Introduction
Hepatocellular carcinoma (HCC) is a heterogeneous
malignant tumor that ranks as the fifth most common primary
malignancy and the third leading cause of cancer-associated
mortality worldwide (1). Several
studies, including studies in North America, Europe, Asia and
Australia, have shown that the prognosis is poor in patients with
HCC (2–4). Despite recent advances in treatment
methods, such as surgical resection and liver transplantation,
curative treatment for advanced tumor stages and patients with
metastasis is often not feasible. As a standard therapeutic option
for intermediate-stage or non-resectable HCC, transarterial
chemoembolization (TACE) has been shown to have benefits (5). According to the Barcelona Clinic Liver
Cancer (BCLC) staging system, TACE has been identified as an
alternative or combination therapy in patients with early or
advanced HCC (6). In patients with
BCLC stage A, TACE combined with radiofrequency ablation (RFA)
exerts better local tumor control compared with that achieved by
RFA alone. For patients with BCLC stage C, sorafenib plus TACE
significantly delays tumor progression and provides improved
survival (7).
Cytokines are a broad and loose category of small
proteins that include chemokines, interferons, interleukins,
lymphokines and tumor necrosis factors (TNFs), and are produced and
released by different cells in the liver; cytokines play an
important role in inflammation and tumor progression (8). As such, TNF-α acts as a master switch
in establishing an intricate link between proinflammation and
disease progression in patients with glioblastoma (9). Through a granulocyte-macrophage
colony-stimulating factor- and interleukin (IL)-5-dependent
mechanism, obesity-mediated low-grade inflammation increases breast
cancer metastasis to the lung (10).
IL-10 is associated with numerous malignancies, including non-small
cell lung cancer (NSCLC), diffuse large B-cell lymphoma, and
cervical and oral cancer (11–15).
Moreover, Zhang et al (16)
identified a hypoxia-inducible factor (HIF)-1α/IL-1β signaling loop
in the tumor microenvironment that results in liver cancer cell
epithelial-mesenchymal transformation (EMT) and metastasis.
Chemokine ligand 5 (CCL5) plays an important role in the process of
chronic liver inflammation, which leads to HCC development by
reducing immune cell infiltration, such as CD4+ and
CD8+ T cells (17).
To date, little is known about the detailed changes
that occur in cytokine profiles after TACE. Due to its rarity, the
majority of previous studies have focused on individual cytokines,
and prior to the present study, few studies have explored changes
in serum cytokines, signaling pathways and disease prognosis based
on microarray data (18–22). The general focus of the present study
was to identify differences in the cytokine expression profiles of
patients with HCC undergoing TACE with a specific interest in
identifying significantly changed biological processes and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathways. The specific
aims of the present study were to investigate the associations
among time-dependent changes in cytokine panels and immune
reactions, and to evaluate the circulating levels of cytokines as
prognostic markers.
Materials and methods
Patients
A total of 60 patients admitted to the Zhongshan
Hospital (Shanghai, China) who received TACE therapy at the Liver
Cancer Institute, and 10 healthy controls (≥18 years old; 5 males
and 5 females), were prospectively recruited between January 2013
and December 2014 and enrolled in the present study (the patient
characteristics are shown in Table
I). All patients possessed a diagnosis of HCC according to
histological evidence or elevated α-fetoprotein (AFP) levels
(>200 ng/ml) with positive radiological findings using CT/MRI.
The primary inclusion criteria were as follows: i) Hepatitis B
virus as the cause of HCC; ii) patients with Child-Pugh class A or
B who had a non-resectable tumor (23); iii) an age ≥18 years; iv) definite
BCLC staging (6); and v) clinical
and laboratory data, including biochemical parameters,
hematological parameters, such as albumin, AFP, alanine
transaminase and bilirubin, and tumor sizes, which were recorded at
baseline and post-TACE. All chemoembolization procedures were
conducted via the right femoral artery and all patients undergoing
treatment via TACE were examined by computed tomography or magnetic
resonance imaging to evaluate the curative effect based on RECIST
1.1 and mRECIST (24), designated as
complete response (CR), partial response (PR), stable disease (SD),
or progressive disease (PD). CR and PR were further summarized into
objective response (OR). Patients were excluded if there was no
clear survival time. A total of 10 patients (≥18 years old; 5 males
and 5 females) were randomly selected from the 60 patients with HCC
to compare their serum cytokine levels with those of healthy
controls, with the samples collected at the pre-intervention time
point. Informed written consent was provided by each patient, and
the present study was approved by the Ethics Committees of
Zhongshan Hospital. The study protocol was conducted in accordance
with the ethical standards specified in the Declaration of
Helsinki.
 | Table I.Characteristics of 60 patients with
hepatocellular carcinoma treated with transarterial
chemoembolization. |
Table I.
Characteristics of 60 patients with
hepatocellular carcinoma treated with transarterial
chemoembolization.
| Characteristic | No. of patients
(%) |
|---|
| Age, years |
|
<50 | 17 (28.3) |
|
≥50 | 43 (71.7) |
| Sex |
|
|
Female | 8 (13.3) |
|
Male | 52 (86.7) |
| Albumin, g/l |
|
|
≥35 | 53 (88.3) |
|
<35 | 7 (11.7) |
| AFP, ng/ml |
|
|
≤20 | 16 (26.7) |
|
>20 | 44 (73.3) |
| Tumor diameter,
cm |
|
| ≤5 | 22 (36.7) |
|
>5 | 38 (63.3) |
| Portal vein
invasion |
|
|
Without | 39 (65.0) |
|
With | 21 (35.0) |
| BCLC stage |
|
| A | 5 (8.3) |
|
B/C | 55 (91.7) |
| Serum IL-10
pre-TACE, pg/ml |
|
|
<1.59 | 30 (50.0) |
|
>1.59 | 30 (50.0) |
Treatment and follow-up laboratory
data
Clinical and laboratory data of enrolled patients
were recorded at baseline (pre-TACE day 0) and at regular intervals
(post-TACE day 3, 7, 14 and 21) during the follow-up period,
including patient age, sex, hematologic parameters,
alanine/aspartate aminotransferases (AST/ALT), albumin, total
bilirubin and portal vein invasion. The 60 patients with HCC were
subsequently divided into 2 groups based on tumor size (≤5 and
>5 cm) and ALT levels (≤40 and >40 U/l) for further
analysis.
Microarray data and processing
Serum samples were collected prior to TACE therapy
and again on days 3, 7, 14 and 21 after the procedure. All samples
were stored at −80°C, and 9 serum samples randomly selected from
the 60 patients with HCC were labelled (Table SI), hybridized and washed using an
Agilent hybridization 5188–5242 kit (Agilent Technologies, Inc.).
Slides were analysed using an Agilent Microarray Scanner with
Feature Extraction G4463AA software (Agilent Technologies, Inc.).
Each serum sample obtained pre-TACE (day 0) was compared with those
obtained on days 3, 7, 14 and 21 and used the limma package of R
3.6.1 software (25) to identify
differentially expressed cytokine genes (DECs). P<0.05 and |log
fold-change (FC)| >1 were counted as detected signals.
Subsequently, the pheatmap package in R 3.6.1 software (25) was used to construct a heat map of
differentially expressed genes. To ensure specificity, the DECs of
all four groups (day 3, 7, 14 and 21 vs. day 0, respectively) were
integrated using venn2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Short time-series expression miner
(STEM)
First, the STEM algorithm and 1.3.12 software was
used to analyse the dataset using the 507 identified cytokine
microarray (http://www.cs.cmu.edu/~jernst/stem) (26). Following the input phase, the STEM
clustering algorithm was executed and displayed the clustering
results. The clustering algorithm that STEM implements takes
advantage of a point-in-time of the dataset and selects a set of
distinct and representative temporal expression profiles called
model profiles (Fig. 1A). Each gene
was assigned to the model profile that most closely matched that
gene's expression profile as determined by the correlation
coefficient. Next, the standard hypothesis testing was used to
determine which model profiles had significantly more genes
assigned to them under the true ordering of the time points in the
permutation runs. The parameters used for STEM clustering were set
at a maximum of 50 model profiles, a maximum unit change between
time points of 2 and a minimum correlation for clustering similar
profiles >0.7. Finally, clusters based on the expression trend
of significant genes were screened out (P<0.05).
Gene Ontology (GO) and signaling
pathways
DECs were determined by means of a significance
analysis of the microarray and model profile data. GO analysis
determined that there were 284 DECs through use of the Database for
Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/) (27) online tool (P<0.05). The signaling
pathways that were likely impacted by the DECs were predicted using
the KEGG pathway map tool (P<0.05) (28).
Protein-protein interaction (PPI)
network
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) 5.18 (https://string-db.org/) is a repository of known and
predicted protein interrelationships (29) and was used to identify possible
interacting proteins between the DECs.
ELISA
Serum samples were collected pre-TACE and on days 3,
7, 14, and 21 after the initiation of treatment from 60 evaluable
patients who underwent transarterial chemotherapy. The cytokine
levels of IL-1B, IL-10, IL-5 and CNTF were analysed using a
multiplex immunoassay Bio-Plex 200 array system and Bio-Plex
Manager v6.0 software [Hangzhou Multisciences (Lianke) Biotech Co.,
Ltd] according to the manufacturer's instructions.
