Introduction
Recently, artificial intelligence (AI) has made
remarkable progress in medicine. Humanity will undergo a dramatic
and irreversible change when AI becomes very advanced, which will
likely occur in this century (1). AI
has exceeded human experts in the field of games with perfect
results (2), revealing novel
strategies or findings. Therefore, as AI may be able to recognize
certain information that conventional procedures cannot, it may
also provide more precise diagnosis in practical medicine.
Additionally, AI may be able to assist clinicians in practical
medicine, reducing time and effort. For example, it has been
reported that using AI-assisted colposcopy may reduce the time and
effort it takes for a gynecologist to become a colposcopy expert,
resulting in more time to improve other skills, training and
activities (3). Moreover, the use of
AI for predicting live births from blastocysts, to a level similar
to that of specialists, may result in time saved for embryologists,
reducing the financial costs of training (4). The aim of the present study was to
investigate the feasibility of applying deep learning, a type of AI
using both image and non-image information simultaneously, for
gynecological clinical practice.
Uterine cervical cancer is a major public health
problem as it is the third most common cancer in women and the
leading cause of cancer-associated mortality among women in Central
America, South-Central Asia, Middle and Western Africa and
Melanesia (5). New methodologies to
prevent cervical cancer should be made available and accessible to
women in all countries (5).
Colposcopy is a well-established procedure for
examining the uterine cervix under magnification (6–8). When
lesions are treated with 3–5% acetic acid, colposcopy can detect
and recognize cervical intraepithelial neoplasia (CIN) (6). Classification systems, such as the
Bethesda system established in 2002, are used to categorize lesions
as low-grade squamous intraepithelial lesions (LSILs) or high-grade
SILs (HSILs) (9,10), previously referred to as CIN1 and
CIN2/CIN3, respectively (9). In
clinical practice, distinguishing HSIL from LSIL in biopsy
specimens is important as further examination or treatment, such as
conization, may be required for HSIL.
In 2003, Burd (11)
revealed that the Human papilloma virus (HPV) is essential to the
transformation of the cervical epithelium. Based on genomic
differences, DNA sequencing has identified >200 types of HPV,
which can be grouped into low-risk (including types 6, 11, 42, 43
and 44) and high-risk (including types 16, 18, 31, 33, 34, 35, 39,
45, 51, 52, 56, 58, 59, 66, 68 and 70) HPV. In the high-risk group,
certain HPV types are less frequently identified in cancers but are
often present in SIL cells. The risk of progression for HPV types
16 and 18 is greater by ~40% compared with that for other HPV types
(11). Thus, HPV types may be
associated with SILs because high-risk HPV may be more detectable
in HSILs compared with LSILs. Information on HPV types may be
beneficial to SIL diagnosis. However, the possibility of combining
colposcopy findings with HPV types has not been previously
explored.
Deep learning with a convolutional neural network
(12,13) to the realm of AI was applied to
develop an original classifier for predicting HSIL or LSIL from
colposcopy images (3) and HPV types.
The aim of the present study was to determine whether AI could
accurately evaluate colposcopy findings (combined with HPV types),
compared with conventional colposcopy findings by gynecologic
oncologists, and also to investigate the feasibility of applying
deep learning (a class of AI using both image and non-image
information simultaneously) in clinical gynecological clinical
practice.
Materials and methods
Patients
The present study used fully de-identified patient
data and was approved by the Institutional Review Board of Shikoku
Cancer Center (approval no. 2017-81). The study was explained to
patients who were not limited by age, had no prior treatment of the
uterine cervix and had advanced lesions of the cervix biopsied at
Shikoku Cancer Center between January 2012 and December 2017.
Patients were directed to a website with additional information,
including an opt-out option for the study. As the present study was
a fully de-identified retrospective study, the Institutional Review
Board of Shikoku Cancer Center approved the informed consent by the
explanation including the opt-out option for patients to choose to
withdraw from this study as informed consent. HPV tests had been
performed in routine examination for patients with abnormal
cervical cytology reports or abnormal colposcopy findings
indicating neoplastic disease diagnosed by gynecological
oncologists at Shikoku Cancer Center. HPV tests were not performed
specifically for this study. Gynecological oncologists determined
the necessity for biopsy in routine conventional practice for
patients with abnormal cervical cytology reports of ASC-US, LSIL,
HSIL, atypical squamous cells cannot ruled out HSIL (ASC-H),
squamous cell carcinoma (SCC), adenocarcinoma in situ (AIS),
atypical glandular cells (AGC) and adenocarcinoma (Adenoca), as
well as for patients with abnormal cervical cytology reports or
abnormal colposcopy findings indicating neoplastic disease such as
LSIL/CIN1, HSIL/CIN2 and HSIL/CIN3. HPV types were tested by either
one of the following commercially available PCR-based assay kits:
Cobas® 4800 system HPV (Roche Diagnostics), which
detects high-risk HPV genotypes such as 16, 18, 31, 33, 35, 39, 45,
51, 52, 56, 58, 59, 66 and 68; or Amplicor® HPV (Roche
Diagnostics), which detects high-risk HPV genotypes such as 16, 18,
31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68. The design of the
study was based on the routine practice at Shikoku Cancer Center. A
total of 253 patients who underwent cervical punch biopsy combined
with HPV typing test and had an image of colposcopy captured were
enrolled in this study.
Images
Colposcopy images of lesions processed with 3%
acetic acid prior to biopsy were captured, cropped and saved in
JPEG format. The image data were input retrospectively for deep
learning.
AI preparation
All de-identified images saved offline were
transferred to the AI system. For the test dataset, 20% of the
images were randomly selected; the remaining images were used as
the training dataset. Next, 80% of the training dataset images were
used to train the AI classifier, and the remaining images were used
as the validation dataset. Thus, these datasets did not overlap.
The AI classifier was trained using a training dataset and
simultaneously validated and tested using the test dataset. The
training datasets were augmented, because colposcopy image
processing of arbitrary degrees of rotation can yield images
resulting in different vector data for the same category.
