Introduction
Osteosarcoma is a very common malignant tumor in
orthopedics and often occurs in children. Surgery is the most
common method for the treatment of osteosarcoma (1). Amputation is mainly to achieve
therapeutic purposes, which is one of the main reasons for the poor
life quality of patients with osteosarcoma (2). Therefore, early diagnosis and
conservative treatment of osteosarcoma is particularly important
for the clinic. At present, researchers world-wide are striving to
explore the possible serum markers in osteosarcoma (3–5), but
still have not made a significant breakthrough. Studies have
pointed out that the median survival time of patients with advanced
osteosarcoma is only ~20 months (6).
Therefore, in-depth understanding of osteosarcoma and the search
for potential diagnostic targets are currently under intense
investigations (7,8).
With the deepening of research, increasing number of
studies have pointed out that the occurrence of osteosarcoma may be
related to microRNAs (miRNAs) (9,10), which
are widely distributed in tissues and cells. They play an important
role in a series of biological activities such as cell
proliferation, apoptosis, metabolism and differentiation (11). As a non-coding single-stranded tiny
molecule RNA, miRNA is composed of 21–25 nucleotides. Through the
combination of incomplete complementary sequences with the 3′
untranslated region (3′UTR) sequence of target mRNA (12), miRNA can inhibit mRNA translation or
lead to direct degradation of miRNA level, indicating the influence
degree of post-transcriptional gene silencing (13). Previous studies have shown that a
variety of miRNA molecules are abnormally expressed in osteosarcoma
tissues and cells and are closely related to invasion,
proliferation, apoptosis and drug resistance of osteosarcoma cells
(14). Among them, miR-144 has been
used as a molecular diagnostic marker for colon cancer in clinic
(15). However, whether miR-144
affects the biological function of osteosarcoma cells remains
unknown and the mechanism is still unclear.
Therefore, this study investigated the influence
mechanism of miRNA-144 on the proliferation and apoptosis of
osteosarcoma cells, in order to provide a reference for early
diagnosis and targeted therapy of osteosarcoma.
Patients and methods
Baseline data
A total of 51 cases of osteosarcoma tissue samples
were collected in the department of orthopedic surgery, Xuzhou
Children's Hospital, Xuzhou Medical University from January 2014 to
February 2017. Further 48 cases of normal bone tissue (from tumor
lesions >5 cm) samples were collected. In the 51 cases of
osteosarcoma, there were 27 males and 24 females. In addition,
serum of 51 patients with osteosarcoma and serum of 48 normal
persons were collected. The average age was 20.63±4.63 years.
Inclusion and exclusion criteria
Inclusion criteria: No radiotherapy, chemotherapy or
immunotherapy before surgery; all patients were treated surgically
in the above hospital and osteosarcoma and normal bone tissue
samples were confirmed by pathology; postoperative tissue samples
were marked and saved immediately in liquid nitrogen; clinical data
is complete; the study was approved by the hospital ethical
committee and and the subjects signed a full informed consent.
Exclusion criteria: Patients with other malignant
tumors; patients with liver and kidney function disease;
hematological systemic disease; psychical disease; patients with
infection before admission; systemic autoimmune diseases.
Culture and transfection of cells
HFOB1.19, MG-63, U2-OS were inoculated into the
culture dish. RPMI-1640 medium containing 15% fetal bovine serum
and 1% penicillin-streptomycin was added to the cell culture
incubator at 37°C, 5% CO2. It was cultured under
constant temperature and saturated humidity. Liquid was changed,
routine trypsin digestion and passage were conducted. Then cells in
logarithmic growth phase were taken for follow-up experiments. The
cells were inoculated into a 6-well plate at a concentration of
lx105 cells/well and cultured until the degree of fusion
reached 60–70%. The cells were divided into 3 groups. miR-144
mimics (50 pmol/l) and 10 µl Lipofectamine 2000 were added to the
mimics group. miR-144 inhibitor (50 pmol/l) and 10 µl Lipofectamine
2000 were added as the inhibitor group. miR-144 negative control
(50 pmol/l) and 10 µl Lipofectamine 2000 were added to the NC group
(negative control fragment), respectively. Cells were transfected
by Lipofectamine 2000 kit. The procedure was carried out in strict
accordance with the kit instructions. The primers were transfected
into cells with the highest differential expression.
Detection method
qRT-PCR detection
Total RNA was extracted from tissues and cultured
cells by using a TRIzol extraction kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The concentration and purity of RNA were
detected by using a NanoDrop 2000 ultraviolet spectrophotometer.
RNA was reverse transcribed into cDNA according to Takara reverse
transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.) and
the synthesized cDNA was stored at −20°C for later use. Primers: U6
as internal reference, miR-144 upstream,
5′-GCTGGGATATCATCATATACTG-3′ and downstream,
5′-CGGACTAGTACATCATCTATACTG-3′; U6 upstream,
5′-CTCGCTTCGGCAGCACA-3′ and downstream, 5′-AACGCTTCACGAATTTGCGT-3′.
The primers were designed and synthesized by Shanghai GenePharma
Co., Ltd. The reaction was carried out on an ABI PRISM 7500
fluorescence ration PCR instrument (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The PCR amplification cyclic conditions
were: Pre-degeneration at 94°C for 30 sec, degeneration at 94°C for
5 sec, annealing and extension at 60°C for 30 sec, for a total of
40 cycles. Each sample was tested 3 times and the relative
expression of the gene was expressed by 2−ΔCT after
numeration.