Statistical analysis
Student's t-test was used for continuous data, and
χ2 test was used for categorical data. The comparison of
multiple groups was performed with ANOVA and Tukey's post hoc test.
The criteria for screening DECs were |logFC|>1.0 and a false
discovery rate <0.05. Spearman's correlation was also used to
measure the correlation between different cytokine levels as well
as between cytokine levels and clinical data. Receiver operating
characteristic (ROC) curves were used by plotting sensitivity
against 1-specificity. The optimal cut-off values for ROC curves
were obtained using the Youden index. The survival curve was
plotted using the Kaplan-Meier method, and the overall survival
(OS) rate between subgroups was compared using the log-rank test.
The Cox proportional hazard regression model was used to evaluate
prognostic factors, and hazard ratio (HR) and 95% confidence
interval was determined. The aforementioned statistical data were
analysed using SPSS v22.0 software (IBM, Corp.) and GraphPad Prism
6 (GraphPad Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of time-series cytokine
genes after TACE
The Agilent cytokine genome microarrays were used to
measure the relative gene expression levels in 9 patients with HCC
pre-TACE and on days 3, 7, 14 and 21 post-TACE. In addition, 9
patients with HCC were selected for microarray analysis based on
objective response, including three OR and six non-OR patients. A
total of 507 cytokine genes were identified and the differentially
expressed genes on days 3, 7, 14 and 21 were compared with day 0.
Given the limited overlap of the differentially expressed genes
among the four datasets (data not shown), only five genes,
including neural differentiation 1 (NEUROD1), triggering receptor
expressed on myeloid cells 1 (TREM1), neuregulin 3 (NRG3),
Charcot-Leyden crystal galectin (CLC) and TNFα-induced protein 6
(TNFAIP6) were identified. Hence, the relevant gene clusters were
identified based on the STEM algorithm. The input parameters c=2
and m=50 were used, where c indicates the units of change and m
indicates the number of candidate profiles. In all, 284 out of 507
differentially expressed genes were significantly clustered in
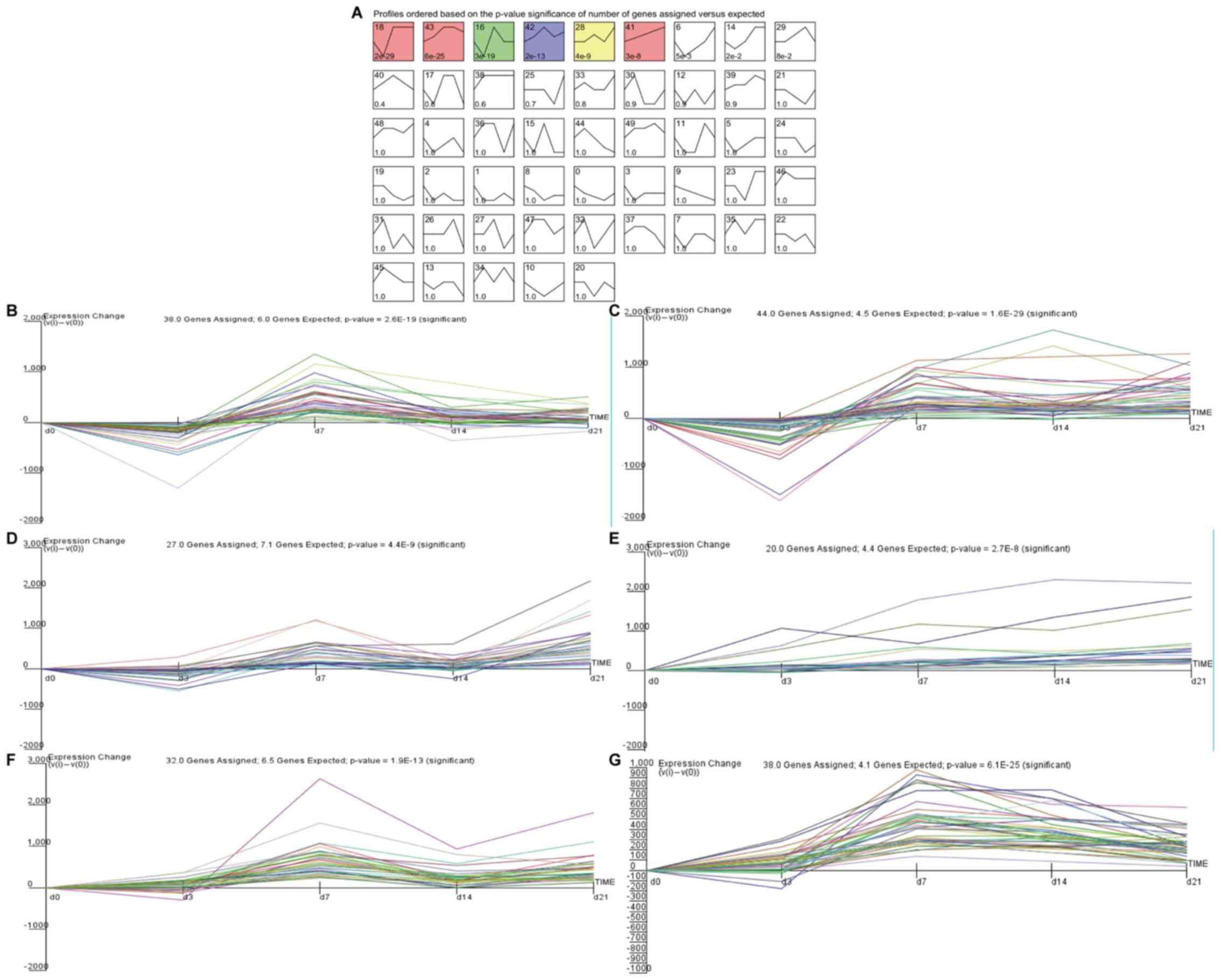
eight models, including profiles 6, 14, 16, 18, 28, 41, 42 and 43.
However, <10 genes were clustered in profiles 6 and 14 and
therefore were eliminated in further analysis (Fig. 1A). Fig.
1 shows the fold-changes and P-values of the gene expression
profiles for the six significant clusters, and their gene symbols
and relative expressions are listed in Table SII. Based on STEM analysis, CTNF and
IL-5 were found to cluster in profile 18, IL-1β and IL-10 were
found to cluster in profile 41, CCL2 and MMP9 were found to cluster
in profile 43, CCR7 and CCR1 were found to cluster in profile 16,
INS and BMP7 were found to cluster in profile 42, and IL-4 and
IL-17RA were found to cluster in profile 28 (Fig. 1B-E and Table II). The trend curves of the gene
signatures clustered in profiles 28, 41, 42 and 41 were similar and
were all gradually upregulated (Fig.
1B-G). However, some cytokines were not significant in this
study, such as IL-6 and IL-8 associated with TACE prognosis. The
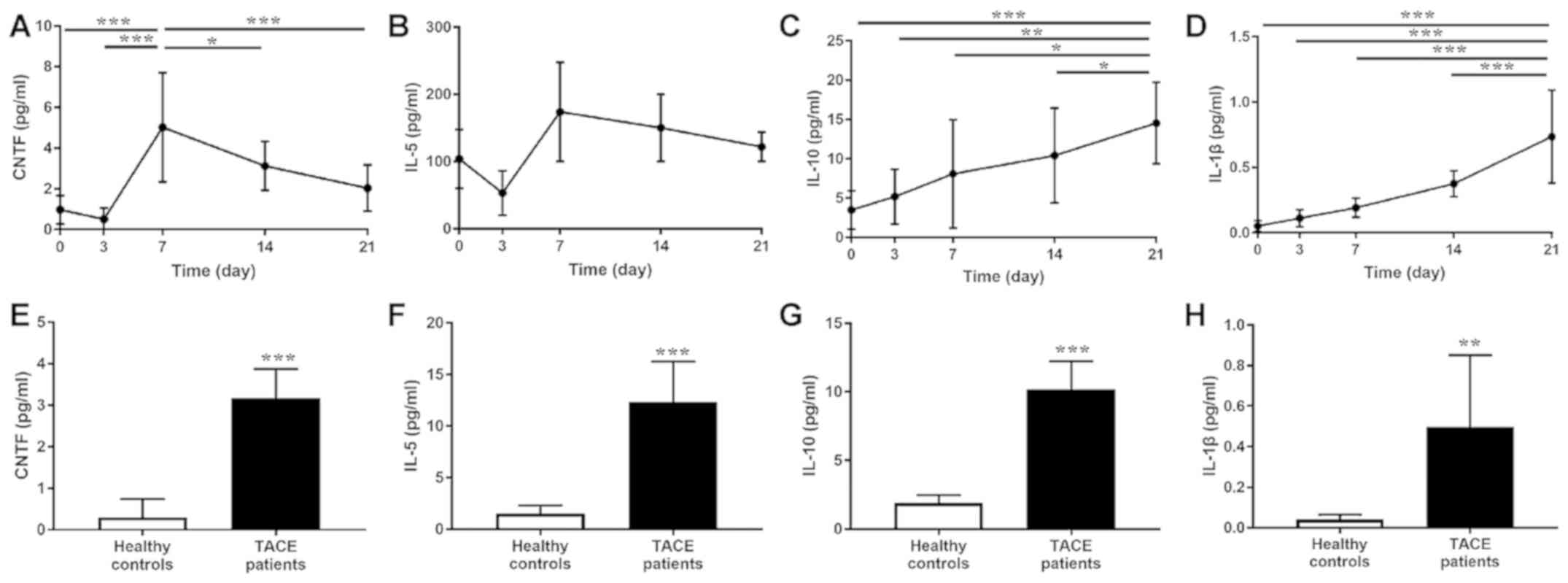
trend result of CTNF, IL-5, IL-1β and IL-10 was identified
according to ELISA data (Fig. 2A-D).