AI classifier
Classifier programs were developed using supervised
deep learning with a convolutional neural network architecture
(12,14) catenated with a HPV type tensor. A
number of convolutional neural networks were tested by varying
image size (50×50, 75×75 and 100×100 pixels), L2 regularization
(15,16) and architectures consisting of a
combination of convolution layers with kernels (17–19),
pooling layers (20–23), flattened layers (24), linear layers (25,26),
rectified linear unit layers (27,28),
catenated layers, batch normalization layers (29) and a softmax layer (30,31)
which demonstrated the probability of LSIL or HSIL from an image
(Table I).
 | Table I.Architecture of the classifier. |
Table I.
Architecture of the classifier.
| Input Image | Input HPV Type |
|---|
| 1.
Convolutional layer | − |
| 2.
Rectified linear unit layer | − |
| 3.
Pooling layer | − |
| 4.
Convolutional layer | − |
| 5.
Rectified linear unit layer | − |
| 6.
Pooling layer | − |
| 7.
Flattening layer | − |
| 8.
Linear layer | − |
| 9.
Rectified linear unit layer | − |
| 10. Linear
layer | 1. HPV type |
| Catenated
layer |
|
| Batch normalization
layer |
|
| Linear layer |
|
| Softmax layer |
|
| Output |
|
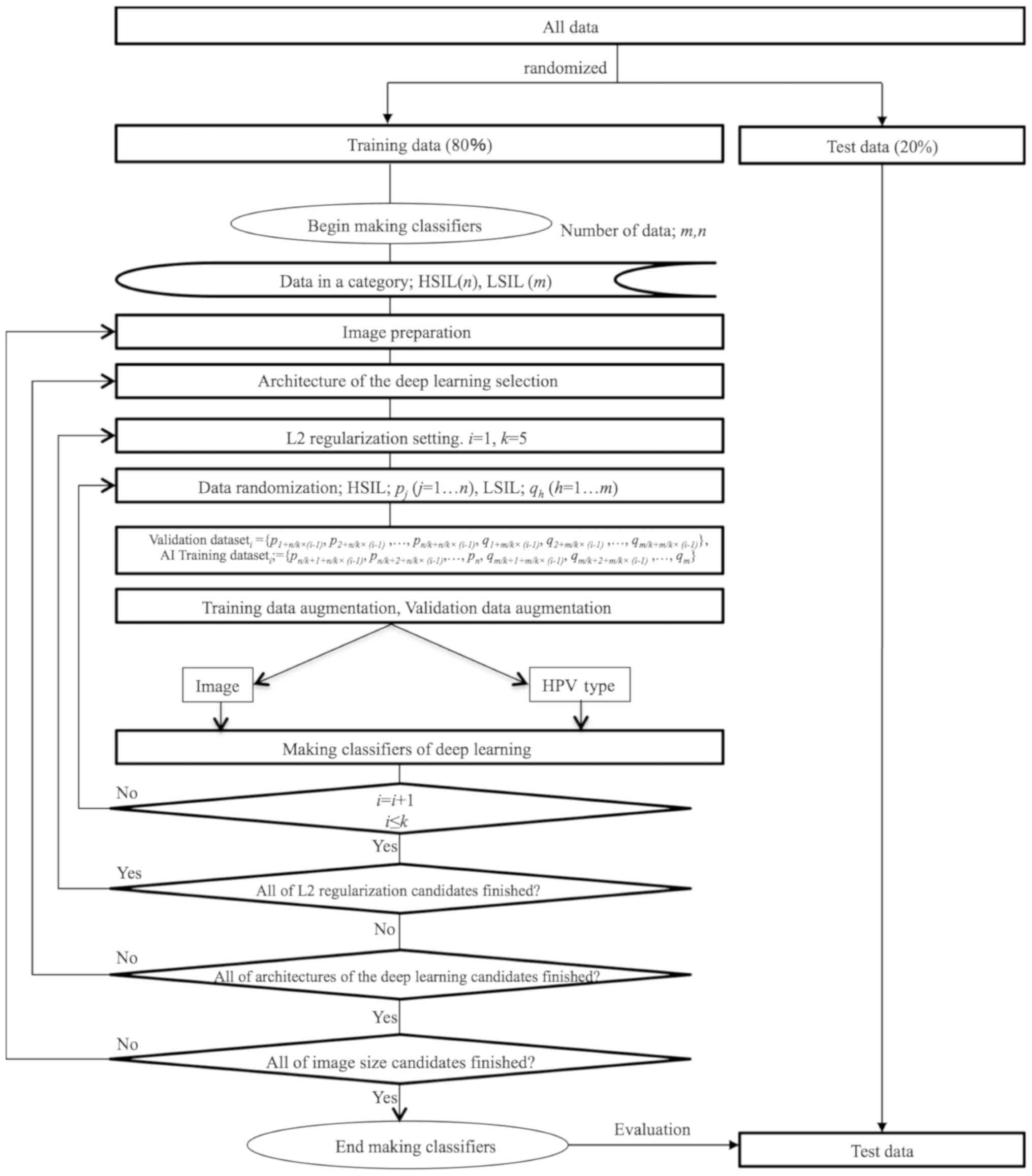
Cross-validation (32–34),
which is a method for model selection, was applied to identify the
optimal machine learning method. The suitable number of images for
the training data was determined using the 5-fold cross-validation
method, which reveals the optimal number of training data and can
be used to avoid overfitting, a modeling error that occurs when a
classifier is too closely fit to a limited set of data points
(35–40). After the optimal number of training
data was calculated, the classifier that exhibited the highest
accuracy was selected. Conventional colposcopy diagnosis and AI
colposcopy diagnosis-catenated HPV types in the test dataset were
compared. A flow chart of the development of the AI classifier is
presented in Fig. 1.
