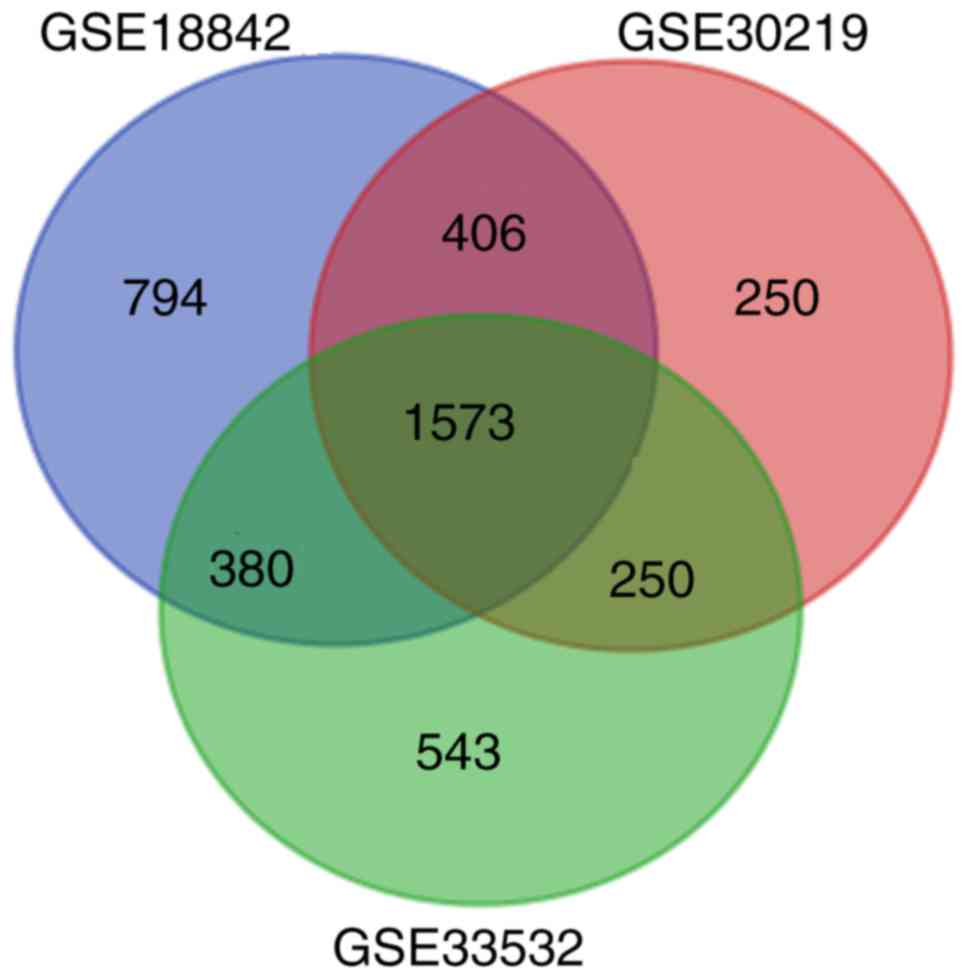
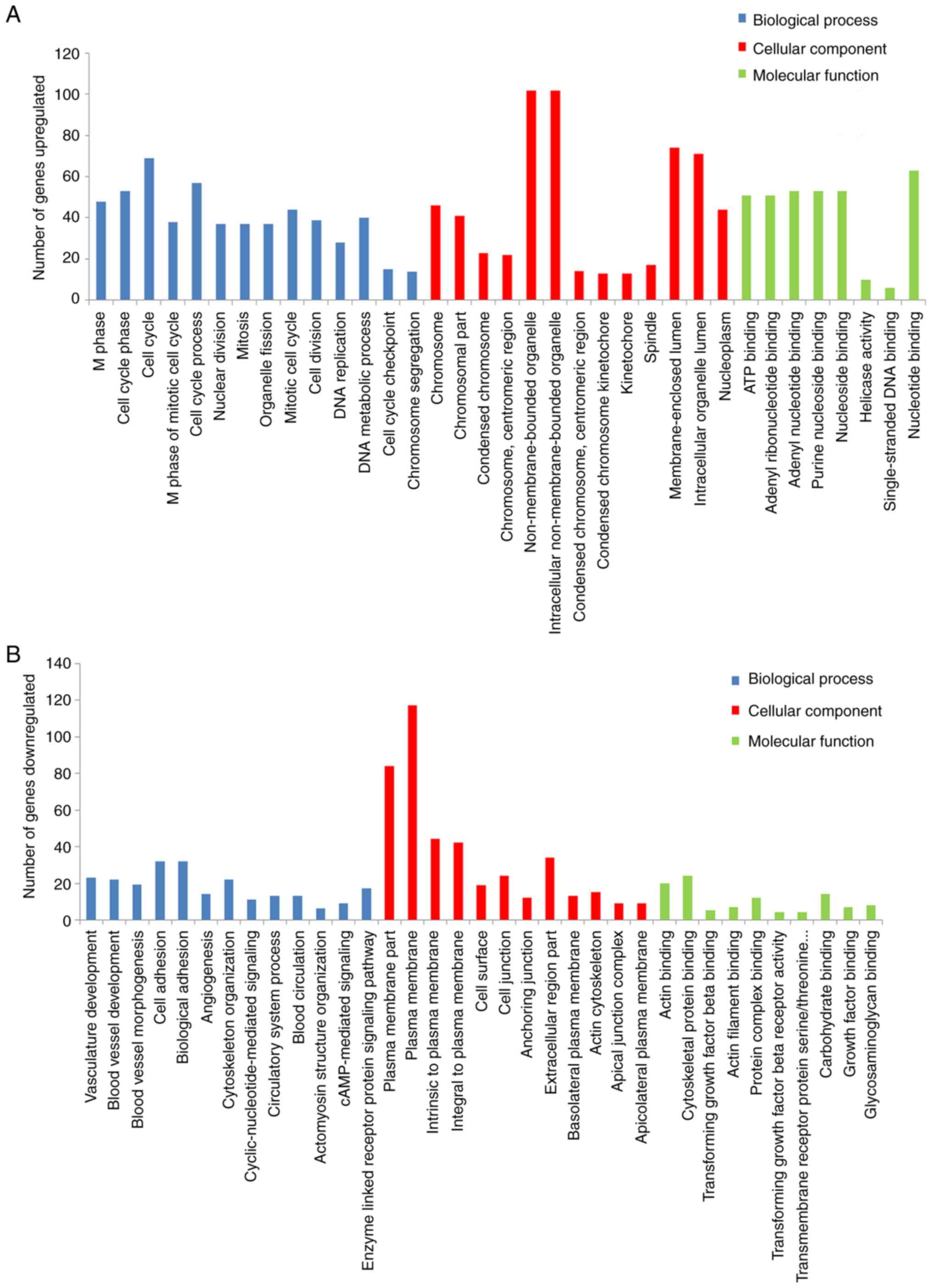
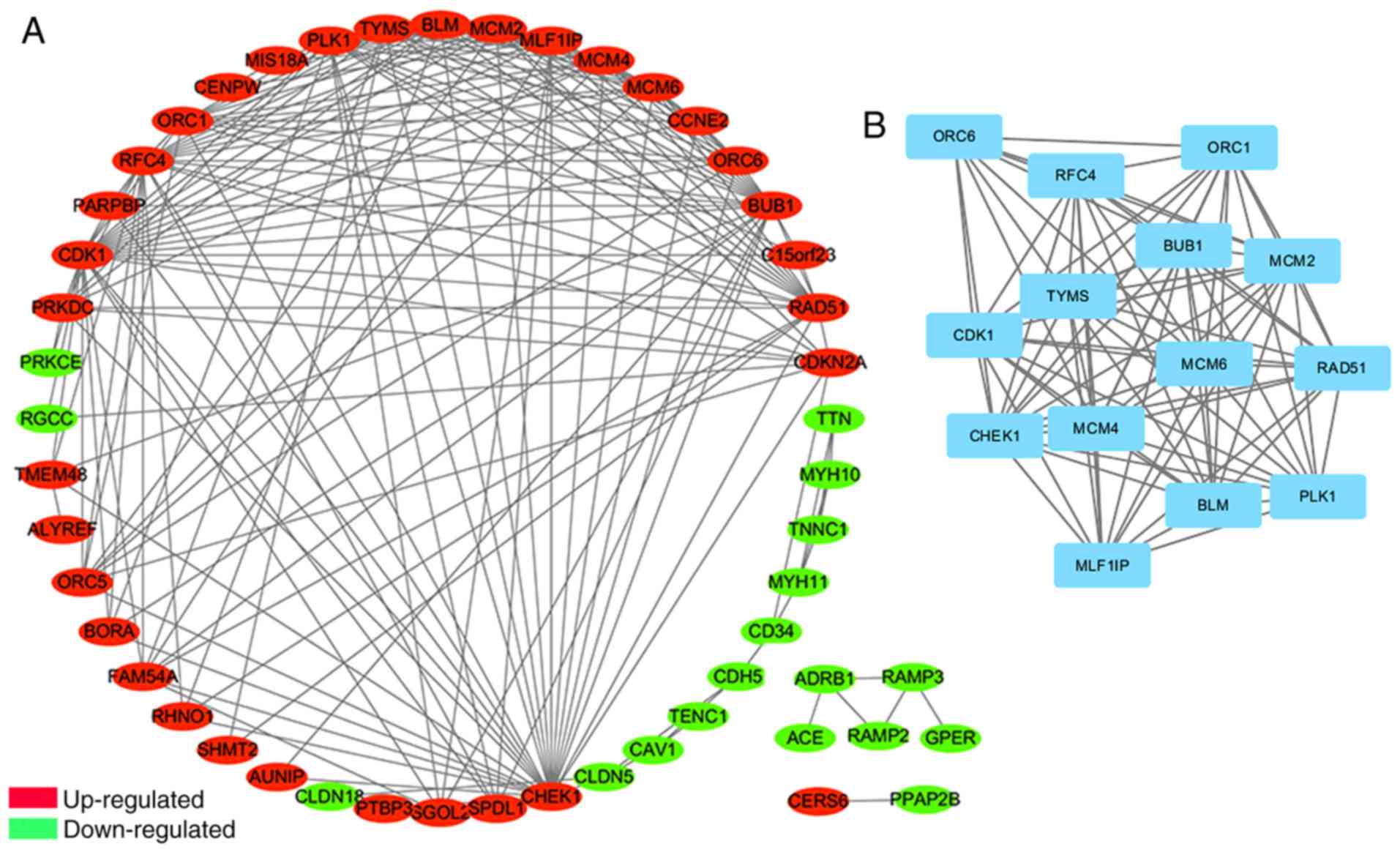
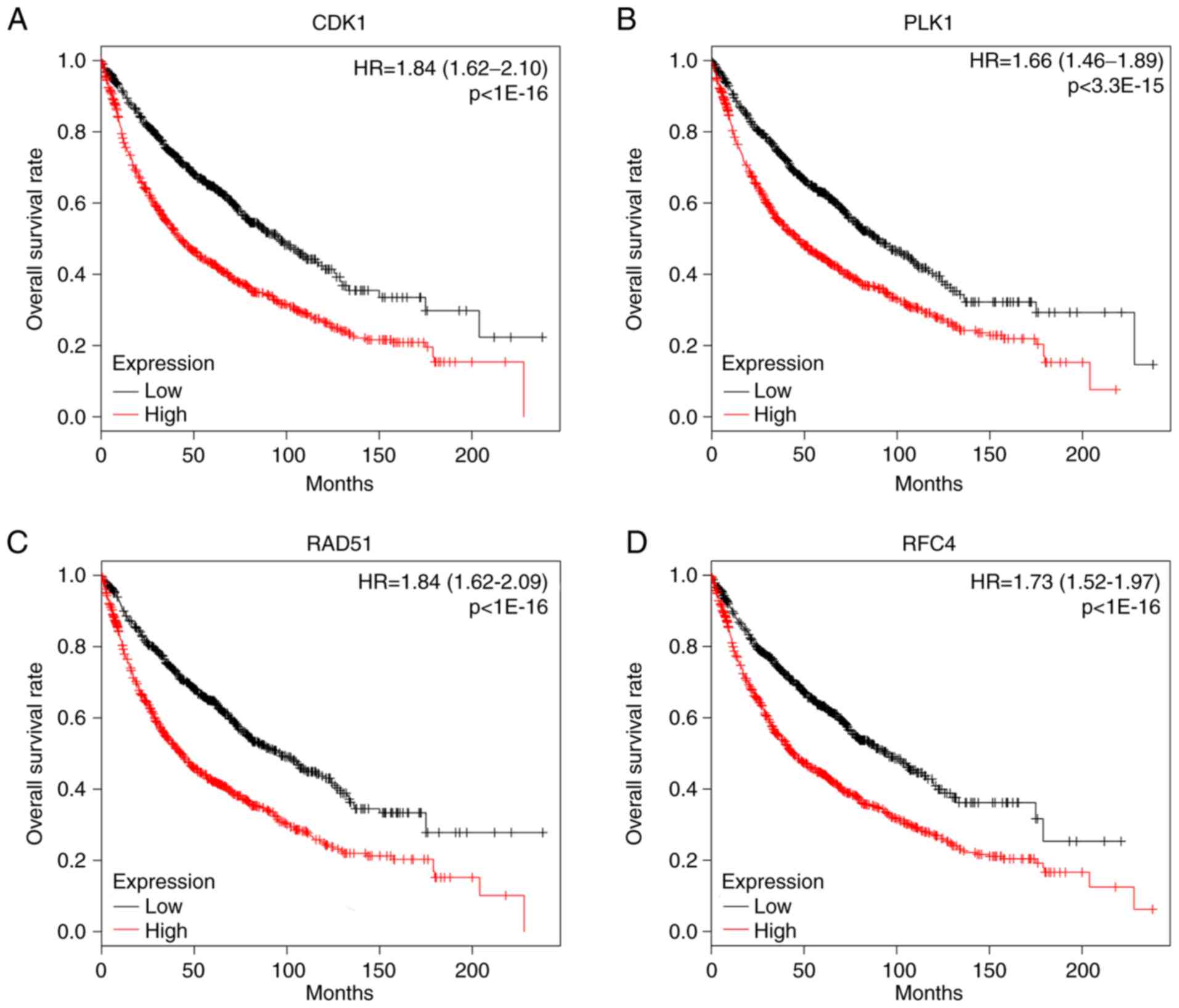
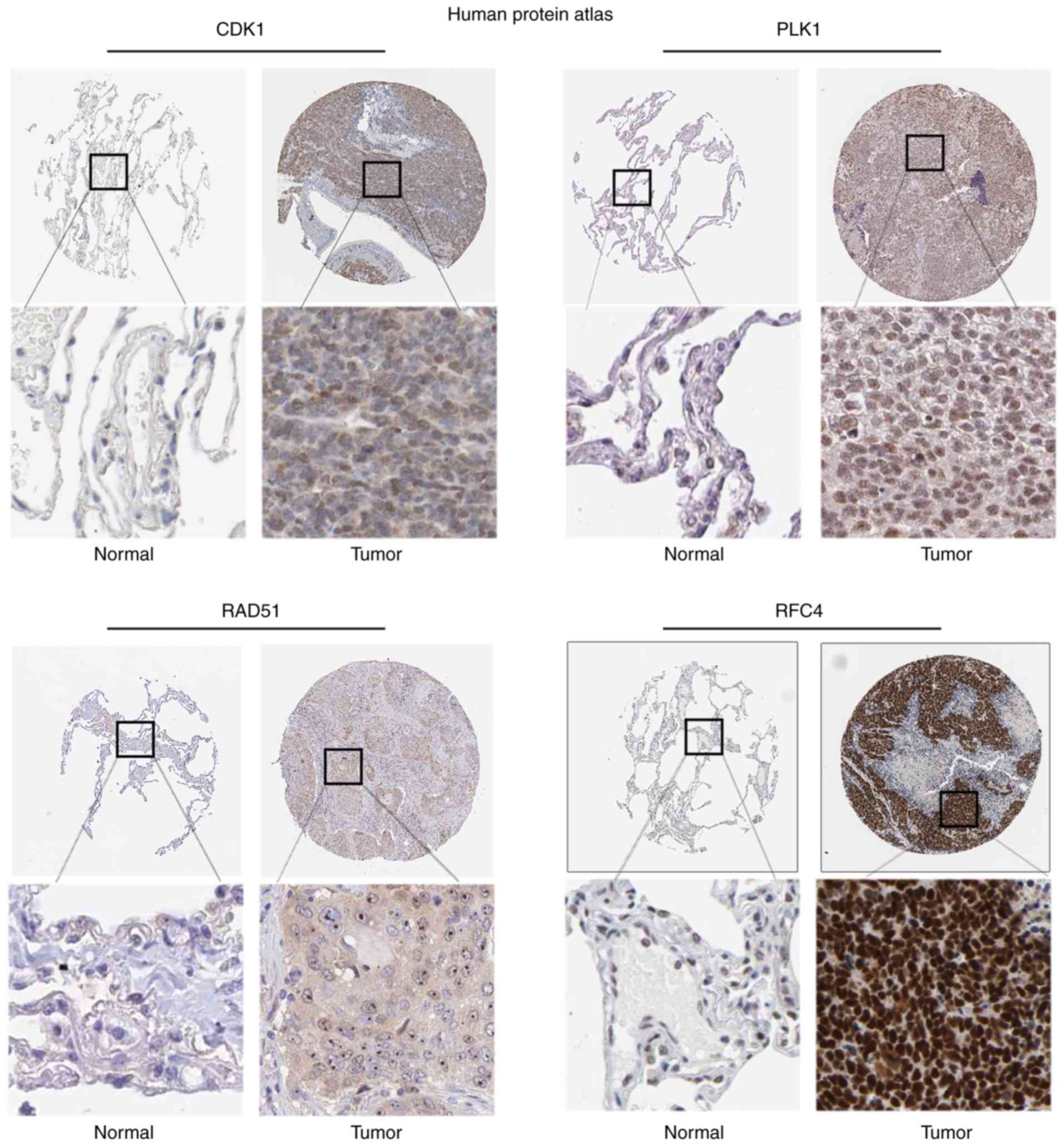
|
1
|
Ansari J, Shackelford RE and El-Osta H:
Epigenetics in non-small cell lung cancer: From basics to
therapeutics. Transl Lung Cancer Res. 5:155–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramshankar V and Krishnamurthy A: Lung