Subsequently, the baseline serum levels of CTNF, IL-5, IL-10 and
IL-1β from patients with HCC who underwent TACE and healthy
controls were compared. The pre-TACE serum levels of CTNF, IL-5,
IL-10 and IL-1β were significantly elevated compared with that in
the healthy controls (Fig.
2E-H).
 | Table II.Degree of gene modulation in the
protein-protein interaction network. |
Table II.
Degree of gene modulation in the
protein-protein interaction network.
| A, Genes in profile
18 |
|---|
|
|---|
| Gene | Degree |
|---|
| TNF | 30 |
| IFNG | 15 |
| IL5 | 13 |
| CCL19 | 10 |
| CXCR2 | 10 |
| TLR1 | 9 |
| CD40LG | 7 |
| CCL1 | 7 |
| CNTF | 7 |
| PRL | 7 |
|
| B, Genes in
profile 43 |
|
| Gene | Degree |
|---|
|
| CCL2 | 15 |
| MMP9 | 14 |
| PGF | 13 |
| ITGAM | 12 |
| PDGFB | 12 |
| FGF13 | 11 |
| FLT4 | 9 |
| MMP3 | 9 |
| PDGFA | 9 |
| FIGF | 8 |
|
| C, Genes in
profile 41 |
|
| Gene | Degree |
|
| IL10 | 13 |
| IL1B | 13 |
| CCL5 | 9 |
| TLR4 | 8 |
| PECAM1 | 8 |
| TNFRSF1A | 7 |
| SELL | 7 |
| CCL17 | 6 |
| LIF | 4 |
| SMAD7 | 4 |
|
| D, Genes in
profile 16 |
|
| Gene | Degree |
|
| CCR7 | 13 |
| ICAM1 | 11 |
| CCR1 | 10 |
| BMP4 | 10 |
| CSF1 | 9 |
| CD80 | 9 |
| NCAM1 | 8 |
| CXCL16 | 8 |
| IFNB1 | 8 |
| IL2RG | 8 |
|
| E, Genes in
profile 42 |
|
| Gene | Degree |
|
| INS | 15 |
| BMP7 | 9 |
| FGF2 | 8 |
| IL1RN | 8 |
| EPO | 8 |
| VCAM1 | 7 |
| ACVR2B | 6 |
| CXCR1 | 6 |
| BMP5 | 6 |
| ACVR1B | 5 |
|
| F, Genes in
profile 28 |
|
| Gene | Degree |
|
| IL4 | 8 |
| IL13 | 6 |
| CXCL1 | 6 |
| ADIPOQ | 5 |
| IL17RA | 5 |
| CCR5 | 4 |
| FGF21 | 4 |
| GPR29 | 4 |
| FGFR3 | 3 |
| PLG | 3 |
GO and KEGG pathway analyses of
time-series cytokine genes after TACE
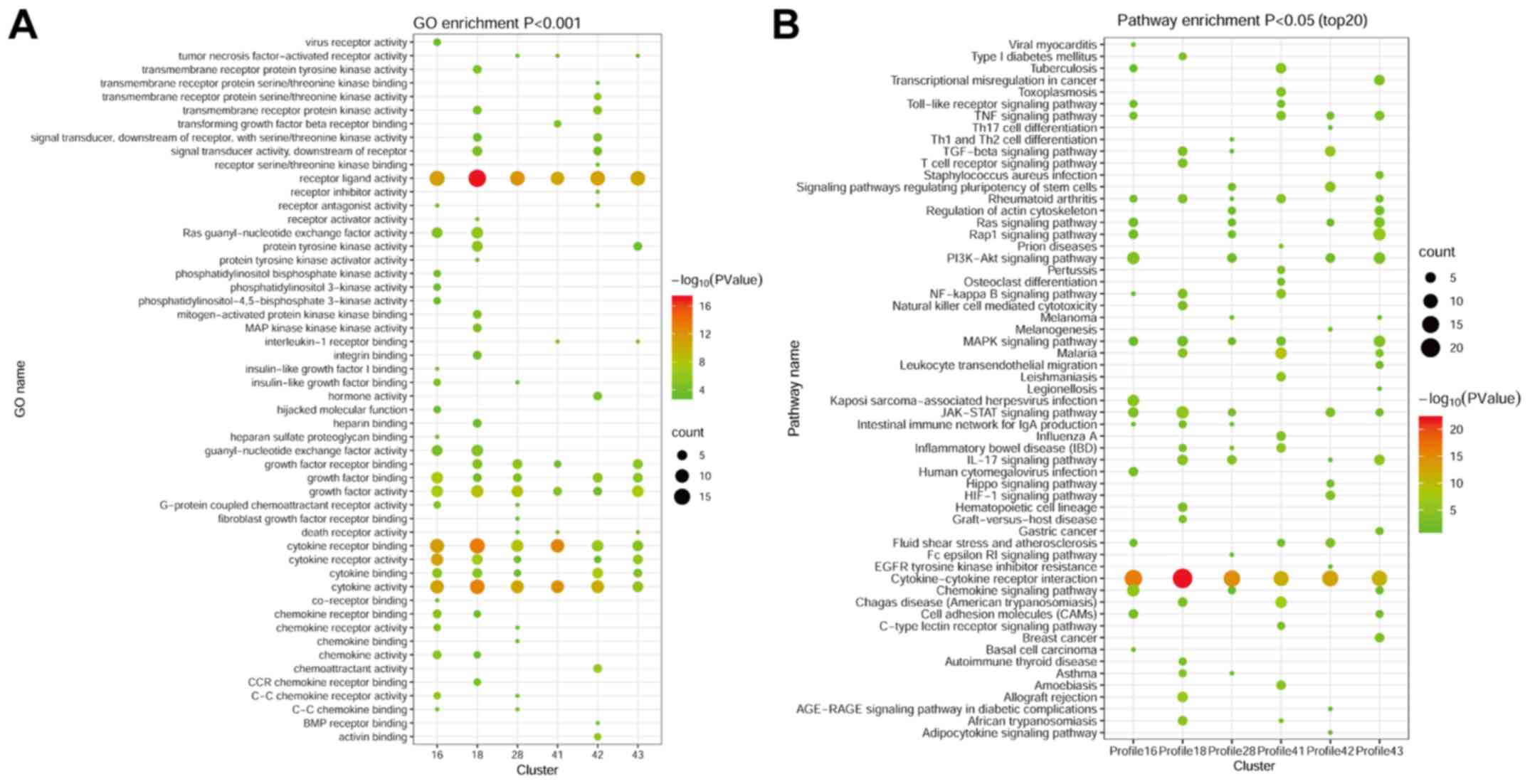
Based on the six significant clusters, GO analysis
was performed. The results demonstrated that the DECs in the six
significant clusters were related to ‘cytokine receptor binding’,
‘receptor ligand activity’ and ‘cytokine activity’ (Fig. 3A). In profile 18, the DECs were also
enriched in ‘growth factor receptor binding’.
According to STEM analysis, KEGG signaling pathways
were examined to determine whether they were dysregulated in the
six significant clusters. ‘Cytokine-cytokine receptor interactions’
and ‘JAK-STAT signaling’ were significantly different in all
profiles, with the exception of profile 41 (Fig. 3B). The IL-17 signaling pathway was
present in four of the six significant clusters (profile 18, 28, 42
and 43) and included IL-5, IL-4, IL-1β, IL-17D and CCL2.
Intriguingly, the signaling pathway exhibited differences in
profile 41, which included cytokines IL-1β and IL-10. Likewise, the
MAPK/RAS and Toll-like receptor signaling pathways, which are
associated with cancer (30), were
detected in profiles 16 and 43, and profiles 16 and 41,
respectively, suggesting that these cytokines could be involved in
inflammation and cancer transformation in HCC. The CNTF, colony
stimulating factor 2 receptor α subunit, interferon-γ, IL-5,
platelet-derived growth factor receptor β (PDGFRB), prolactin (PRL)
and IL-27 receptor subunit α genes were differentially expressed
and enriched in the JAK-STAT signaling pathway in profile 18.
Similar to profile 18, the DECs and pathways clustered in profile
16 also included inflammatory signaling pathways (the JAK-STAT and
NF-kB pathways), as well as CCR1, CCR7, ICAM1 and BMP4 (Table SIII).