Development environment
The following development environment was used: A
Macintosh running OS X 10.14.5 (Apple, Inc.) and Mathematica
12.0.0.0 (Wolfram Research, Inc.).
Statistical analysis
The laboratory and AI classifier data were compared
using Mathematica 12.0.0.0 (Wolfram Research, Inc.). The Cochran
Armitage test, Cohen's κ, χ2 test and Fisher's exact
test were used. P<0.05 was considered to indicate a
statistically significant difference.
Results
The number of patients with cytology reports were
stratified as follows: HSIL, 149; LSIL, 75; AUC-US, 43; ASC-H, 38;
SCC, 18; AGC, 2; AIS; 2; Adenoca, 2; NILM, 8. Mean ± standard
deviation, median and range of patient age in the HSIL vs. LSIL
groups were 31.66±5.01 vs. 33.75±8.94, 32 vs. 33 and 19–46 vs.
19–62, respectively. The pathological diagnoses and the
corresponding number of patients who underwent punch biopsy were as
follows: HSIL, 213; LSIL, 97; squamous cell carcinoma, 12;
adenocarcinoma, 5; adenocarcinoma in situ, 2; and
microinvasive squamous cell carcinoma, 1 (Table II). The HPV types of the patients
were as follows: Type 16, 87; type 18, 8; type 16 and 18, 4;
high-risk HPV but not type 16 or 18, 159; and HPV negative, 13. A
total of 57 patients (17.2%) did not receive a HPV type test.
Conventional colposcopy diagnoses based on pathological results and
HPV types are presented in Tables
III and IV. A total of 20/273
patients had no image data.
 | Table II.Patients with pathological results
confirmed by punch biopsy and different HPV types. |
Table II.
Patients with pathological results
confirmed by punch biopsy and different HPV types.
|
| Pathological
results |
|---|
|
|
|
|---|
| HPV type | HSIL | LSIL | Microinvasive
SCC | Invasive SCC | Adenocarcinoma
in situ | Adenocarcinoma |
|---|
| Not available | 3 | 54 | 0 | 0 | 0 | 0 |
| HPV-negative | 6 | 6 | 0 | 0 | 0 | 1 |
| High risk but not
type 16 or 18 | 123 | 33 | 1 | 2 | 0 | 0 |
| Type 16 | 75 | 2 | 0 | 8 | 0 | 2 |
| Type 18 | 5 | 2 | 0 | 0 | 2 | 1 |
| Type 16+18 | 1 | 0 | 0 | 2 | 0 | 1 |
 | Table III.Patients with pathological results
confirmed by punch biopsy and conventional colposcopy diagnosis by
gynecologic oncologists. |
Table III.
Patients with pathological results
confirmed by punch biopsy and conventional colposcopy diagnosis by
gynecologic oncologists.
|
| Colposcopy
diagnosis |
|---|
|
|
|
|---|
| Pathological
results | CIN1 (LSIL) | CIN2 (HSIL) | CIN3 (HSIL) | Cervicitis | Invasive
cancer |
|---|
| HSIL | 32 | 63 | 114 | 1 | 3 |
| LSIL | 70 | 17 | 5 | 5 | 0 |
| Microinvasive
SCC | 0 | 0 | 1 | 0 | 0 |
| Invasive SCC | 0 | 0 | 4 | 0 | 8 |
| Adenocarcinoma
in situ | 0 | 0 | 2 | 0 | 0 |
| Adenocarcinoma | 0 | 0 | 1 | 0 | 4 |
 | Table IV.Patients with all types of HPV and
the conventional colposcopy diagnosis by gynecologic
oncologists. |
Table IV.
Patients with all types of HPV and
the conventional colposcopy diagnosis by gynecologic
oncologists.
|
| Colposcopy
diagnosis |
|---|
|
|
|
|---|
| HPV type | CIN1 (LSIL) | CIN2 (HSIL) | CIN3 (HSIL) | Cervicitis | Invasive
cancer |
|---|
| Not available | 48 | 9 | 0 | 0 | 0 |
| HPV-negative | 4 | 5 | 1 | 2 | 1 |
| High risk but not
type 16 or 18 | 40 | 46 | 70 | 2 | 1 |
| Type 16 | 9 | 18 | 50 | 1 | 9 |
| Type 18 | 1 | 2 | 5 | 1 | 1 |
| Type 16 + 18 | 0 | 0 | 1 | 0 | 3 |
A total of 253 patients with colposcopy images,
pathological LSIL or HSIL and known HPV types were finally enrolled
in this study. The ages of patients with pathological HSIL and LSIL
were 31.66±4.83 and 30.12±5.10 (mean ± standard deviation),
respectively (data not shown). The median age (range) of
pathological HSIL and LSIL were 32 (19–46) and
33 (19–62), respectively (data not shown). HPV
types were reclassified as follows: Type 16 or 18 were considered
high-risk HPV, and not type 16 or 18 was considered to represent
low-risk HPV/HPV-negative due to the limited number of available
HPV types and pathological results. HPV-negative described the
absence of high-risk HPV, not type 16 or 18.
The numbers of patients with type 16 and/or 18,
high-risk HPV but not type 16 or 18 and HPV-negative were 85, 156
and 12, respectively (Table II).
The numbers of patients with HSIL and LSIL were 210 and 43,
respectively. Among the 210 pathological HSIL cases, the numbers of
type 16 and/or 18, high-risk HPV but not type 16 or 18 and
HPV-negative were 81, 123 and 6, respectively. Among the 43
pathological LSIL cases, the numbers of type 16 and/or 18,
high-risk HPV but not type 16 or 18 and HPV-negative were 4, 33 and
6, respectively. The HPV types were associated with the
pathological results (P<6.70×10−6; Cochran Armitage
test). HPV-negative results were observed in 2.85% (6/210) and
14.00% (6/43) of HSIL and LSIL, respectively. In the present study,
type 16- or 18-positive HPV in pathological HSIL and LSIL were
observed in 38.6 and 9.3% of cases, respectively. The incidence of
type 16 and/or 18 positivity in pathological HSIL was significantly
higher, compared with that in LSIL (P<0.0005; Fisher's exact
test with Yates's correction).
