
cancer. Biomed Pharmacother. 85:248–255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Han L, Zhang J and Xia Q:
Down-regulated NRSN2 promotes cell proliferation and survival
through PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Dig Dis
Sci. 60:3011–3018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeong YJ, Choi Y, Shin JM, Cho HJ, Kang
JH, Park KK, Choe JY, Bae YS, Han SM, Kim CH, et al: Melittin
suppresses EGF-induced cell motility and invasion by inhibiting
PI3K/Akt/mTOR signaling pathway in breast cancer cells. Food Chem
Toxicol. 68:218–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuo YY, Jim WT, Su LC, Chung CJ, Lin CY,
Huo C, Tseng JC, Huang SH, Lai CJ, Chen BC, et al: Caffeic acid
phenethyl ester is a potential therapeutic agent for oral cancer.
Int J Mol Sci. 16:10748–10766. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fernandez-Pol S, Ma L, Ohgami RS and Arber
DA: Immunohistochemistry for p53 is a useful tool to identify cases
of acute myeloid leukemia with myelodysplasia-related changes that
are TP53 mutated, have complex karyotype, and have poor prognosis.
Mod Pathol. 30:382–392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gupta A, Shridhar K and Dhillon PK: A
review of breast cancer awareness among women in India: Cancer
literate or awareness deficit? Eur J Cancer. 51:2058–2066. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brodsky AS, Xiong J, Yang D, Schorl C,
Fenton MA, Graves TA, Sikov WM, Resnick MB and Wang Y:
Identification of stromal ColXalpha1 and tumor-infiltrating
lymphocytes as putative predictive markers of neoadjuvant therapy
in estrogen receptor-positive/HER2-positive breast cancer. BMC
Cancer. 16:2742016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan H, Zhang H, Yang W, Fu Y, Gu Y, Du M,
Cheng D and Shi H: Breast-specific gamma imaging with
Tc-99m-sestamibi in the diagnosis of breast cancer and its
semiquantitative index correlation with tumor biologic markers,
subtypes, and clinicopathologic characteristics. Nucl Med Commun.
37:792–799. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Adamczyk A, Niemiec J, Ambicka A,
Mucha-Małecka A, Ryś J, Mituś J, Wysocki WM, Cichocka A and
Jakubowicz J: Survival of breast cancer patients according to
changes in expression of selected markers between primary tumor and
lymph node metastases. Biomark Med. 10:219–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Araki K and Miyoshi Y: Mechanism of
resistance to endocrine therapy in breast cancer: The important
role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative
breast cancer. Breast Cancer. 25:392–401. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gutsche K, Randi EB, Blank V, Fink D,
Wenger RH, Leo C and Scholz CC: Intermittent hypoxia confers
pro-metastatic gene expression selectively through NF-κB in
inflammatory breast cancer cells. Free Radic Biol Med. 101:129–142.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guerrero-Zotano A, Mayer IA and Arteaga
CL: PI3K/AKT/mTOR: Role in breast cancer progression, drug
resistance, and treatment. Cancer Metastasis Rev. 35:515–524. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Z, Qian J, Li J and Zhu C: Knockdown of
lncRNA-HOTAIR downregulates the drug-resistance of breast cancer
cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp
Ther Med. 18:435–442. 2019.PubMed/NCBI
|
|
26
|
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X,
Lin L, Yao H, Su F, Li D, et al: A cytoplasmic NF-κB interacting
long noncoding RNA blocks IκB phosphorylation and suppresses breast
cancer metastasis. Cancer Cell. 27:370–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen Q, Cohen B, Zheng W, Rahbar R, Martin
B, Murakami K, Lamorte S, Thompson P, Berman H, Zúñiga-Pflücker JC,
et al: Notch shapes the innate immunophenotype in breast cancer.
Cancer Discov. 7:1320–1335. 2017. View Article : Google Scholar : PubMed/NCBI
|