WB detection
Total protein was extracted by RIPA lysis from each
group of cells after culture. Protein concentration was detected by
BCA. The protein concentration was adjusted to 4 µg/µl. Then
electrophoresis was used with 12% SDS-PAGE and transferred to PVDF
film after ionization, then dyed with ponceau working fluid,
soaking and washing in PBST for 5 min, and blocked with 5% skim
milk powder for 2 h. First antibody (1:1,000) was added and sealed
overnight at 4°C. The first antibody was removed by washing the
film, and horseradish peroxidase conjugated goat anti-rabbit second
antibody (1:5,000) was added, incubated at 37°C for 1 h, and rinsed
3 times with PBS for 5 min each time. Development was carried out
in a darkroom and the excess liquid on the film was dried with
filter paper. The ECL was illuminated and developed. The protein
bands were scanned and the gray values were analyzed in the
Quantity One. Relative expression level of its protein = the gray
value of the target protein band/the gray value of the β-actin
protein band.
Detection of cell proliferation-8
(CCK-8)
After 24 h of transfection, the cells were collected
and adjusted to 4×106 cells. The cells were diluted into
a cell suspension of 5×105 cells/ml and 100 µl of the
cells were collected and inoculated into a 96-well plate. After
cultured for 24, 48, 72 and 96 h, 10 µl CCK solution and 90 µl
basic medium were added to each well. Detection solution was
prepared according to CCK-8 solution: Culture solution, 1:9. 100 µl
of the test solution was added to each well, incubated for 2 h and
cultured at 37°C for 2 h. Then the OD value of each group was
measured under the absorbance of 570 nm by using enzyme-labelling
measuring instrument.
Detection of apoptosis by flow
cytometry
The cells were removed after transfection for 48 h
and digested by trypsin, and then washed twice with PBS after
digestion. Binding buffer (100 µl) was added to collocate a
suspension of 1×106 cells/ml. Annexin V-FITC and PI were
added and incubated at room temperature for 5 min away from light,
and then placed in flow cytometer tube. FACSCanto flow cytometry
(Becton-Dickinson) was used to complete the detection within 1 h.