cancer detection by screening-presenting circulating miRNAs as a
promising next generation biomarker breakthrough. Asian Pac J
Cancer Prev. 14:2167–2172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Naruke T, Goya T, Tsuchiya R and Suemasu
K: Prognosis and survival in resected lung carcinoma based on the
new international staging system. J Thorac Cardiovasc Surg.
96:440–447. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ginsberg RJ and Rubinstein LV: Randomized
trial of lobectomy versus limited resection for T1 N0 non-small
cell lung cancer. Lung Cancer Study Group. Ann Thorac Surg.
60:615–622; discussion 622–623. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldstraw P, Crowley J, Chansky K, Giroux
DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V and Sobin L;
International Association for the Study of Lung Cancer
International Staging Committee; Participating Institutions, : The
IASLC Lung Cancer Staging Project: Proposals for the revision of
the TNM stage groupings in the forthcoming (seventh) edition of the
TNM Classification of malignant tumours. J Thorac Oncol. 2:706–714.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu X, Yang X, Zhang Z and Wang D:
Meta-analysis of serum tumor markers in lung cancer. Zhongguo Fei
Ai Za Zhi. 13:1136–1140, (In Chinese). PubMed/NCBI
|
|
7
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopaedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meister M, Belousov A, Xu EC, Schnabel P,
Warth A and Hoffman H: Intra-tumor heterogeneity of gene expression
profiles in early stage non-small cell lung cancer. J Bioinf Res
Stud. 1:12014.
|
|
13
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jakopovic M, Thomas A, Balasubramaniam S,
Schrump D, Giaccone G and Bates SE: Targeting the epigenome in lung
cancer: Expanding approaches to epigenetic therapy. Front Oncol.