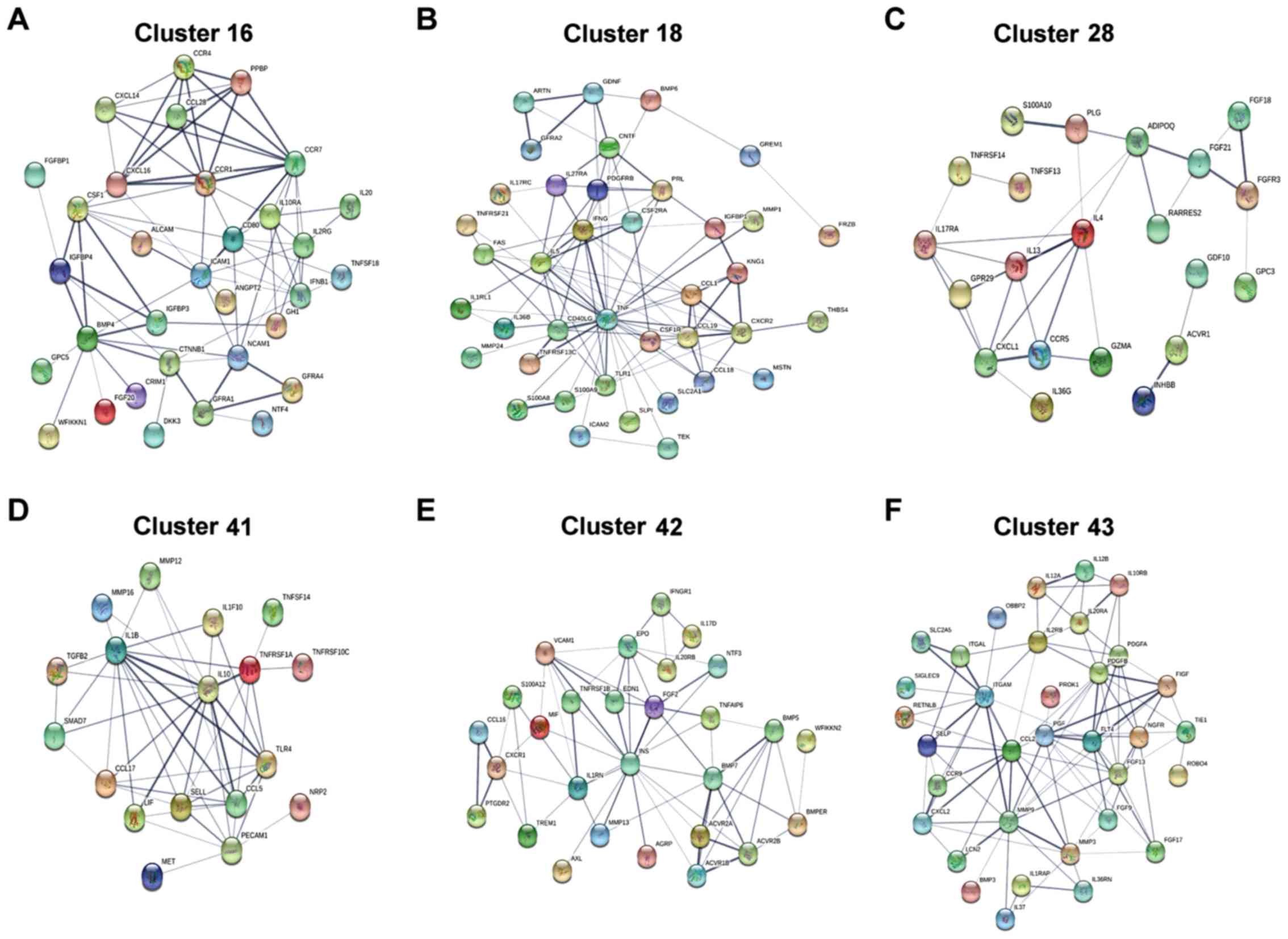
PPI network
To examine the associations among the DECs in each
significant cluster, a PPI network was constructed (Fig. 4). The STRING database was used to
identify possible interactions of each cytokine expression profile.
The results demonstrated that the 32, 39, 21, 18, 28 and 34
cytokines identified in profiles 16, 18, 28, 41, 42 and 43,
respectively, were interacting with each other. In each profile,
the high degree of cytokines, which were based on the degree and
the association with tumors, including CTNF, IL-5, IL-10 and IL-1β,
were selected. Additionally, the cytokines with the highest
association in each respective cluster were CCR7, TNF, IL-4, IL-10,
INS and CCL2 (Table II).
Correlation between liver function and
the post-treatment cytokine CNTF levels
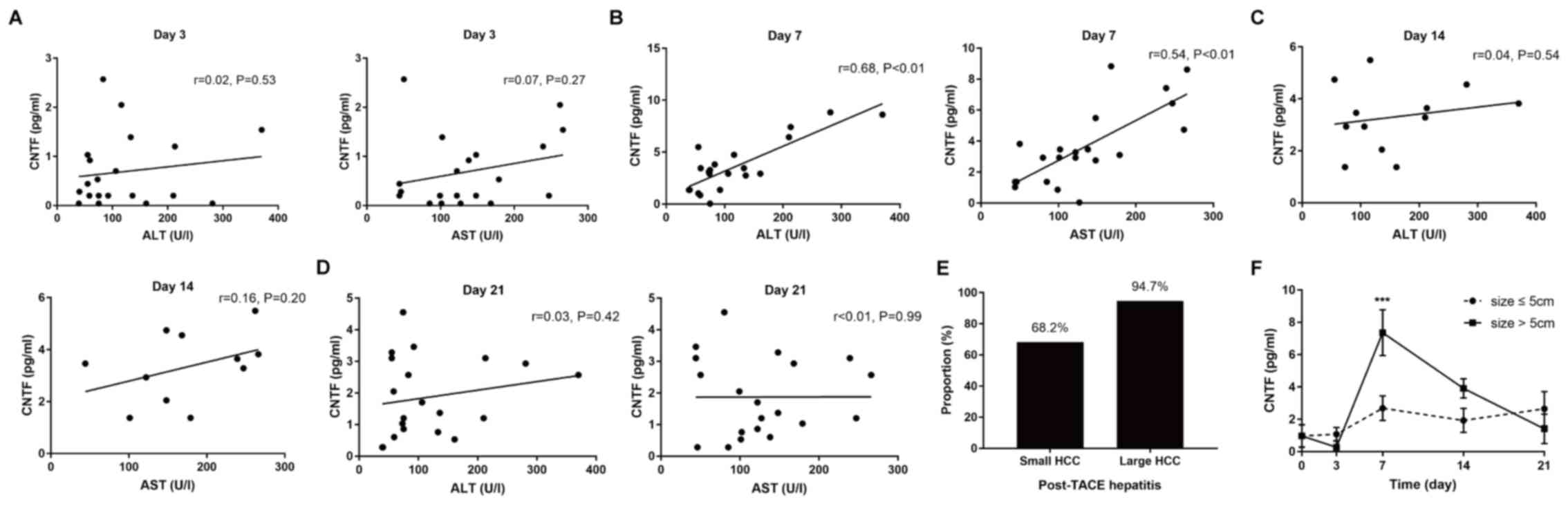
The association between the level of ALT or AST and
the CNTF cytokine levels during treatment was investigated in
patients with HCC undergoing TACE. There was no significant
correlation between serum ALT (P=0.53) or AST (P=0.27) levels and
CNTF levels on post-treatment day 3. Positive correlations with
CNTF levels appeared over time and were significant on day 7 for
the ALT (r=0.68; P<0.01) and AST (r=0.54; P<0.01) levels.
However, on days 14 and 21 after TACE, there was no longer a
correlation between the level of ALT or AST and the CNTF levels
(Fig. 5A-D). Subsequently the
changes in ALT and CNTF levels between small and large HCC tumors
were measured. In 60 patients with HCC, those with large tumors
(>5 cm) exhibited more frequent episodes of severe hepatitis
(ALT elevation >40 U/l) compared with those with small tumors
[36/38 (94.7%) vs. 15/22 (68.2%); Fig.
5E and F]. Patients with large tumors (>5 cm) had a higher
baseline and greater increases in CNTF day 7 after TACE, whereas
patients with small HCC (≤5 cm) exhibited only minor rises in CNTF
levels. This characteristic change in CNTF that occurred following
TACE suggests that there is an increase in CNTF levels in patients
with tumors >5 cm, on day 7.
Pretreatment serum levels of IL-10 and
post-treatment serum levels of IL-1β predict an objective response
to TACE therapy
Subsequently it was investigated whether the serum
levels of IL-10 and IL-1β had predictive value regarding the trend
curve from the microarray data observed for profile 41 (Fig. 1E). The correlations between serum AFP
and IL-10/IL-1β were investigated, and there was a significant
correlation between the serum AFP and pre-TACE IL-10 levels
(Fig. 6A and B). However, tumor
diameter was positively associated with IL-1β levels, and patients
with large tumors (>5 cm) had higher baseline levels and greater
increases in post-TACE levels of IL-1β (Fig. 6C and D). This characteristic pattern
of IL-1β release might be associated with larger tumors, which have
increased hypoxic areas. However, there was not a significant
association between the serum levels of IL-10 and IL-1β and BCLC
stage, including BCLC stage A (n=5) and B/C (n=55) (Fig. 6E and F) on day 0 pre-treatment and
day 3 post-treatment.