The experiment was repeated three times. Annexin V single-staining
indicated early apoptosis of cells. Annexin V/PI double staining
indicated advanced apoptosis of cells.
Main instruments and reagents
UV-Spectrophotometer (NanoDrop 2000; Beijing Keyi
Xingye Technology Development Co., Ltd.), 7500 fluorescence
quantitative PCR instrument (ABI), flow cytometry (FACSCanto II;
American BD Company), RNA extraction kit (TRIzol and reverse
transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.),
CCK-8 kit, (C0037; Beyotime Biotechnology), RIPA reagent, BCA
protein kit, ECL luminescence kit, trypsin, Lipofectamine 2000
Transfection Reagent (89901, 23225, 32209, 90059, 11668030;
American Thermo Scientific), Bax, caspase-3, Bcl-2 HPR-labeled goat
anti-mouse IgG secondary antibody (AF820, AF835, AF810; R&D
Systems, Inc.), Annexin V/PI apoptosis detection kit (40302ES20;
Shanghai Yu Sheng Biotechnology Co., Ltd.).
Statistical methods
Statistical analysis was performed by using SPSS
20.0 (Beijing Easybio Technology Co., Ltd.). The data was drawn by
using GraphPad Prism 7 (Shenzhen Soft Network Co., Ltd.).
Measurement data were expressed as mean number ± standard deviation
(mean ± SD). Measurement data were compared by using independent
sample t-test among groups. Multiple time-point data were compared
by repeated measures analysis of variance, expressed as F. At
P<0.05, ROC was used to plot the diagnostic value of miR-144 in
patients with osteosarcoma. Pearson's test analyzed the correlation
between miR-144 and serum in the tissue. P<0.05 indicates a
statistically significant difference.
Results
Expression of miR-144 in osteosarcoma
tissues and normal bone tissues in osteosarcoma and normal human
serum
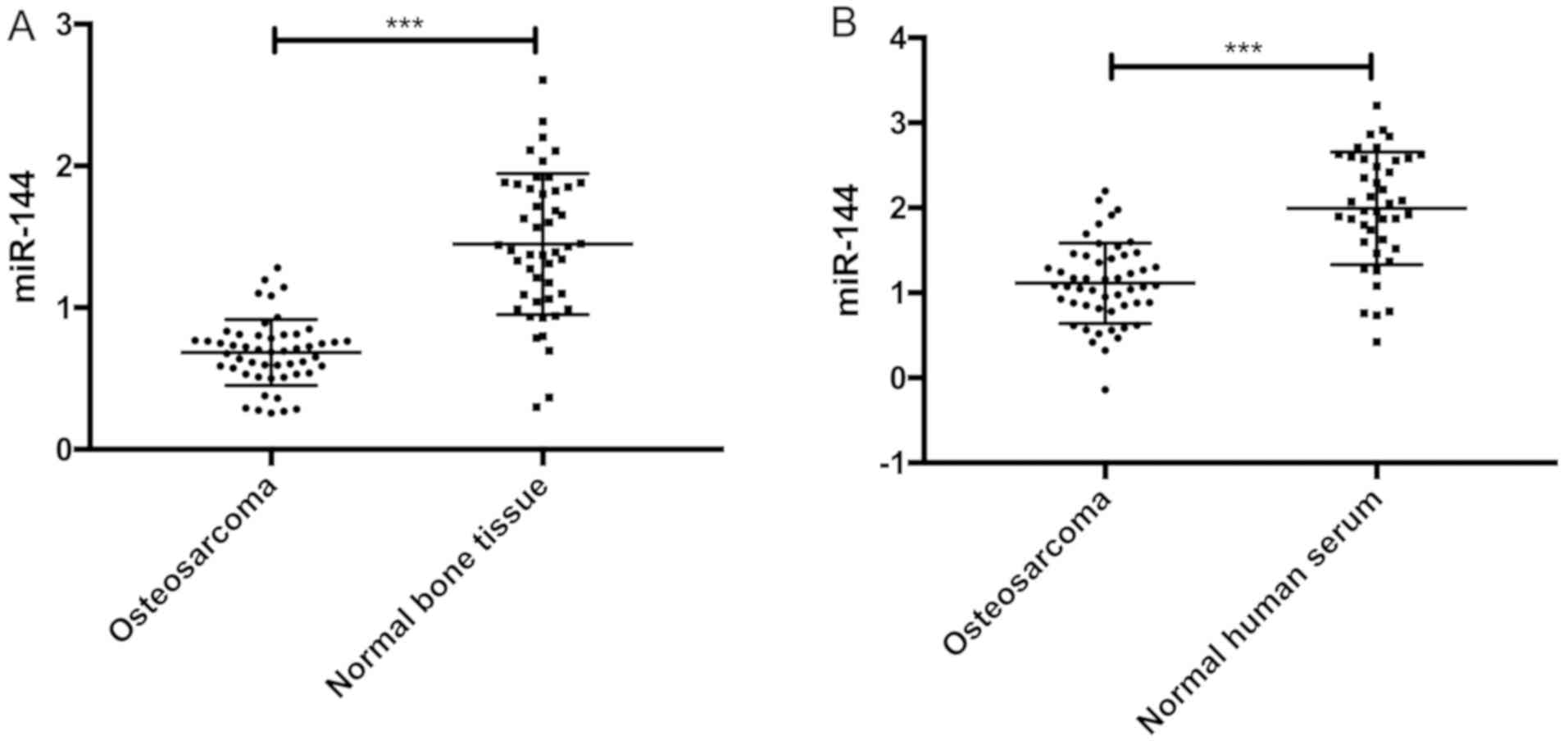
The relative expression of miR-144 in osteosarcoma
tissues and normal bone tissues were 0.763±0.214 and 1.546±0.483,
respectively. The relative expression of miR-144 in osteosarcoma
tissues was significantly lower than that in normal bone tissue
(t=10.530, P<0.001). The relative expression of miR-144 in
osteosarcoma and normal human serum were 1.163±0.514 and
1.974±0.612, respectively. The relative expression of miR-144 in