Among the 210 pathological HSIL cases, 177 patients
received a conventional colposcopy diagnosis by gynecologists of
CIN2 (HSIL) or CIN3 (HSIL), 29 were diagnosed with CIN1 (LSIL),
three were diagnosed with invasive cancer and one was diagnosed
with cervicitis. Among the 43 pathological LSIL cases, 13 received
a conventional colposcopy diagnosis by gynecologists of HSIL, 25
were diagnosed with LSIL and 5 with cervicitis. The accurate
diagnoses of HSIL and LSIL were 202 out of 253 (0.798). The
accuracy, sensitivity, specificity, positive predictive value,
negative predictive value and Youden's J index of the conventional
colposcopy diagnosis for pathological HSIL were 0.828 (202/244),
0.859 (177/206), 0.658 (25/38), 0.932 (177/190), 0.463 (25/54) and
0.517, respectively.
Among the 85 cases with HPV type 16 and/or 18, 71
received a conventional colposcopy diagnosis by gynecologists of
CIN2 (HSIL) or CIN3 (HSIL), 10 were diagnosed with CIN1 (LSIL), two
with cervicitis and two with invasive cancer. Among the 156 cases
with HPV type 16 and/or 18, 113, 40, 2 and 1 received a
conventional colposcopy diagnosis by gynecologists of CIN2 (HSIL)
or CIN3 (HSIL), CIN1 (LSIL), cervicitis and invasive cancer,
respectively. Among the 12 HPV-negative cases 6, 4 and 2 received a
conventional colposcopy diagnosis by gynecologists of CIN2 (HSIL)
or CIN3 (HSIL), CIN1 (LSIL) and cervicitis, respectively. There are
no relationships between HPV types and colposcopy.
The highest accuracy for HSIL of the best AI
classifier combined with HPV types for a test dataset was 0.941
(48/51) when the number of the augmented training dataset was
1,212, the value of L2 regularization was 0.02, and the image size
was 50×50 pixels. The accuracy, sensitivity, specificity, positive
predictive value, negative predictive value, Youden's J index
(41), the area under the receiver
operating characteristic curve (AUC) ± standard error, the 95%
confidence interval of the AUC and Cohen's k (42) coefficients of HSIL for the AI
colposcopy combined with HPV types and pathological results are
presented in Table V.
 | Table V.The results of the best AI classifier
combined with HPV types and conventional colposcopy for 51 test
datasets (20% of all qualified datasets). |
Table V.
The results of the best AI classifier
combined with HPV types and conventional colposcopy for 51 test
datasets (20% of all qualified datasets).
| Variable | AI | Conventional
colposcopy |
|---|
| Accuracy | 0.941 (48/51) | 0.843 (43/51) |
| Sensitivity | 0.956 (43/45) | 0.844 (38/45) |
| Specificity | 0.833 (5/6) | 0.833 (5/6) |
| Positive predictive
value | 0.977 (43/44) | 0.974 (38/39) |
| Negative predictive
value | 0.714 (5/7) | 0.500 (6/12) |
| Youden's J
index | 0.789 | 0.677 |
| AUC (± standard
error) | 0.963±0.026 | N/A |
| Cohen's κ | 0.769 | 0.473 |
The comparison of the conventional colposcopy
diagnosis by gynecological oncologists and the best AI classifier
for the test dataset is presented in Table VI. As the AI classifier was not
trained for cervicitis or invasive cancer, when the colposcopy
diagnosis was limited to HSIL and LSIL by ignoring colposcopy
diagnoses of cervicitis and invasive cancer, the Cohen's k
coefficient of the colposcopy diagnosis and the AI classifier was
0.407. The agreement of the two methods was moderate (43), but not significant (P=0.077).
 | Table VI.Comparison of the diagnosis of
conventional colposcopy by gynecological oncologists and the best
AI classifier and the pathological results for patients with HSIL
or LSIL in the test dataset. |
Table VI.
Comparison of the diagnosis of
conventional colposcopy by gynecological oncologists and the best
AI classifier and the pathological results for patients with HSIL
or LSIL in the test dataset.
|
| AI diagnosis (image
combined with HPV type) | Pathological
diagnosis |
|---|
|
|
|
|
|---|
| Colposcopy
Diagnosis | HSIL | LSIL | HSIL | LSIL |
|---|
| HSIL | 36 | 2 | 38 | 0 |
| LSIL | 6 | 4 | 5 | 5 |
| Cervicitis | 1 | 1 | 1 | 1 |
| Invasive
cancer | 1 | 0 | 1 | 0 |
The comparison of the conventional colposcopy
diagnosis by gynecological oncologists and the pathological results
for the test dataset is presented in Table VI. The accurate number of HSIL and
LSIL by conventional colposcopy for the test dataset was 43 of 51
(0.843). The conventional colposcopy results for the test dataset
and for all of the datasets were not significantly different, and
the time required for classification was <0.2 sec per
patient.
Discussion
In the present study, a classifier was developed
using deep learning with convolutional neural networks using images
of cervical SILs combined with HPV types to predict the
pathological diagnosis. The accuracy for the test dataset achieved
by the classifier and by gynecological oncologists was 0.941 and
0.843, respectively; the latter accuracy was calculated
tentatively, and these two accuracies could not be compared as the
AI was trained for HSIL and LSIL classes, whereas colposcopy could
identify lesions such as cervicitis, invasive cancer and
adenocarcinoma. The numbers of accurate HSIL and LSIL diagnoses by
conventional colposcopy for the test dataset were 43 out of 51 and
for all datasets were and 202 out of 253. Compared with the
classifier, the conventional colposcopy results for the test
dataset and for all of the datasets were not significantly
different, which suggested that the AI classifier using deep
learning with convolutional neural networks using images of
cervical SILs combined with HPV types was not inferior to
conventional colposcopy performed by gynecologic oncologists.