3:2612013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Singhal S, Vachani A, Antin-Ozerkis D,
Kaiser LR and Albelda SM: Prognostic implications of cell cycle,
apoptosis, and angiogenesis biomarkers in non-small cell lung
cancer: A review. Clin Cancer Res. 11:3974–3986. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Voortman J, Checińska A and Giaccone AG:
The proteasomal and apoptotic phenotype determine bortezomib
sensitivity of non-small cell lung cancer cells. Mol Cancer.
6:732007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang Y, Ikezoe TT, Saito T, Kobayashi M,
Koeffler HP and Taguchi H: Proteasome inhibitor PS-341 induces
growth arrest and apoptosis of non-small cell lung cancer cells via
the JNK/c-Jun/AP-1 signaling. Cancer Sci. 95:176–180. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang CL, Yokomise H and Miyatake A:
Clinical significance of the p53 pathway and associated gene
therapy in non-small cell lung cancers. Future Oncol. 3:83–93.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kastan MB and Bartek J: Cell-cycle
checkpoints and cancer. Nature. 432:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Malumbres M and Barbacid M: Mammalian
cyclin-dependent kinases. Trends Biochem Sci. 30:630–641. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Q, Su L, Liu N, Zhang L, Xu W and
Fang H: Cyclin dependent kinase 1 inhibitors: A review of recent
progress. Curr Med Chem. 18:2025–2043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma PS, Sharma R and Tyagi R:
Inhibitors of cyclin dependent kinases: Useful targets for cancer
treatment. Curr Cancer Drug Targets. 8:53–75. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Malumbres M, Pevarello P, Barbacid M and
Bischoff JR: CDK inhibitors in cancer therapy: What is next? Trends
Pharmacol Sci. 29:16–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yim H: Current clinical trials with
polo-like kinase 1 inhibitors in solid tumors. Anticancer Drugs.
24:999–1006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baumann P and West SC: Role of the human
RAD51 protein in homologous recombination and double-stranded-break
repair. Trends Biochem Sci. 23:247–251. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Venkitaraman AR: Cancer susceptibility and
the functions of BRCA1 and BRCA2. Cell. 108:171–182. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Raderschall E, Stout K, Freier S, Suckow
V, Schweiger S and Haaf T: Elevated levels of Rad51 recombination
protein in tumor cells. Cancer Res. 62:219–225. 2002.PubMed/NCBI
|
|
32
|
Davies AA, Masson JY, McIlwraith MJ,
Stasiak AZ, Stasiak A, Venkitaraman AR and West SC: Role of BRCA2
in control of the RAD51 recombination and DNA repair protein. Mol
Cell. 7:273–282. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnson A, Yao NY, Bowman GD, Kuriyan J
and O'Donnell M: The replication factor C clamp loader requires
arginine finger sensors to drive DNA binding and proliferating cell
nuclear antigen loading. J Biol Chem. 281:35531–35543. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim HS and Brill SJ: Rfc4 interacts with
Rpa1 and is required for both DNA replication and DNA damage
checkpoints in Saccharomyces cerevisiae. Mol Cell Biol.
21:3725–3737. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiang J, Fang L, Luo Y, Yang Z, Liao Y,
Cui J, Huang M, Yang Z, Huang Y, Fan X, et al: Levels of human
replication factor C4, a clamp loader, correlate with tumor
progression and predict the prognosis for colorectal cancer. J
Transl Med. 12:3202014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin J, Marquardt G, Mullapudi N, Wang T,
Han W, Shi M, Keller S, Zhu C, Locker J and Spivack SD: Lung cancer
transcriptomes refined with laser capture microdissection. Am J
Pathol. 184:2868–2884. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hallou A, Jennings J and Kabla AJ: Tumour
heterogeneity promotes collective invasion and cancer metastatic
dissemination. R Soc Open Sci. 4:1610072017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fend F and Raffeld M: Laser capture
microdissection in pathology. J Clin Pathol. 53:666–672. 2000.
View Article : Google Scholar : PubMed/NCBI
|