osteosarcoma serum was significantly lower than that in normal
human serum (t=6.947, P<0.001) (Fig.
1).
Relationship between miR-144 in
osteosarcoma tissues and clinicopathological features of
patients
There was no significant difference in the
expression of miR-144 in osteosarcoma tissues in terms of sex, age,
grade of malignancy, pathological types, or degree of pathological
differentiation (P>0.05) (Table
I).
 | Table I.Relationship between miR-144 in
osteosarcoma tissues and clinicopathological features of
patients. |
Table I.
Relationship between miR-144 in
osteosarcoma tissues and clinicopathological features of
patients.
| Category | n (51) | miR-144 | F/t-value | P-value |
|---|
| Sex |
|
| 1.759 | 0.085 |
| Male | 27 | 0.672±0.194 |
|
|
|
Female | 24 | 0.763±0.173 |
|
|
| Age (years) |
|
| 0.889 | 0.378 |
| ≤15 | 26 | 0.815±0.139 |
|
|
|
>15 | 25 | 0.778±0.158 |
|
|
| Grade of
malignancy |
|
| 1.464 | 0.242 |
| Grade
1 | 13 | 0.724±0.139 |
|
|
| Grade
2 | 20 | 0.703±0.143 |
|
|
| Grade
3 | 18 | 0.636±0.156 |
|
|
| Pathological
types |
|
| 2.270 | 0.093 |
|
Osteoblastic type | 12 | 0.633±0.158 |
|
|
|
Chondroblastic type | 13 | 0.648±0.162 |
|
|
|
Fibroblastic type | 16 | 0.673±0.160 |
|
|
| Mixed
type | 10 | 0.795±0.156 |
|
|
| Degree of
pathological differentiation |
|
| 0.931 | 0.401 |
| Well
differentiated | 12 | 0.825±0.196 |
|
|
|
Moderately differentiated | 25 | 0.767±0.185 |
|
|
| Poorly
differentiated | 14 | 0.741±0.178 |
|
|
Correlation of miR-144 expression
between serum and tumor tissues
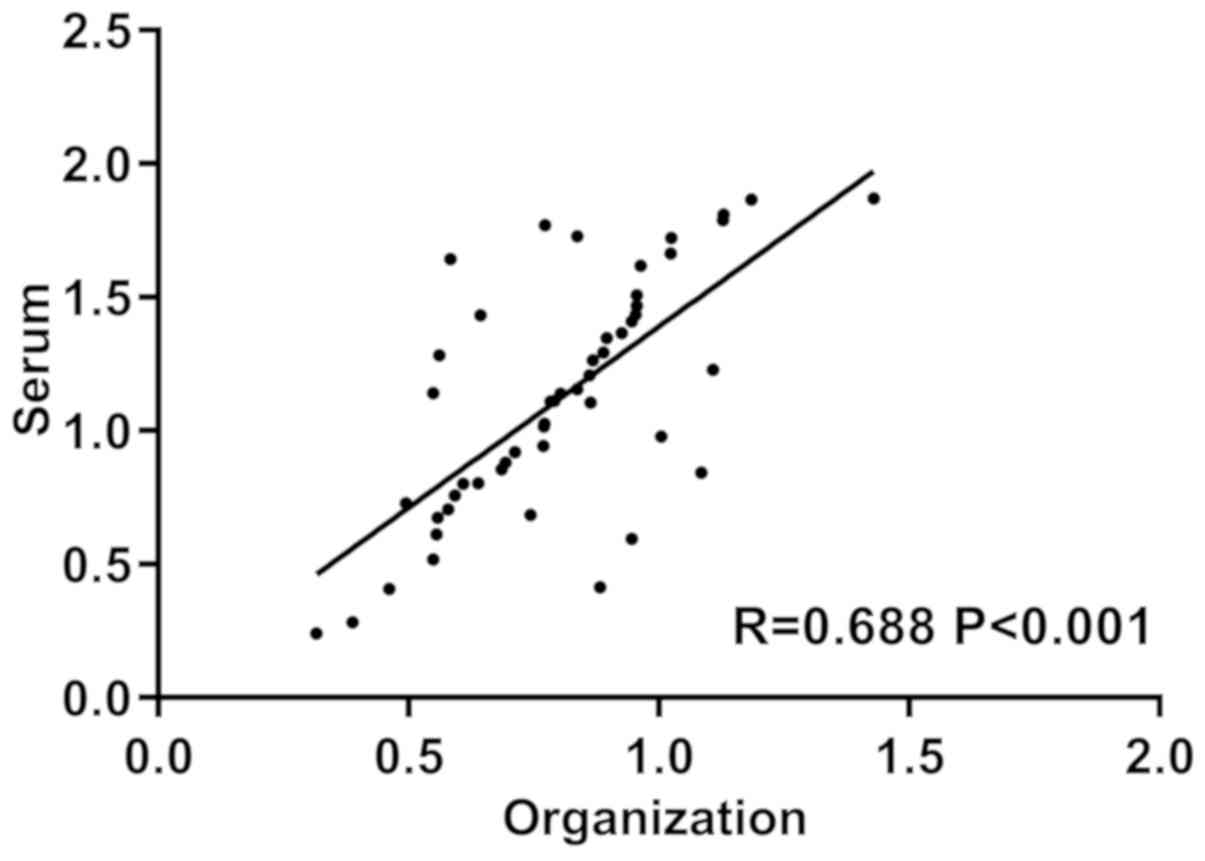
We analyzed the relationship between serum and
cancer tissues and miR-144 expression by Pearsons correlation and
found that the serum of patients was positively correlated with the
cancer tissue miR-144 expression (r=0.688, P<0.001) (Fig. 2).
Diagnostic value of miR-144 in
patients with osteosarcoma
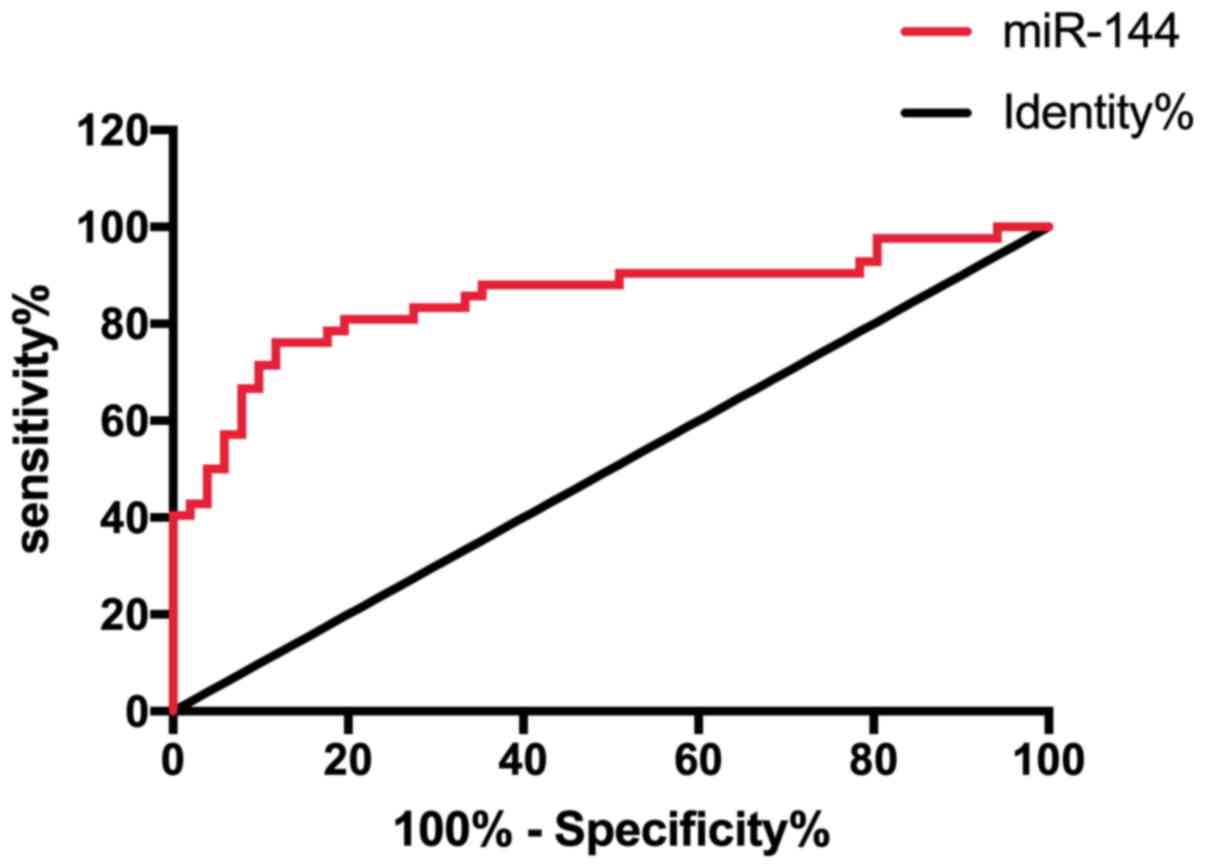
ROC curve was drawn to analyze the diagnostic value
of miR-144 in patients with osteosarcoma through expression of
miR-144 in patients with osteosarcoma and normal population. It was
found that the area under the miR-144 curve was 0.852, 95% CI,
0.768–0.936 (Fig. 3 and Table II).
 | Table II.ROC curve data. |
Table II.