In the present study, 12 cases of pathological HSIL
and LSIL were HPV-negative, although both HPV type information and
colposcopy images were used for analysis. These cases may have
represented false negatives as HPV infection is essential to the
transformation of cervical epithelial cells (11) and the HPV detection kits that were
commercially available and widely used result in <3.1% of false
negatives in pathological HSIL in Japan as stated by the
manufacturer. The data not excluded as only a small number of
HPV-negative cases were identified. Previous studies have reported
8–13% of false negative results for HPV detection (44–46). Lee
et al (47) reported that the
classic nested PCR and Sanger DNA sequencing technology for routine
HPV testing exhibited that a true negative HPV PCR invariably
indicated the absence of precancerous cells in the cytology
samples.
The accuracy of 0.941 was an acceptable result of
the classifier for deep learning. In medicine, several studies have
used AI for deep learning with convolutional neural networks
(48,49). The accuracy values of AI with deep
learning have been published and include 0.997 for the
histopathological diagnosis of breast cancer (50), 0.980 for the morphological quality of
blastocysts and evaluation by an embryologist (51), 0.640–0.880 for predicting live birth
from a blastocyst image of patients by age (4,52), 0.650
for predicting live birth without aneuploidy from a blastocyst
image (53), 0.823 (3), 0.720 (54) and 0.500 (55) for colposcopy, 0.830 to 0.900 for the
early diagnosis of Alzheimer's disease (56), 0.830 for urological dysfunctions
(57) and 0.830 for the diagnostic
imaging of orthopedic trauma (58).
A number of studies have reported a limitation of conventional
colposcopy. A study of the accuracy of biopsy under colposcopy
reported a total biopsy failure rate, comprising both non-biopsy
and incorrect selection of biopsy site, of 0.200 in CIN1, 0.110 in
CIN2 and 0.090 in CIN3 (59). The
colposcopic impression of high-grade CIN had a specificity of 0.880
and a sensitivity of 0.540, as determined by nine expert
colposcopists in 100 cervigrams (60). The sensitivity of an online
colpophotographic assessment of HSIL by 20 colposcopists was 0.390
(61). Thus, conventional colposcopy
does not provide good sensitivity, even when performed by
colposcopy specialists. By contrast, the accuracy and sensitivity
reported in this study for predicting HSIL from colposcopy images
combined with HPV types using deep learning were 0.941 and 0.956,
respectively, which appears to be satisfactory. Since the
classifier was not trained in colposcopy findings such as mosaic
acetowhite epithelium and punctuation, it may recognize certain
morphological features of cervical SILs by itself. It is also
possible that the AI classifier may recognize features that
colposcopists do not, such as relative or absolute brightness of
acetowhite, complexity of the shape of the lesion, quantitative
marginal evaluation of borders and distribution of punctuation
density. The pathological results in the present study were
obtained and defined by punch biopsy as it was not recommended for
patients with CIN1 (LSIL) diagnosed by colposcopy to undergo
conization or hysterectomy. The advanced lesion would have been
revealed if the pathological results were defined by conization or
hysterectomy rather than by punch biopsy; thus, both conventional
colposcopy and the AI classifier may have demonstrated different
results. When AI is used for advanced diseases, such as squamous
cell carcinoma and adenocarcinoma, the pathological diagnosis
should be provided by conization or hysterectomy.
It is important for clinicians to distinguish HSIL
from LSIL in biopsy specimens in clinical practice as further
examination or treatment, such as conization, may be required for
HSIL. The clinician should consider biopsy when a reliable
classifier indicates HSIL in clinical practice. The classifier
developed in the present study may help untrained gynecologists
avoid or reduce the risk of misidentifying HSIL. When the
performance of the AI classifier is further improved in accuracy,
sensitivity and specificity for classifying SILs, gynecologists may
be able to obtain more precise classification without requiring a
colposcopy specialist.
Several reasons for obtaining high accuracy by AI
were considered in the present study. First, the association
between the pathological results, colposcopy diagnosis and HPV
types was important. The pathological results were affected by the
HPV types. However, no association was identified between HPV types
and the results of colposcopy. Thus, HPV types and colposcopy were
associated with pathological results, but not with each other. In
our preliminary study, the accuracy achieved by deep learning with
only images of colposcopy was 0.823 (data not shown). Thus, the
association among the pathological results, colposcopy diagnosis
and HPV type may be a reason for high accuracy.
Second, AI has the ability to use images and
non-image data simultaneously. However, AI is not established to
digitize images to numerical data indicating the features of the
images for multivariate analysis; AI, including deep learning, can
acquire numerical data to indicate the features of an image and use
the numerical data indicating the features of colposcopy images and
the numeric tensor data of HPV types. This is an important feature
of AI, which may be the second reason for high accuracy in this
study from the perspective of computer science.
Third, in the present study, the neural network
architecture including a batch normalization layer (29) was adequate. Neural network
architecture is a key component of deep learning. A batch
normalization layer was added following catenating information from
colposcopy images and HPV types. This method makes normalization a
part of the model architecture and performs the normalization for
each training mini-batch. Batch normalization allows the use of
high learning rates. This architecture may be the third reason for
high accuracy in the present study.
The architecture of the neural network has been
progressing. The LeNet study in 1998 (62) consisted of 5 layers. AlexNet in 2012
(30) consisted of 14, and Google
Net in 2014 (26) was constructed
from a combination of micronetworks. ResNet-50 in 2015 (63) consisted of modules with a shortcut
process. Squeeze-and-excitation networks were first published in
2017 (64). However, AI for image
recognition remains in development. Image information is one of the
parameters requiring further investigation. Only 15×15 pixels have
been used to detect cervical cancer (65); thus, image size remains an issue. In
a previous colposcopy study, the reported accuracy for images of
150×150 pixels was higher compared with that for images of 300×300
or 32×32 pixels (55). In the
present study, 111×111, 70×70 and 50×50 pixel images were tested. A
size of 50×50 pixels, which exhibited the best performance in the
present study, falls within the acceptable range. Regularization
values that are routinely used in developing AI of deep learning
are also an important hyperparameter for constructing a good
classifier that avoids overfitting (35–40).