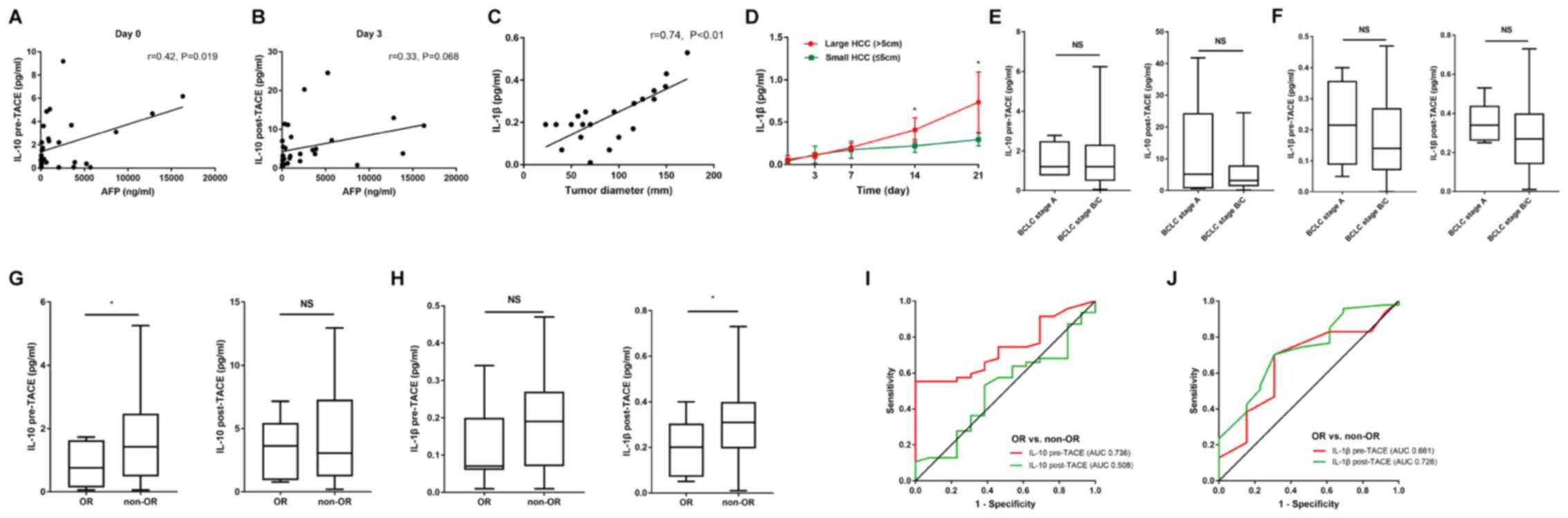 | Figure 6.Preinterventional serum levels of
IL-10 and post-interventional serum levels of IL-1β predict the
objective response after TACE. Spearman's correlation analysis
between (A) IL-10 levels and pre-TACE AFP levels, (B) IL-10 levels
and AFP levels on day 3 after TACE, and (C) IL-1β levels and tumor
diameter. (D) Patients with large HCC tumors showed a more
prominent early-phase increase in IL-1β levels after therapy
compared with those with small tumors. Serum levels of (E) IL-10
and (F) IL-1β pre-and post-TACE treatment were unaltered between
patients with BCLC stage A and B/C. Serum levels of (G) IL-10 and
(H) IL-1β pre- and post-TACE treatment in patients with and without
OR. ROC curve analysis for (I) IL-10 and (J) IL-1β pre- and
post-TACE treatment in patients with and without OR. OR patients,
n=13; and non-OR patients, n=47. *P<0.05; ns, P>0.05. BCLC,
Barcelona Clinic Liver Cancer; OR, objective response; ROC,
receiver operating characteristic; AUC, area under the curve, TACE,
transarterial chemoembolization; AFP, α-fetoprotein; NS,
non-significant; HCC, hepatocellular carcinoma; IL,
interleukin. |
The cohort was further divided into two groups:
Patients with an OR (complete or partial remission; n=13) and
patients who showed a non-OR (stable or progressive disease; n=47).
In the analysis, the pretreatment levels of IL-10 and
post-treatment levels of IL-1β were significantly higher in
patients who showed a non-OR after TACE compared with that in
patients with an OR, while the post-treatment levels of IL-10 and
pretreatment levels of IL-1β were not significantly different
between OR and non-OR patients (Fig. 6G
and H). In line with this finding, ROC curve analysis of the
differences between patients with either an OR or non-OR revealed
that the area under the curve (AUC) value of 0.736 for pre-TACE
IL-10 was higher than that for post-TACE IL-10 (AUC, 0.508) and
that the AUC value of 0.726 for post-TACE IL-1β was higher than
that for pre-TACE IL-1β (AUC, 0.661) (Fig. 6I and J).
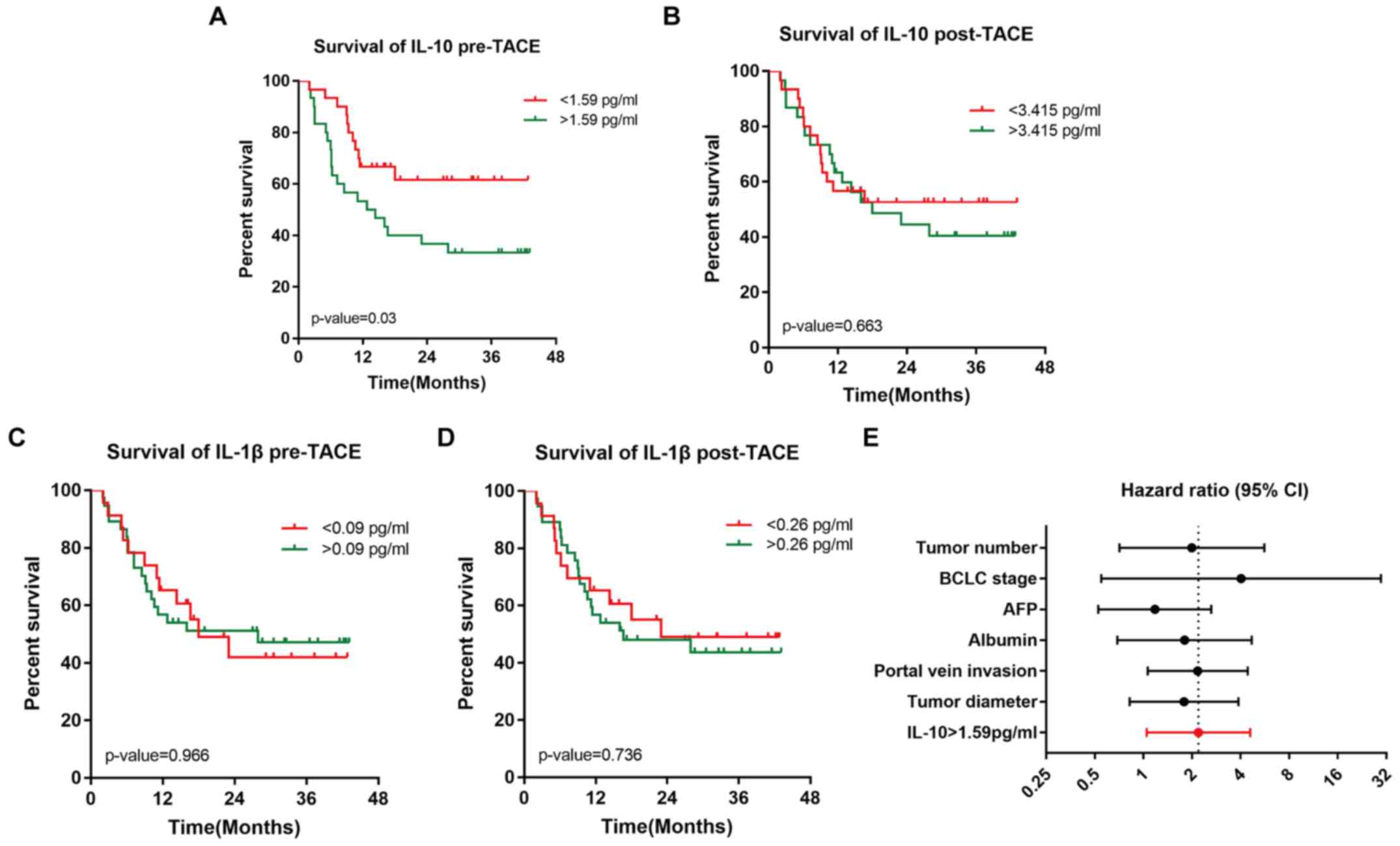
Serum levels of pretreatment IL-10
predict OS time after TACE
Based on the predictive value of the pretreatment
serum levels of IL-10 and post-treatment serum levels of IL-1β, the
serum levels of IL-10 and IL-1β was subsequently evaluated to
determine whether they influenced patient OS. Based on the Youden
index of the ROC curve analysis, which is used to identify the
ideal cut-off values for cytokines, Kaplan-Meier analysis showed
that patients with pretreatment serum levels of IL-10 above the
ideal cut-off value (1.59 pg/ml) experience reduced survival
compared to patients with levels below the ideal cut-off values
[mOS: 13.55 months vs. undefined (median OS not reached); P=0.03]
(Fig. 7A). By contrast, the ideal
cut-off values for post-treatment IL-10, pretreatment IL-1β and
post-treatment IL-1β levels did not effectively identify patients
with a poor prognosis (Fig.