ROC curve data.
| Index | AUC | 95% CI | Specificity | Sensitivity | Youden index | Cut-off |
|---|
| VEGF | 0.852 | 0.768–0.936 | 88.24% | 73.81% | 62.05% | >1.612 |
Expression of miR-144 in osteosarcoma
cell line
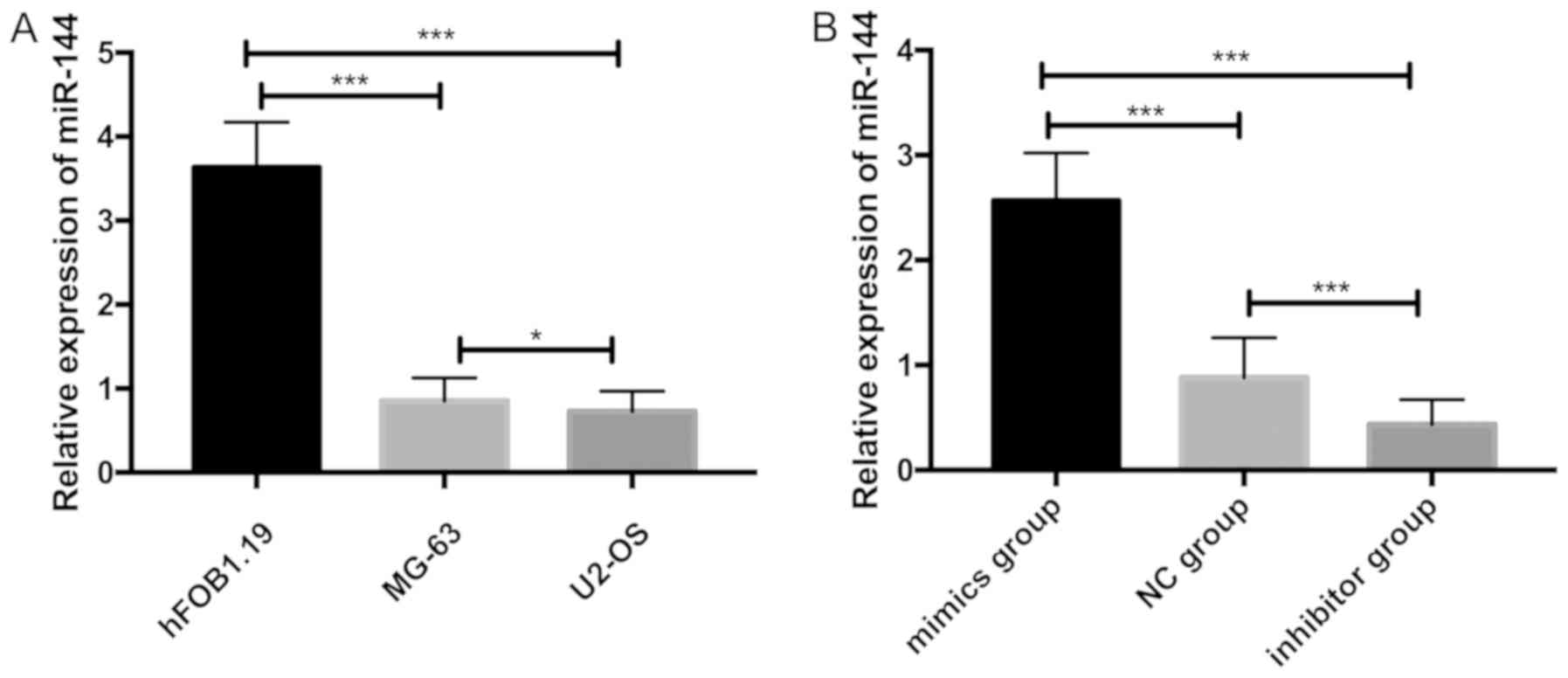
The expression of miR-144 in each group was
detected. It was found that the relative expression levels of
miR-144 in hFOB1.19, MG-63 and U2-OS were 3.637±0.534, 0.852±0.274,
0.726±0.242, respectively. The relative expression of miR-144 in
MG-63 and U2-OS cells was significantly lower than that in hFOB1.19
cells (P<0.001), while the relative expression of miR-144 in
U2-OS cells was significantly lower than that in MG-63 cells
(P<0.05). U2-OS cells were transfected with miR-144-mimics. The
results showed that the relative expression of miR-144 in U2-OS
cells of mimics group was significantly higher than that of
inhibitor group and NG group (P<0.001) and the relative
expression of miR-144 in U2-OS cells of inhibitor group was
significantly lower than that in mimics group and NG group
(P<0.001) (Fig. 4).