Selecting the appropriate number of training datasets is also very
important; in addition, the validation dataset prevents
overfitting. Generally, more varied patterns of images may be
needed for datasets as 500–1,000 images are reportedly prepared for
each class during image classification with deep learning (52,66). The
classifier that uses both image and HPV types may require more
images combined with HPV types, which may result in improvement in
the classifier with deep learning.
In the present study, a classifier was developed
based on deep learning that used both HPV types and images of
uterine cervical SILs to predict pathological HSIL/LSIL. The
accuracy of the classifier was 0.941. Although further study using
more datasets and modified neural network architecture and/or
hyperparameters is required to validate the classifier, the results
of the present study demonstrated that AI may have a potential for
clinical use in colposcopy examinations and may provide benefits to
patients and clinicians.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the
current study are not publicly available since data sharing is not
approved by the Institutional Review Board of Shikoku Cancer Center
(approval no. 2017-81) but are available from the corresponding
author on reasonable request.
Author's contributions
YM designed the study, programmed the AI, produced
the AI classifiers, performed statistical analysis and wrote the
manuscript. YM and YN designed the AI architecture. KT performed
clinical intervention, data entry and collection, designed the
study and critically reviewed the manuscript. YN critically
reviewed the manuscript. TM designed the study and critically
reviewed the manuscript.
Ethics approval and consent to
participate
The protocol for this retrospective study used fully
de-identified patient data and was approved by the Institutional
Review Board of Shikoku Cancer Center (approval no. 2017-81). This
study was explained to patients, who were also directed to a
website with additional information, including an opt-out option
that informed them of their right to not participate in this study.
Written informed consent for this study design was not required,
according to the guidance of the Ministry of Education, Culture,
Sports, Science and Technology of Japan.
Patient consent for publication
Not applicable.
Competing interests
YM, YN and TM declare that they have no competing
interests. KT declares receipt of personal funding from Taiho
Pharmaceuticals, Chugai Pharma, AstraZeneca, Nippon Kayaku, Eisai,
Ono Pharmaceutical, Terumo Corporation and Daiichi Sankyo.
References
|
1
|
Müller VC and Bostrom N: Future progress
in artificial intelligence: A survey of expert opinion. In:
Fundamental Issues of Artificial Intelligence. Springer; Cham: pp.
555–572. 2016
|
|
2
|
Silver D, Schrittwieser J, Simonyan K,
Antonoglou I, Huang A, Guez A, Hubert T, Baker L, Lai M, Bolton A,
et al: Mastering the game of go without human knowledge. Nature.
550:354–359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miyagi Y, Takehara K and Miyake T:
Application of deep learning to the classification of uterine
cervical squamous epithelial lesion from colposcopy images. Mol
Clin Oncol. 11:583–589. 2019.PubMed/NCBI
|
|
4
|
Miyagi Y, Habara T, Hirata R and Hayashi
N: Feasibility of predicting live birth by combining conventional
embryo evaluation with artificial intelligence applied to a
blastocyst image in patients classified by age. Reprod Med Biol.
18:344–356. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arbyn M, Castellsagué X, de Sanjosé S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Ann Oncol. 22:2675–2686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
García-Arteaga JD, Kybic J and Li W:
Automatic colposcopy video tissue classification using higher order
entropy-based image registration. Comput Biol Med. 41:960–970.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kyrgiou M, Tsoumpou I, Vrekoussis T,
Martin-Hirsch P, Arbyn M, Prendiville W, Mitrou S, Koliopoulos G,
Dalkalitsis N, Stamatopoulos P and Paraskevaidis E: The up-to-date
evidence on colposcopy practice and treatment of cervical
intraepithelial neoplasia: The Cochrane colposcopy and cervical
cytopathology collaborative group (C5 group) approach. Cancer Treat
Rev. 32:516–523. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
O'Neill E, Reeves MF and Creinin MD:
Baseline colposcopic findings in women entering studies on female
vaginal products. Contraception. 78:162–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waxman AG, Chelmow D, Darragh TM, Lawson H
and Moscicki AB: Revised terminology for cervical histopathology
and its implications for management of high-grade squamous
intraepithelial lesions of the cervix. Obstet Gynecol.
120:1465–1471. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Darragh TM, Colgan TJ, Thomas Cox J,
Heller DS, Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM,
Stoler MH, et al: Members of the LAST project work groups. The
lower anogenital squamous terminology standardization project for
HPV-associated lesions: Background and consensus recommendations
from the College of American Pathologists and the American society
for colposcopy and cervical pathology. Int J Gynecol Pathol.
32:76–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burd EM: Human papillomavirus and cervical
cancer. Clin Microbiol Rev. 16:1–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rumelhart D, Hinton G and Williams R:
Learning representations by back-propagating errors. Nature.
323:533–536. 1986. View
Article : Google Scholar
|
|
13
|
Bengio Y, Courville A and Vincent P:
Representation learning: A review and new perspectives. IEEE Trans
Pattern Anal Mach Intell. 35:1798–1828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmidhuber J: Deep learning in neural
networks: An overview. Neural Netw. 61:85–117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Srivastava N, Hinton G, Krizhevsky A,
Sutskever I and Salakhutdinov R: Dropout: A simple way to prevent
neural networks from overfitting. J Mach Lean Res. 15:1929–1958.
2014.
|
|
16
|
Nowlan SJ and Hinton GE: Simplifying
neural networks by soft weight-sharing. Neural Comput. 4:473–493.
1992. View Article : Google Scholar
|
|
17
|
Bengio Y: Learning deep architectures for
AI. Foundations and trends® in machine learning.