7B-D).
To further investigate prognostic factors, the
results of the Kaplan-Meier analysis for OS were further analysed
using univariate and multivariate Cox regression models. Fig. 7E summarizes the univariate factor
analysis, and the HR for IL-10 was 2.193 (P=0.037; 95% CI,
1.048–4.589; Table III).
Subsequently parameters with P<0.70 from the univariate Cox
regression models (IL-10, tumor diameter, portal vein invasion,
BCLC stage B/C vs. A, albumin and AFP) were removed and
multivariate Cox analysis (Table
IV) was performed. Importantly, the prognostic value of the
pretreatment serum level of IL-10 was independent of those
parameters.
 | Table III.Univariate Cox regression analysis to
predict overall survival. |
Table III.
Univariate Cox regression analysis to
predict overall survival.
|
| Univariate Cox
regression |
|---|
|
|
|
|---|
| Parameter | P-value | Hazard ratio (95%
CI) |
|---|
| Serum IL-10
>1.59 pg/ml | 0.037 | 2.193
(1.048–4.589) |
| Tumor diameter
(cm) | 0.144 | 1.785
(0.821–3.882) |
| Portal vein
invasion | 0.033 | 2.171
(1.064–4.431) |
| BCLC stage | 0.171 | 4.031
(0.548–29.636) |
| Albumin (g/l) | 0.230 | 1.799
(0.690–4.691) |
| AFP (ng/ml) | 0.690 | 1.178
(0.526–2.636) |
 | Table IV.Multivariate Cox regression analysis
to predict overall survival. |
Table IV.
Multivariate Cox regression analysis
to predict overall survival.
|
| Multivariate Cox
regression |
|---|
|
|
|
|---|
| Parameter | P-value | Hazard ratio (95%
CI) |
|---|
| Serum IL-10
>1.59 pg/ml | 0.049 | 2.249
(1.005–5.031) |
| Tumor diameter
(cm) | 0.855 | 0.918
(0.369–2.288) |
| Portal vein
invasion | 0.115 | 1.870
(0.858–4.074) |
| BCLC stage | 0.229 | 3.682
(0.441–30.762) |
| Albumin (g/l) | 0.680 | 1.241
(0.445–3.459) |
| AFP (ng/ml) | 0.889 | 1.066
(0.435–2.608) |
Discussion
TACE is considered one of the most effective
treatments for non-resectable liver cancer (31). At present, numerous studies have
reported that changes in cytokines occur in patients with liver
cancer after TACE, and these changes may be related to acute
inflammatory reactions, acute liver injury and prognostic factors
(18,19). However, the potential mechanisms
involving these cytokines in patients with a post-TACE prognosis
have not been completely elucidated. In the present study, the
cytokine expression profiles of 9 patients with HCC at different
time points after TACE using a cytokine mRNA microarray were
analysed. Subsequently, the cytokine mRNA expression profiles
post-TACE on days 3, 7, 14 and 21 were compared to the profiles
obtained preintervention (day 0). Venn 2.1 was used for
differential gene integration. The data demonstrated that five
genes, including NEUROD1, TREM1, NRG3, CLC and TNFAIP6, were
present in four groups: Day 3 vs. 0, day 7 vs. 0, day 14 vs. 0 and
day 21 vs. 0. Borromeo et al (32) reported that NEUROD1 plays crucial
roles in promoting malignant behaviour and survival in patients
with SCLC. TREM2, a novel pattern recognition receptor family
member, is generally regarded to be an enhancer of immune responses
(33). According to
pharmacogenomics, NRG3 rs1649942 genetic variants have been
validated to affect epithelial ovarian cancer first-line treatment
outcomes, which confirmed patients carrying the NRG3 rs1649942 A
allele presented a significantly longer OS time (34). In addition, CLC and TNFAIP6, which
are cytokine receptors, are involved in disease progression and the
immune response (35) in a variety
of tumors, such as gastric cancer (36), breast cancer (37) and urothelial carcinomas (38).
STEM was used to reveal temporal gene expression
profiles and identify significant patterns from the day before TACE
(day 0) to day 21 post-TACE that were >2-fold different
(P<0.05). A total of 6 separate and significant time-varied
expression patterns among 507 genes were identified, including
profiles 16, 18, 28, 41, 42 and 43. DECs were identified, and their
functions were predicted through GO and signaling pathway
analyses.
The clustered profile 18 showed initial
downregulation before TACE and subsequent upregulation after TACE
(Fig. 1). These results might
suggest that genes that are downregulated during the initial phase
after TACE might be turned off and could therefore be responsible
for hindering the HCC-like phenotype and hepatic injury after a
tumor-intervening event. The CNTF, colony stimulating factor 2
receptor α subunit, interferon-γ, IL-5, PDGFRB, PRL and IL-27
receptor subunit α genes were differentially expressed and enriched
in the JAK-STAT signaling pathway in profile 18 (Table SIII). Previous studies showed that
JAK-STAT signaling was closely associated with cytokine signalling,
as it regulates essential cellular mechanisms, such as
proliferation, invasion, survival, inflammation and immunity
(39,40). In addition, CNTF is an important
hepatoprotective agent in carbon tetra chloride
(CCl4)-induced hepatic injury (41). In a retrospective cohort study, IL-5,
PDGFRB and PRL predicted survival benefits in patients with
advanced HCC treated with sorafenib (42). Similar to profile 18, the DECs and
pathways clustered in profile 16 also included inflammatory
signaling pathways (the JAK-STAT and NF-kB pathways), as well as
CCR1, CCR7, ICAM1 and BMP4 (Table
SIII). CCR1 and CCR7 play important roles in head and neck
squamous cell carcinoma progression and regional lymph node
metastasis and recurrence. CCR1 were highly expressed in primary
carcinomas and CCR7 was associated with disease-free survival time
(43). The trend curves of the gene
signatures clustered in profiles 28, 41, 42 and 41 were similar and
were all gradually upregulated (Fig.
1B-G). The DECs clustered in profile 28, including CXCL1, IL-4,
IL-13 and IL-17RA, and in profile 43, including LCN2, CCL2, CXCL2,
MMP3 and MMP9, were enriched in the IL-17 signaling pathway
(Table SIII). A previous study
demonstrated a relationship between the IL-17/IL-17R axis and the
tumor inflammatory microenvironment in patients with NSCLC.
Immunoreactivity for IL-17A, IL-17F, IL-17RA and IL- 17RC was
significantly elevated, while IL-17E was reduced (44). In profile 41, IL-1β, IL-10 and CCL5
were differentially expressed and enriched in the NF-kB, TNF and
Toll-like receptor signaling pathways. Meanwhile, JAK-STAT and
IL-17 signalling pathways were not associated in profile 41. In
line with previous results, IL-1β, IL-10 and CCL5 were involved in
multiple signaling pathways that affect the development and
progression of tumors. IL-1β caused expansion of the bone
metastatic niche and led to tumor proliferation in breast cancer
(45). IL-10 and integrin pathways
have been reported to be strongly associated with head and neck
cancer progression (46). In
addition, it has been reported that CCl5/CCR1/β-catenin/Slug via
mesenchymal stem cells promotes colorectal cancer development
(47). In addition to these common
signaling pathways, HIF-1 signaling was also significantly enriched
in profile 42. It was previously demonstrated that HIF-1, a key
transcription factor involved in hypoxia and inflammation, induces
HCC cells to undergo EMT (48,49).
To better understand the role of cytokine expression
during TACE, ELISA was performed and demonstrated that the temporal
variations in cytokines were similar to the results obtained by
STEM. The CNTF gene, which clustered in profile 18, was acutely
increased after TACE but decreased after day 7 and reached baseline
levels thereafter, while the trend for IL-5 was not statistically
significant. Hu et al (50)
reported that CNTF regulates PI3K and AMP-activated protein kinase
(AMPK) and is therefore involved in metabolic diseases and
hepatocarcinogenesis. CNTF acts as a hepatoprotective agent against
CCl4-induced hepatic injury, and a correlation between
the post-treatment day 7 serum aminotransferase and CNTF levels was
observed in the present study. In agreement with previous data
(41) the proportion of released
CNTF was positively correlated with the degree of hepatic damage,
which inhibited lipoprotein secretion and impaired the ability to
act as transport vehicles for lipids from the liver to the
circulation. According to the trend curve and signaling pathway of
profile 41, the survival and prognostic value of IL-1β and IL-10
was investigated further in clinical features. The results of the
present study demonstrated that the pre-TACE serum levels of IL-10
predicted the local tumor response and OS time of a patient, while
the post-TACE serum levels of IL-1β only indicated the local tumor
response. In addition, multivariate Cox regression analysis
revealed that the prognostic value of the pre-TACE IL-10 levels was
independent of tumor diameter, BCLC stage, AFP, portal vein
invasion and liver function. However, studies of the effect of
IL-10 on tumor progression have previously concentrated on
antitumor immune effects (51).