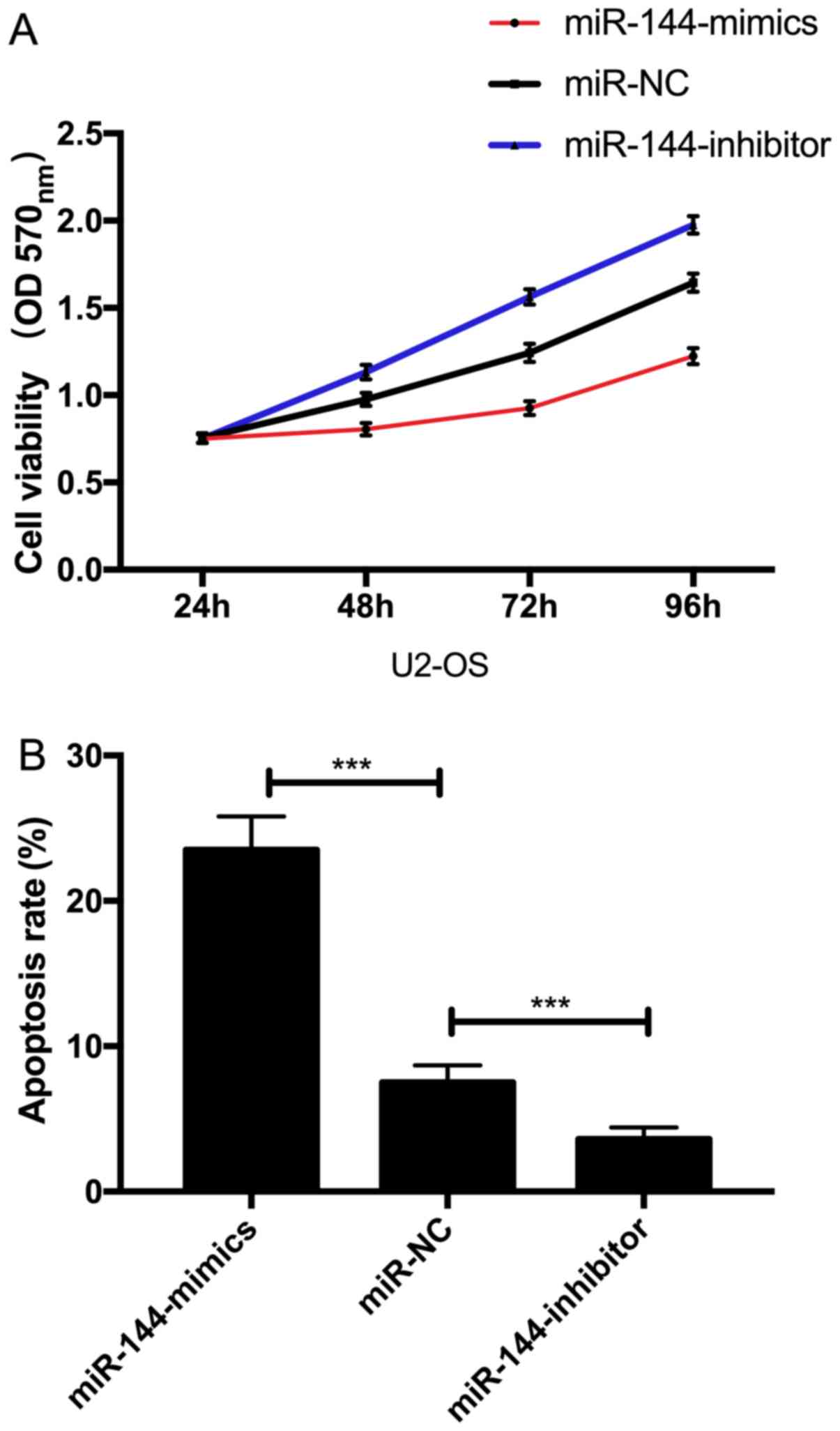
Effect of miR-144 on proliferation and
apoptosis of U2-OS cells
The proliferation and apoptosis of cells after
transfection was detected. The results showed that the
proliferation ability of U2-OS cells transfected with
miR-144-mimics was significantly inhibited and the apoptosis rate
was significantly increased (P<0.05) (Fig. 5).
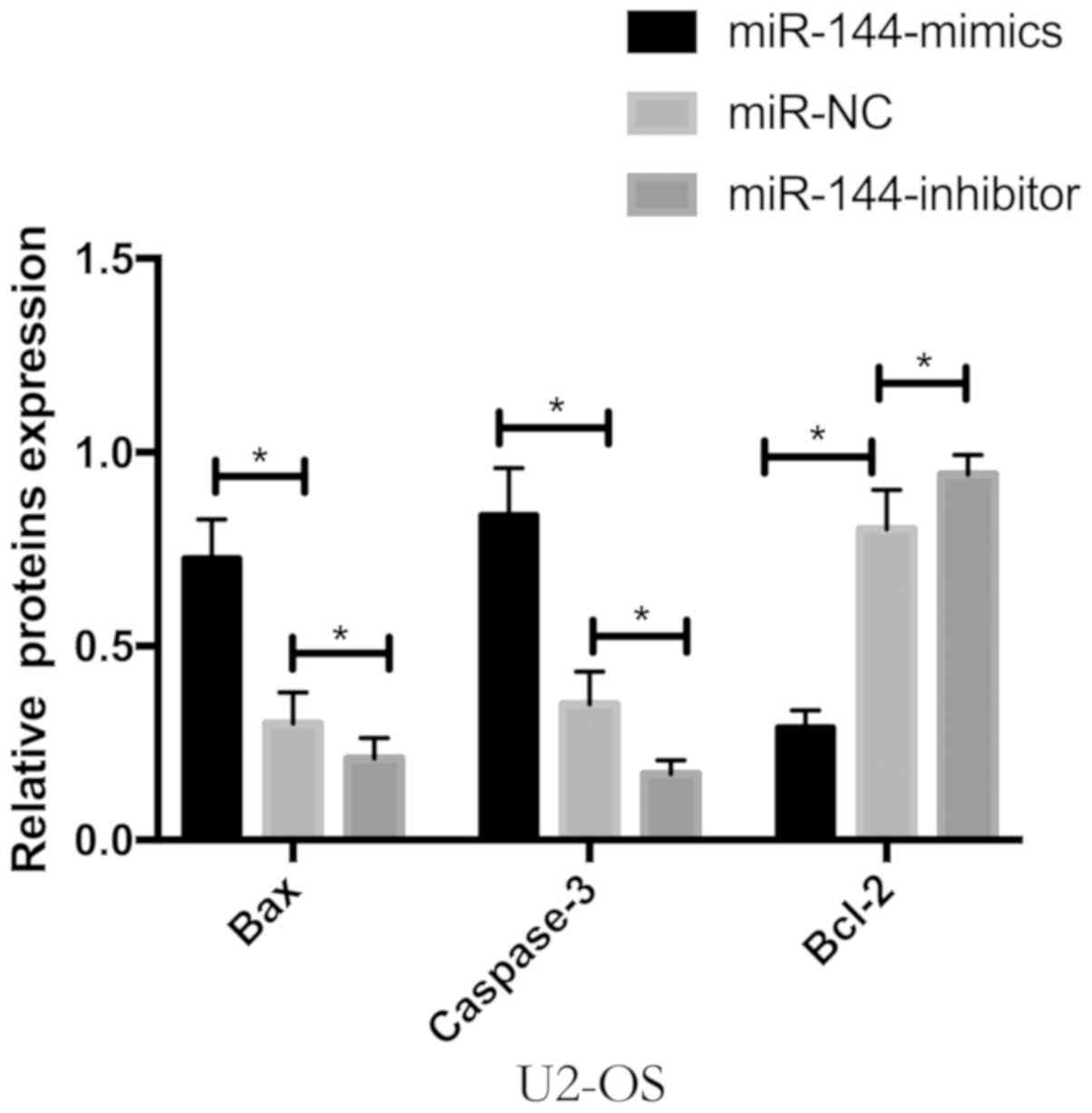
Detection of apoptosis protein by
WB
WB detection was performed. It was found that the
expression of Bcl-2 protein was significantly decreased and the
expression of Bax and caspase-3 protein was significantly increased
by the expression of Bax, caspase-3 and Bcl-2 proteins in U2-OS
cells transfected with miR-144 (P<0.05) (Fig. 6).
Discussion
Osteosarcoma, the most common of primary malignant
bone tumors, originates from formation of mesenchymal cells by
normal bone and is characterized by the direct formation of
immature bone or osteoid tissue by tumor cells (16). Classic osteosarcoma is a rare, highly
malignant tumor with an estimated incidence of 3 cases/million
population/year. Osteosarcoma mainly occurs in long bones and
rarely occurs in soft tissues. A few years ago, all patients with
osteosarcoma were treated with surgery, but the cure rate was
<10% (17). Almost all patients
died within one year after diagnosis and the disability rate and
mortality were high (18). With the
gradual development of molecular biology, miRNA has become a
molecular biology indicator for the treatment strategy and
prognosis of osteosarcoma (19). The
occurrence and deterioration mechanisms of osteosarcoma has not
been elucidated, but the proliferation and apoptosis of tumor cells
is one of the mechanisms of osteosarcoma (20). Therefore, it is of great significance
to explore the molecular biological markers closely related to the
occurrence and deterioration of osteosarcoma for the diagnosis and
molecular therapy of osteosarcoma.
miR-144 plays an important role in a variety of
diseases (21). Studies by Zhao
et al (22) showed that
downregulation of miR-144 is associated with osteosarcoma cell
growth and invasion by regulating TAGLA expression. Studies by Guo
et al (23) showed that the
downregulation of miR-144 increase the proliferation of bladder
cancer cells by targeting EZH2 and regulating Wnt signaling
conduction. The results confirmed that the expression levels of
miR-144 were downregulated in bladder cancer, which was similar to
the results of our study, indicating miR-144 is downregulated in
cancer tissues. In this study, the expression of miR-144 in