2:1–127. 2009. View Article : Google Scholar
|
|
18
|
Mutch J and Lowe DG: Object class
recognition and localization using sparse features with limited
receptive fields. Int J Comput Vision. 80:45–57. 2008. View Article : Google Scholar
|
|
19
|
Neal RM: Connectionist learning of belief
networks. Art Intell. 56:71–113. 1992. View Article : Google Scholar
|
|
20
|
Ciresan DC, Meier U, Masci J, Maria
Gambardella L and Schmidhuber J: Flexible, high performance
convolutional neural networks for image classification. IJCAI'11
Proceedings of the Twenty-Second international joint conference on
artificial intelligence. 2:1237–1242. 2011.
|
|
21
|
Scherer D, Müller A and Behnke S:
Evaluation of pooling operations in convolutional architectures for
object recognition. Artificial Neural Networks-ICANN 2010.
Diamantaras K, Duch W and Iliadis LS: Lecture Notes in Computer
Science Springer; Heidelberg: pp. 92–101. 2010, View Article : Google Scholar
|
|
22
|
Huang FJ and LeCun Y: Large-scale learning
with SVM and convolutional for generic object categorization.
Computer Vision and Pattern Recognition, 2006 IEEE Computer Society
Conference. 1:284–291. 2006.
|
|
23
|
Jarrett K, Kavukcuoglu K, Ranzato M and
LeCun Y: What is the best multi-stage architecture for object
recognition? Computer vision. 12th IEEE international conference on
computer vision. 2146–2153. 2009.
|
|
24
|
Zheng Y, Liu Q, Chen E, Ge Y and Zhao JL:
Time series classification using multi-channels deep convolutional
neural networks. Web-Age Information Management. WAIM 2014. Lecture
notes in computer science. Li F, Li G, Hwang S, Yao B and Zhang Z:
Springer; Cham: pp. 298–310. 2014
|
|
25
|
Mnih V, Kavukcuoglu K, Silver D, Rusu AA,
Veness J, Bellemare MG, Graves A, Riedmiller M, Fidjeland AK,
Ostrovski G, et al: Human-level control through deep reinforcement
learning. Nature. 518:529–533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szegedy C, Liu W, Jia Y, Sermanet P, Reed
S, Anguelov D, Erhan D, Vanhoucke V and Rabinovich A: Going deeper
with convolutions. Proceedings of the IEEE conference on computer
vision and pattern recognition. 1–9. 2015.
|
|
27
|
Glorot X, Bordes A and Bengio Y: Deep
sparse rectifier neural networks. Proceedings of the fourteenth
international conference on artificial intelligence and statistics
PMLR. 15:315–323. 2011.
|
|
28
|
Nair V and Hinton G: Rectified linear
units improve restricted Boltzmann machines. Proceedings of the
27th International Conference on Machine Learning Haifa. 807–814.
2010.
|
|
29
|
Ioffe S and Szegedy C: Batch
Normalization: Accelerating Deep Network Training by Reducing
Internal Covariate Shift. 32nd International Conference on Machine
Learning Lille: 2015
|
|
30
|
Krizhevsky A, Sutskever I and Hinton GE:
ImageNet Classification with Deep Convolutional Neural Networks.
25th International Conference on Neural Information Processing
Systems. 1097–1105. 2012.
|
|
31
|
Bridle JS: Probabilistic interpretation of
feedforward classification network outputs, with relationships to
statistical pattern recognition. Neurocomputing. Soulié FF and
Hérault J: Springer; Berlin: pp. 227–236. 1990, View Article : Google Scholar
|
|
32
|
Kohavi R: A study of cross-validation and
bootstrap for accuracy estimation and model selection. Proceedings
of the 14th international joint conference on artificial
intelligence. 2:1137–1143. 1195.
|
|
33
|
Schaffer C: Selecting a classification
method by cross-validation. Mach Lear. 13:135–143. 1993. View Article : Google Scholar
|
|
34
|
Refaeilzadeh P, Tang L and Liu H:
Cross-Validation. Encyclopedia of Database Systems. Liu L and Özsu
MT: Springer; Boston: pp. 532–538. 2009
|
|
35
|
Yu L, Chen H, Dou Q, Qin J and Heng PA:
Automated melanoma recognition in dermoscopy images via very deep
residual networks. IEEE Trans Med Imaging. 36:994–1004. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Caruana R, Lawrence S and Giles CL:
Overfitting in neural nets: Backpropagation, conjugate gradient,
and early stopping. Advances in neural information processing
systems. 13:402–408. 2001.
|
|
37
|
Baum EB and Haussler D: What size net
gives valid generalization? Neural Computation. 1:151–160. 1989.
View Article : Google Scholar
|
|
38
|
Geman S, Bienenstock E and Doursat R:
Neural networks and the bias/variance dilemma. Neural Computation.
4:1–58. 1992. View Article : Google Scholar
|
|
39
|
Krogh A and Hertz JA: A simple weight
decay can improve generalization. In Advances in neural information
processing systems. 4:950–957. 1992.
|
|
40
|
Moody JE: The effective number of
parameters: An analysis of generalization and regularization in
nonlinear learning systems. Advances in Neural Information
Processing Systems. Moody JE, Hanson SJ and Lippmann RP: Morgan
Kaufmann Publishers Inc.; San Francisco, CA: pp. 847–854. 1992
|
|
41
|
Youden WJ: Index for rating diagnostic
tests. Cancer. 3:32–35. 1950. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cohen J: A coefficient of agreement for
nominal scales. Educ Psychol Meas. 20:37–46. 1960. View Article : Google Scholar
|
|
43
|
McHugh ML: Interrater reliability: The
kappa statistic. Biochem Med (Zagreb). 22:276–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Poljak M, Kovanda A, Kocjan BJ, Seme K,
Jancar N and Vrtacnik-Bokal E: The abbott RealTime high risk HPV
test: Comparative evaluation of analytical specificity and clinical
sensitivity for cervical carcinoma and CIN 3 lesions with the
Hybrid Capture 2 HPV DNA test. Acta Dermatovenerol Alp Pannonica
Adriat. 18:94–103. 2009.PubMed/NCBI
|
|
45
|
Tjalma WA, Fiander A, Reich O, Powell N,
Nowakowski AM, Kirschner B, Koiss R, O'Leary J, Joura EA, Rosenlund
M, et al: Differences in human papillomavirus type distribution in
high-grade cervical intraepithelial neoplasia and invasive cervical
cancer in Europe. Int J Cancer. 132:854–867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
De Sanjose S, Quint WG, Alemany L, Geraets
DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin
HR, et al: Human papillomavirus genotype attribution in invasive
cervical cancer: A retrospective cross-sectional worldwide study.