IL-10 is an immunomodulatory cytokine that has previously been
shown to participate in T-cell inactivation and the impairment of
adaptive immunity (52–54). Regarding its prognostic value, high
IL-10 expression was found to be a marker of longer survival
prognosis in breast cancer in addition to decreased tumor cell
migration (55). In HCC, IL-10 and
IL-12 have been shown to be involved in the progression of chronic
inflammation leading to HCC, and they could therefore be considered
biomarkers that reflect the degree of inflammation in HCC
development (56). Meanwhile, the
pre-TACE serum levels of IL-10 exhibited better survival prediction
compared with post-TACE levels. One possible reason for this result
is that TACE may disturb the tumor microenvironment, which
contributed to hypoxia for killing tumor cells. In the current
study, the role of the serum IL-10 level as a good prognostic
marker seems to be dependent on the tissue microenvironment.
However, there are many limitations to the present
study. First, a cytokine microarray was used to detect differential
gene expression. In accordance to a previous study, this study also
reported that some cytokines were undetectable in a number of
patients, and many differences were not significant, such as IL-6
and IL-8 associated with TACE prognosis (19). Second, due to lack of an accepted
interventional animal model and limited number of healthy controls,
the effect of TACE on cytokine levels in normal liver tissue could
not be evaluated. Meanwhile, as TACE is a minimally invasive
surgery, it is difficult to obtain a large number of liver tissue
samples for further study, such as immunohistochemistry and western
blot analyses. Third, the present study was conducted using a small
cohort of patients, and only four cytokines were identified using
ELISA. Moreover, determining the mechanisms underlying the effects
of cytokines in HCC after TACE requires further study.
In summary, a time series of cytokine expression
profiles in patients with HCC undergoing TACE was identified. Early
phase increases in CNTF serum levels observed after TACE may exert
hepatoprotective effects and are associated with post-treatment
hepatic injury. Changes in the expression of IL-1β and IL-10 during
the process of TACE are associated with long-term outcomes and
prognostic survival. These data provide insight into the respective
contributions to different biological functions of cytokines
released after TACE and their clinical value.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81772590 and 81572395),
Shanghai Municipality: Shanghai Outstanding Academic Leaders Plan
(grant no. 2013-48) and Shanghai Program of Shanghai Academic
Research Leader (grant no. 14XD1401100).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Author' contributions
FQ, LW and PH designed the study. FQ, LW, PH and ZZ
recruited the patients. FQ performed the experiments. JX, ZZ and BY
designed the stufy and performed the assessment of the radiological
TACE response. FQ and ZZ performed statistical analyses and
generated the figures and tables. FQ and JX drafted the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from each patient, and
the present study was approved by The Ethics Committees of
Zhongshan Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
STEM
|
Short Time-series Expression Miner
|
|
TACE
|
transarterial chemoembolization
|
References
|
1
|
Tang A, Hallouch O, Chernyak V, Kamaya A
and Sirlin CB: Epidemiology of hepatocellular carcinoma: Target
population for surveillance and diagnosis. Abdom Radiol (NY).
43:13–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoyos S, Escobar J, Cardona D, Guzmán C,
Mena Á, Osorio G, Pérez C, Restrepo JC and Correa G: Factors
associated with recurrence and survival in liver transplant
patients with HCC-a single center retrospective study. Ann Hepatol.
14:58–63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vitale A, Peck-Radosavljevic M, Giannini
EG, Vibert E, Sieghart W, Van Poucke S and Pawlik TM: Personalized
treatment of patients with very early hepatocellular carcinoma. J
Hepatol. 66:412–423. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patel T: Increasing incidence and
mortality of primary intrahepatic cholangiocarcinoma in the United
States. Hepatology. 33:1353–1357. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sacco R, Tapete G, Simonetti N, Sellitri
R, Natali V, Melissari S, Cabibbo G, Biscaglia L, Bresci G and
Giacomelli L: Transarterial chemoembolization for the treatment of
hepatocellular carcinoma: A review. J Hepatocell Carcinoma.
4:105–110. 2007. View Article : Google Scholar
|
|
6
|
Han K and Kim JH: Transarterial
chemoembolization in hepatocellular carcinoma treatment: Barcelona
clinic liver cancer staging system. World J Gastroenterol.
21:10327–10335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raoul JL, Forner A, Bolondi L, Cheung TT,
Kloeckner R and de Baere T: Updated use of TACE for hepatocellular
carcinoma treatment: How and when to use it based on clinical
evidence. Cancer Treat Rev. 72:28–36. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Opal SM and DePalo VA: Anti-inflammatory
cytokines. Chest. 117:1162–1172. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ramaswamy P, Goswami K, Dalavaikodihalli
Nanjaiah N, Srinivas D and Prasad C: TNF-α mediated MEK-ERK
signaling in invasion with putative network involving NF-κB and
STAT-6: A new perspective in glioma. Cell Biol Int. 43:1257–1266.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Quail DF, Olson OC, Bhardwaj P, Walsh LA,
Akkari L, Quick ML, Chen IC, Wendel N, Ben-Chetrit N, Walker J, et
al: Obesity alters the lung myeloid cell landscape to enhance
breast cancer metastasis through IL5 and GM-CSF. Nat Cell Biol.
19:974–987. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shih CM, Lee YL, Chiou HL, Hsu WF, Chen
WE, Chou MC and Lin LY: The involvement of genetic polymorphism of
IL-10 promoter in non-small cell lung cancer. Lung Cancer.
50:291–297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lech-Maranda E, Baseggio L, Bienvenu J,
Charlot C, Berger F, Rigal D, Warzocha K, Coiffier B and Salles G:
Interleukin-10 gene promoter polymorphisms influence the clinical
outcome of diffuse large B-cell lymphoma. Blood. 103:3529–3534.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao HY, Zou P and Zhou H: Genetic
association of interleukin-10 promoter polymorphisms and
susceptibility to diffuse large B-cell lymphoma: A meta-analysis.
Gene. 519:288–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsumoto K, Oki A, Satoh T, Okada S,
Minaguchi T, Onuki M, Ochi H, Nakao S, Sakurai M, Abe A, et al:
Interleukin-10 −1082 gene polymorphism and susceptibility to
cervical cancer among Japanese women. Jpn J Clin Oncol.
40:1113–1116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vairaktaris E, Yapijakis C, Serefoglou Z,
Derka S, Vassiliou S, Nkenke E, Vylliotis A, Spyridonidou S, Neukam
FW, Schlegel KA and Patsouris E: The interleukin-10 (−1082A/G)
polymorphism is strongly associated with increased risk for oral
squamous cell carcinoma. Anticancer Res. 28:309–314.
2008.PubMed/NCBI
|
|
16
|
Zhang J, Zhang Q, Lou Y, Fu Q, Chen Q, Wei
T, Yang J, Tang J, Wang J, Chen Y, et al: Hypoxia-inducible
factor-1α/interleukin-1β signaling enhances hepatoma
epithelial-mesenchymal transition through macrophages in a
hypoxic-inflammatory microenvironment. Hepatology. 67:1872–1889.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mohs A, Kuttkat N, Reissing J, Zimmermann
HW, Sonntag R, Proudfoot A, Youssef SA, de Bruin A, Cubero FJ and
Trautwein C: Functional role of CCL5/RANTES for HCC progression
during chronic liver disease. J Hepatol. 66:743–753. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim MJ, Jang JW, Oh BS, Kwon JH, Chung KW,
Jung HS, Jekarl DW and Lee S: Change in inflammatory cytokine
profiles after transarterial chemotherapy in patients with
hepatocellular carcinoma. Cytokine. 64:516–522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loosen SH, Schulze-Hagen M, Leyh C, Benz
F, Vucur M, Kuhl C, Trautwein C, Tacke F, Bruners P, Roderburg C
and Luedde T: IL-6 and IL-8 serum levels predict tumor response and
overall survival after TACE for primary and secondary hepatic
malignancies. Int J Mol Sci. 19:E17662018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Almeida BR, Barros BCSC, Araújo ACL,
Alcantara C and Suzuki E: Paracoccidioides species present distinct
fungal adherence to epithelial lung cells and promote different
IL-8 secretion levels. Med Microbiol Immunol; 2019, View Article : Google Scholar
|
|
21
|
Markov N and Simon HU: IL-37: A new player
in the chronic rhinosinusitis arena. J Allergy Clin Immunol. Oct
28–2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moaaz M, Youssry S, Elfatatry A and El
Rahman MA: Th17/Treg cells imbalance and their related cytokines
(IL-17, IL-10 and TGF-β) in children with autism spectrum disorder.