osteosarcoma was significantly lower than that in normal bone
tissue and the expression of miR-144 in serum of osteosarcoma
patients was also significantly lower than normal level, thus, the
expression trend in serum was consistent with that in tissues. Cao
et al (24) showed that,
miR-144 was significantly reduced in hepatocellular carcinoma
tissue and cell lines. Forced overexpression of miR-144
significantly reduced cell proliferation and increased apoptosis of
cells, which was similar to our results. It indicated that miR-144
may also play an important role in the occurrence and deterioration
of osteosarcoma. Subsequently, we conducted correlation analysis
based on the expression of serum of patients and the expression of
miR-144 in the cancer tissues, and the results showed that the
serum of patients was positively correlated with the expression of
miR-144 in the cancer tissues. The ROC curve was drawn to analyze
the diagnostic value of miR-144 in patients with osteosarcoma
through the expression of miR-144 in patients with osteosarcoma and
normal population. It was found that the area under the miR-144
curve was 0.852, 95% CI, 0.768–0.936. This indicated that miR-144
can be used as a diagnostic indicator for patients with
osteosarcoma. Then the expression of miR-144 was examined in
osteosarcoma cell lines and it was found that the relative
expression of miR-144 was the lowest in U2-OS. By further
transfection, the relative expression of miR-144 in U2-OS cells of
mimics group was significantly higher than that in inhibitor and NG
groups. The proliferation ability of U2-OS cells transfected with
miR-144-mimics was significantly inhibited and the apoptosis rate
was significantly increased. In the study of Wang et al
(25), miR-144 was also
significantly downregulated in osteosarcoma cell lines and clinical
specimens. The decrease of miR-144 expression in osteosarcoma was
closely related to disease progression and metastasis. It indicated
that miR-144 can be used as a potential target for the treatment of
osteosarcoma. Overexpression of miR-144 can inhibit cell
proliferation, promote apoptosis of cells and it has been mutually
verified with our previous experiments. Finally, WB detection was
performed. The detection results showed that the pro-apoptotic
protein Bax and caspase-3 were elevated and the anti-apoptotic
protein Bcl-2 was decreased by the expression of Bax, caspase-3 and
Bcl-2 proteins in U2-OS cells transfected with miR-144. Therefore,
there is a targeted regulation relationship between miR-144 and
Bax, and caspase-3 and Bcl-2.