Lancet Oncol. 11:1048–1056. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee SH, Vigliotti JS, Vigliotti VS and
Jones W: From human papillomavirus (HPV) detection to cervical
cancer prevention in clinical practice. Cancers (Basel).
6:2072–2099. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Miyagi Y, Fujiwara K, Oda T, Miyake T and
Coleman RL: Development of new method for the prediction of
clinical trial results using compressive sensing of artificial
intelligence. J Biostat Biometric App. 3:2032018.
|
|
49
|
Abbod MF, Catto JW, Linkens DA and Hamdy
FC: Application of artificial intelligence to the management of
urological cancer. J Urol. 178:1150–1156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Litjens G, Sánchez CI, Timofeeva N,
Hermsen M, Nagtegaal I, Kovacs I, Hulsbergen-Van De Kaa C, Bult P,
Van Ginneken B and van der Laak J: Deep learning as a tool for
increased accuracy and efficiency of histopathological diagnosis.
Sci Rep. 6:262862016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Khosravi P, Kazemi E, Zhan Q, Toschi M,
Malmsten JE, Hickman C, Meseguer M, Rosenwaks Z, Elemento O,
Zaninovic N and Hajirasouliha I: Robust automated assessment of
human blastocyst quality using deep learning. BioRxiv.
3948822018.
|
|
52
|
Miyagi Y, Habara T, Hirata R and Hayashi
N: Feasibility of deep learning for predicting live birth from a
blastocyst image in patients classified by age. Reprod Med Biol.
18:190–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Miyagi Y, Habara T, Hirata R and Hayashi
N: Feasibility of artificial intelligence for predicting live birth
without aneuploidy from a blastocyst image. Reprod Med Biol.
18:204–211. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Simões PW, Izumi NB, Casagrande RS, Venson
R, Veronezi CD, Moretti GP, da Rocha EL, Cechinel C, Ceretta LB,
Comunello E, et al: Classification of images acquired with
colposcopy using artificial neural networks. Cancer Inform.
13:119–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sato M, Horie K, Hara A, Miyamoto Y,
Kurihara K, Tomio K and Yokota H: Application of deep learning to
the classification of images from colposcopy. Oncol Lett.
15:3518–3523. 2018.PubMed/NCBI
|
|
56
|
Ortiz A, Munilla J, Gorriz JM and Ramirez
J: Ensembles of deep learning architectures for the early diagnosis
of the Alzheimer's disease. Int J Neural Syst. 26:16500252016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gil D, Johnsson M, Chamizo JMG, Paya AS
and Fernandez DR: Application of artificial neural networks in the
diagnosis of urological disfunctions. Expert Syst Appl.
36:5754–5760. 2009. View Article : Google Scholar
|
|
58
|
Olczak J, Fahlberg N, Maki A, Razavian AS,
Jilert A, Stark A, Sköldenberg O and Gordon M: Artificial
intelligence for analyzing orthopedic trauma radiographs. Acta
Orthop. 88:581–586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sideri M, Garutti P, Costa S, Cristiani P,
Schincaglia P, Sassoli de Bianchi P, Naldoni C and Bucchi L:
Accuracy of colposcopically directed biopsy: Results from an online
quality assurance programme for colposcopy in a population-based
cervical screening setting in Italy. Biomed Res Int.
2015:6140352015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sideri M, Spolti N, Spinaci L, Sanvito F,
Ribaldone R, Surico N and Bucchi L: Interobserver variability of
colposcopic interpretations and consistency with final histologic
results. J Lower Genital Tract Dis. 8:212–216. 2004. View Article : Google Scholar
|
|
61
|
Massad LS, Jeronimo J, Katki HA and
Schiffman M; National Institutes of Health/American Society for
Colposcopy and Cervical Pathology Research Group, : The accuracy of
colposcopic grading for detection of high-grade cervical
intraepithelial neoplasia. J Lower Genital Tract Dis. 13:137–144.
2009. View Article : Google Scholar
|
|
62
|
LeCun Y, Haffner P, Bottou L and Bengio Y:
Object recognition with gradient-based learning. Shape, contour and
grouping in computer vision. Lecture Notes in Computer Science.
Springer, Berlin, Heidelberg. 1681:319–345. 1999.
|
|
63
|
He K, Zhang X, Ren S and Sun J: Deep
residual learning for image recognition. Proceedings of the IEEE
conference on computer vision and pattern recognition. 770–778.
2016.PubMed/NCBI
|
|
64
|
Hu J, Shen L and Sun G:
Squeeze-and-excitation networks. Proceedings of the IEEE conference
on computer vision and pattern recognition. 7132–7141. 2018.
|
|
65
|
Kudva V, Prasad K and Guruvare S:
Automation of detection of cervical cancer using convolutional
neural networks. Crit Rev Biomed Eng. 46:135–145. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Esteva A, Kuprel B, Novoa RA, Ko J,
Swetter SM, Blau HM and Thrun S: Dermatologist-level classification
of skin cancer with deep neural networks. Nature. 542:115–118.
2017. View Article : Google Scholar : PubMed/NCBI
|