J Neuroimmunol. 337:5770712019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou L, Wang LY, Zhang XM, Zeng NL, Chen
TW, Li R, Huang YC and Tang YL: Semi-quantitative assessment of the
presence and Child-Pugh class of hepatitis B related cirrhosis by
using liver lobe-based dynamic contrast-enhanced MRI. Clin Radiol.
71:1289–1295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sato Y, Watanabe H, Sone M, Onaya H,
Sakamoto N, Osuga K, Takahashi M and Arai Y; Japan Interventional
Radiology in Oncology Study Group-JIVROSG, : Tumor response
evaluation criteria for HCC (hepatocellular carcinoma) treated
using TACE (transcatheter arterial chemoembolization): RECIST
(response evaluation criteria in solid tumors) version 1.1 and
mRECIST (modified RECIST): JIVROSG-0602. Ups J Med Sci. 118:16–22.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jalal H, Pechlivanoglou P, Krijkamp E,
Alarid-Escudero F, Enns E and Hunink MG: An overview of R in health
decision sciences. Med Decis Making. 37:735–746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ernst J and Bar-Joseph Z: STEM: A tool for
the analysis of short time series gene expression data. BMC
Bioinformatics. 7:1912006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dolcino M, Tinazzi E, Puccetti A and
Lunardi C: In systemic sclerosis, a unique long non coding RNA
regulates genes and pathways involved in the three main features of
the disease (vasculopathy, fibrosis and autoimmunity) and in
carcinogenesis. J Clin Med. 8:2019. View Article : Google Scholar
|
|
31
|
Silva JP, Berger NG, Tsai S, Christians
KK, Clarke CN, Mogal H, White S, Rilling W and Gamblin TC:
Transarterial chemoembolization in hepatocellular carcinoma with
portal vein tumor thrombosis: A systematic review and
meta-analysis. HPB (Oxford). 19:659–666. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Borromeo MD, Savage TK, Kollipara RK, He
M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH
and Johnson JE: ASCL1 and NEUROD1 reveal heterogeneity in pulmonary
neuroendocrine tumors and regulate distinct genetic programs. Cell
Rep. 16:1259–1272. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang W, Lv B, Yang B, Chen Y, Yuan F, Ma
L, Chen S, Zhang S and Xia J: TREM2 acts as a tumor suppressor in
hepatocellular carcinoma by targeting the PI3K/Akt/β-catenin
pathway. Oncogenesis. 8:92019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pinto R, Assis J, Nogueira A, Pereira C,
Coelho S, Brandão M, Dias J, Alves S, Pereira D and Medeiros R:
Pharmacogenomics in epithelial ovarian cancer first-line treatment
outcome: Validation of GWAS-associated NRG3 rs1649942 and BRE
rs7572644 variants in an independent cohort. Pharmacogenomics J.
19:25–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Al-Nashmi M, Taha S, Alsharoqi I and
Bakhiet M: Interleukin 1 receptor antagonist and
2′-5′-oligoadenylate synthetase-like molecules as novel biomarkers
for multiple sclerosis patients in Bahrain. Mult Scler Relat
Disord. 18:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gu Z, Li Y, Yang X, Yu M, Chen Z, Zhao C,
Chen L and Wang L: Overexpression of CLC-3 is regulated by XRCC5
and is a poor prognostic biomarker for gastric cancer. J Hematol
Oncol. 11:1152018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang H, Ma L, Wang Y, Zuo W, Li B, Yang Y,
Chen Y, Chen L, Wang L and Zhu L: Activation of ClC-3 chloride
channel by 17β-estradiol relies on the estrogen receptor α
expression in breast cancer. J Cell Physiol. 233:1071–1081. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chan TC, Li CF, Ke HL, Wei YC, Shiue YL,
Li CC, Yeh HC, Lee HY, Huang SK, Wu WJ and Li WM: High TNFAIP6
level is associated with poor prognosis of urothelial carcinomas.
Urol Oncol. 37:293.e11–293.e24. 2019. View Article : Google Scholar
|
|
39
|
Pencik J, Pham HT, Schmoellerl J, Javaheri
T, Schlederer M, Culig Z, Merkel O, Moriggl R, Grebien F and Kenner
L: JAK-STAT signaling in cancer: From cytokines to non-coding
genome. Cytokine. 87:26–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wubetu GY, Utsunomiya T, Ishikawa D,
Yamada S, Ikemoto T, Morine Y, Iwahashi S, Saito Y, Arakawa Y,
Imura S, et al: High STAT4 expression is a better prognostic
indicator in patients with hepatocellular carcinoma after
hepatectomy. Ann Surg Oncol. 21 (Suppl 4):S721–S728. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cui MX, Jiang JF, Min GN, Han W and Wu YJ:
Ciliary neurotrophic factor analogue aggravates
CCl4-induced acute hepatic injury in rats. Can J Physiol
Pharmacol. 95:620–623. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hayashi T, Yamashita T, Terashima T, Suda
T, Okada H, Asahina Y, Hayashi T, Hara Y, Nio K, Sunagozaka H, et
al: Serum cytokine profiles predict survival benefits in patients
with advanced hepatocellular carcinoma treated with sorafenib: A
retrospective cohort study. BMC Cancer. 17:8702017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
González-Arriagada WA, Lozano-Burgos C,
Zúñiga-Moreta R, González-Díaz P and Coletta RD:
Clinicopathological significance of chemokine receptor (CCR1, CCR3,
CCR4, CCR5, CCR7 and CXCR4) expression in head and neck squamous
cell carcinomas. J Oral Pathol Med. 47:755–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang Q, Ma XC, Yang X, Wang W, Li Y, Lv
Z, Corrigan CJ, Chen Y and Ying S: Expression of IL-17A, E, and F
and their receptors in non-small-cell lung cancer. J Biol Regul
Homeost Agents. 32:1105–1116. 2018.PubMed/NCBI
|
|
45
|
Tulotta C, Lefley DV, Freeman K, Gregory
WM, Hanby AM, Heath PR, Nutter F, Wilkinson JM, Spicer-Hadlington
AR, Liu X, et al: Endogenous production of IL1B by breast cancer
cells drives metastasis and colonization of the bone
microenvironment. Clin Cancer Res. 25:2769–2782. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bornstein S, Schmidt M, Choonoo G, Levin
T, Gray J, Thomas CR Jr, Wong M and McWeeney S: IL-10 and integrin
signaling pathways are associated with head and neck cancer
progression. BMC Genomics. 17:382016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen K, Liu Q, Tsang LL, Ye Q, Chan HC,
Sun Y and Jiang X: Human MSCs promotes colorectal cancer
epithelial-mesenchymal transition and progression via
CCL5/β-catenin/Slug pathway. Cell Death Dis. 8:e28192017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Q, Bai X, Chen W, Ma T, Hu Q, Liang
C, Xie S, Chen C, Hu L, Xu S and Liang T: Wnt/β-catenin signaling
enhances hypoxia-induced epithelial-mesenchymal transition in
hepatocellular carcinoma via crosstalk with hif-1α signaling.
Carcinogenesis. 34:962–973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen W, Ma T, Shen XN, Xia XF, Xu GD, Bai
XL and Liang TB: Macrophage-induced tumor angiogenesis is regulated
by the TSC2-mTOR pathway. Cancer Res. 72:1363–1372. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hu X, Zhao Y, He X, Li J, Wang T, Zhou W,
Wan D, Wang H and Gu J: Ciliary neurotrophic factor receptor alpha
subunit-modulated multiple downstream signaling pathways in hepatic
cancer cell lines and their biological implications. Hepatology.
47:1298–1308. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mumm JB, Emmerich J, Zhang X, Chan I, Wu
L, Mauze S, Blaisdell S, Basham B, Dai J, Grein J, et al: IL-10
elicits IFNγ-dependent tumor immune surveillance. Cancer Cell.
20:781–796. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang S, Gao X, Shen G, Wang W, Li J, Zhao
J, Wei YQ and Edwards CK: Interleukin-10 deficiency impairs
regulatory T cell-derived neuropilin-1 functions and promotes Th1
and Th17 immunity. Sci Rep. 6:242492016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huber S, Gagliani N, Esplugues E, O'Connor
W Jr, Huber FJ, Chaudhry A, Kamanaka M, Kobayashi Y, Booth CJ,
Rudensky AY, et al: Th17 cells express interleukin-10 receptor and
are controlled by Foxp3− and Foxp3+ regulatory CD4+ T
cells in an interleukin-10-dependent manner. Immunity. 34:554–565.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dennis KL, Blatner NR, Gounari F and
Khazaie K: Current status of interleukin-10 and regulatory T-cells
in cancer. Curr Opin Oncol. 25:637–645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ahmad N, Ammar A, Storr SJ, Green AR,
Rakha E, Ellis IO and Martin SG: IL-6 and IL-10 are associated with
good prognosis in early stage invasive breast cancer patients.
Cancer Immunol Immunother. 67:537–549. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
El-Emshaty HM, Nasif WA and Mohamed IE:
Serum cytokine of IL-10 and IL-12 in chronic liver disease: The
immune and inflammatory response. Dis Markers. 2015:7072542015.
View Article : Google Scholar : PubMed/NCBI
|