In this study, we preliminarily proved that miR-144
can promote apoptosis by regulating Bax, caspase-3 and Bcl-2
proteins. However, further study is required.
In conclusion, miR-144 may be involved in the
occurrence and deterioration of osteosarcoma. In future, it is
expected to become a potential indicator for the diagnosis and
treatment of osteosarcoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, ZL and HQ conceived and designed the study, and
drafted the manuscript. XZ, WJ, XC, QG and DL collected, analyzed
and interpreted the experimental data. ZL revised the manuscript
for important intellectual content. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Xuzhou Children's Hospital, Xuzhou Medical University (Xuzhou,
China). Signed informed consents were obtained from the patients
and/or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ottaviani G and Jaffe N: Epidemiology of
osteosarcoma. Pediatric and Adolescent Osteosarcoma. Jaffe N,
Bruland SO and Bielack S: Springer; Boston, MA: pp. 3–13. 2010
|
|
2
|
Anderson ME: Recent advances in
osteosarcoma survival. Orthop Clin North Am. 47:283–292. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment methods and successful
cooperative approaches. J Clin Oncol. 33:30292015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lindsey BA, Markel JE and Kleinerman ES:
Osteosarcoma overview. Rheumatol Ther. 4:25–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harrison DJ, Geller DS, Gill JD, Lewis VO
and Gorlick R: Existing and future treatments for osteosarcoma.
Expert Rev Anticancer Ther. 18:39–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duan Z, Gao Y, Shen J, Choy E, Cote G,
Harmon D, Bernstein K, Lozano-Calderon S, Mankin H and Hornicek FJ:
miR-15b modulates multidrug resistance in human osteosarcoma in
vitro and in vivo. Mol Oncol. 11:151–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sampson VB, Yoo S, Kumar A, Vetter NS and
Kolb EA: Potential targets for microRNAs and osteosarcoma. Front
Pediatr. 3:692015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulda V, Svaton M, Mukensnabl P, Hrda K,
Dvorak P, Houdek Z, Houfkova K, Vrzakova R, Babuska V, Pesek M, et
al: Predictive relevance of miR-34a, miR-224 and miR-342 in
patients with advanced squamous cell carcinoma of the lung
undergoing palliative chemotherapy. Oncol Lett. 15:592–599.
2018.PubMed/NCBI
|
|
10
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peng L, Chen Y, Ma N and Chen X: NARRMDA:
Negative-aware and rating-based recommendation algorithm for
miRNA-disease association prediction. Mol Biosyst. 13:2650–2659.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grosswendt S and Rajewsky N: Essentials of
miRNA-dependent control of mRNA translation and decay, miRNA
targeting principles, and methods for target identification.
Essentials of Noncoding RNA in Neuroscience: Ontogenetics,
Plasticity of the Vertebrate Brain. Academic Press; London: pp.
19–38. 2017, View Article : Google Scholar
|
|
13
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44:D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang
S and Liu X: Long noncoding RNA MALAT1 as a potential therapeutic
target in osteosarcoma. J Orthop Res. 34:932–941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sheng S, Xie L, Wu Y, Ding M, Zhang T and
Wang X: miR-144 inhibits growth and metastasis in colon cancer by
downregulating SMAD4. Biosci Rep. 39:BSR201818952019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brown HK, Tellez-Gabriel M and Heymann D:
Cancer stem cells in osteosarcoma. Cancer Lett. 386:189–195. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Picci P: Osteosarcoma (osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rickel K, Fang F and Tao J: Molecular
genetics of osteosarcoma. Bone. 102:69–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Z, Yu X and Shen J: Long non-coding
RNAs: Emerging players in osteosarcoma. Tumour Biol. 37:2811–2816.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C and Lin J: Long noncoding RNA
ZEB1-AS1 activates ZEB1 as an oncogene of osteosarcoma by
epigenetic inheritance. Am J Transl Res. 8:40952016.PubMed/NCBI
|
|
21
|
Chen S, Li P, Li J, Wang Y, Du Y, Chen X,
Zang W, Wang H, Chu H, Zhao G, et al: miR-144 inhibits lung cancer
cell proliferation and induces apoptosis and autophagy by targeting
TIGAR. Cell Physiol Biochem. 35:997–1007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao M, Huang J, Gui K, Xiong M, Cai G, Xu
J, Wang K, Liu D, Zhang X and Yin W: The downregulation of miR-144
is associated with the growth and invasion of osteosarcoma cells
through the regulation of TAGLN expression. Int J Mol Med.
34:1565–1572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang
Z, Qiu F and Lin J: miR-144 downregulation increases bladder cancer
cell proliferation by targeting EZH2 and regulating Wnt signaling.
FEBS J. 280:4531–4538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao T, Li H, Hu Y, Ma D and Cai X: miR-144
suppresses the proliferation and metastasis of hepatocellular
carcinoma by targeting E2F3. Tumour Biol. 35:10759–10764. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang W, Zhou X and Wei M: MicroRNA-144
suppresses osteosarcoma growth and metastasis by targeting ROCK1
and ROCK2. Oncotarget. 6:10297–10308. 2015.PubMed/NCBI
|