Introduction
Lung cancer is one of the most prevalent types of
malignancy worldwide, ranking as the first and second leading
causes of cancer-associated mortality in males and females,
respectively (1). Based on a cancer
statistic in 2013, there were 228,190 newly diagnosed cases of lung
cancer, which consisted of 118,080 males and 110,110 females in the
United States of America. Among these cases, it was estimated that
87,260 male (73.9%) and 72,220 female (65.6%) patients succumbed to
this malignancy (2). Therefore, this
serious situation mandates the necessity to identify novel
therapeutic targets for the clinical diagnosis and treatment of
lung cancer.
Recently, high-throughput transcriptome analysis has
revealed that >90% of the transcriptome is transcribed into
non-coding RNAs, among which long non-coding RNAs (lncRNAs) have
been implicated in the malignant behaviors of lung cancer (3,4).
Currently, a body of evidence has established the implication of
lncRNAs in lung cancer (5,6). For instance, lncRNA HNF1A-AS1 is
significantly more highly expressed in lung cancer compared with
the matched non-tumor tissues, and its expression level is
significantly associated with Tumor-Node-Metastasis (TNM) stage
(7), tumor size and lymph node
metastasis, leading to a poorer overall survival rate (8).
In previous studies regarding cancer biomarkers
screening, several key lncRNAs have been identified to be
significantly downregulated using microarray analysis of renal cell
carcinoma (RCC) and adjacent non-tumor tissues (9–11). One of
these significantly dysregulated lncRNAs was BX357664 (9), which was later predicted by the Coding
Potential Assessment Tool to have no protein-coding potential
(12). The BX357664 was initially
named CR613822, which was implicated to have functional roles in
human cancer. The information regarding CR613822 was uploaded to
the NCBI nucleotide database but was deleted by the uploader
shortly after. Meanwhile, the updated details referred to the
BX357664 gene (12). Furthermore, the
length of the sequence of BX357664 is 650 nucleotides (nt)
(12). By referring to the definition
of lncRNA, the present study operated under the assumption that
BX357664 is an lncRNA, and thereafter focused on the functional
roles of BX357664 in human cancers. Notably, one pioneer study
revealed that BX357664 regulated cell proliferation and
epithelial-to-mesenchymal transition via inhibition of
TGF-β1/p38/HSP27 signaling in RCC (13). This observation reinforced the
hypothesis that BX357664 may serve a critical role in human
carcinogenesis.
At present, the functional roles of BX357664 in
human cancer remain largely unknown. As one part of a larger
project focusing on lncRNAs in lung cancer, the present study aimed
to investigate the roles of BX357664 in lung cancer cell
proliferation, migration and apoptosis. Since BX357664 is a novel
lncRNA, its expression profile was initially determined in clinical
lung cancer and in a series of lung cancer cell lines. The
gain-of-function and loss-of-function experiments were then
investigated in lung cancer cell lines. The results of the present
study may provide novel insight into the molecular targeted
treatment and diagnosis of lung cancer in a clinical setting.
Materials and methods
Human samples
The present study was approved by the Ethics
Committee of the Central Hospital of Zhuzhou City (Zhuzhou, China)
and written informed consent was obtained from all participants.
Lung cancer specimens were randomly selected from 100 patients
(male:female, 63:37; age range, 45–70 years; median age, 58 years)
who had undergone surgery at the Central Hospital of Zhuzhou City.
None of the patients had received chemotherapy or radiotherapy
prior to surgery. Tumor tissues and their adjacent non-cancerous
tissues were dissected from each case and immediately frozen in
liquid nitrogen until use for the subsequent RNA extraction.
Cell culture and transfection
The Human lung cancer A549, H1975 and H-125 cell
lines were purchased from American Type Culture Collection
(Manassas, VA, USA) and applied for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis to detect the expression of BX357664. Another two lung
cancer cell lines, 95D and SPC-A-1, and the normal human 293T cell
line were commercially available from the Cell Bank of Chinese
Academy of Sciences (Beijing, China) and used to investigate the
transcript level of BX357664. Due to the special characteristics of
293T cells and lack of normal lung epithelia cells, 293T cells were
used as a control. All cells were cultured in the DMEM supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) at 37°C in 5% CO2 atmosphere.
When cells grew to a confluence of 80%, the constructed pcDNA3.1
BX357664 plasmid or the vector (pcDNA3.1, an empty plasmid;
Invitrogen; Thermo Fisher Scientific, Inc.), the whole sequence of
BX357664 were transfected with 100 ng pcDNA-BX357664 empty vector
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) by normal PCR, according to the manufacturer's
protocols, and ligation (T4 ligase; New England BioLabs, Inc.,
Ipswich, MA, USA), according to the manufacturer's protocol.
Subsequently, BX357664 plasmid was dissolved in water at 1,000
ng/µl and a total of 2 µg DNA was transfected using Lipofectamine
2000, according to the manufacturer's protocols, into a well in
six-well plates (1×104 cells). After 6 h transfection,
the culture medium was replaced and 48 h later, subsequent analysis
was performed.
RNA extraction and RT-qPCR
Total RNA was extracted from clinical tissues and
cultured A549 and 95D cells using TRIzol Reagent (Takara
Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocol. RNA quality and quantity were determined
by Nanodrop 2000 (Thermo Fisher Scientific, Inc.). A total of 500
ng RNAs were reversely transcribed into cDNA using the Transcriptor
First Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd.)
(37°C for 15 min and 85°C for 5 sec). The expression levels of
BX357664 relative to GAPDH control transcripts were calculated by
qPCR using the ABI 7900 Fast Real-Time PCR system (SeqGen, Inc.,
Torrance, CA, USA). The SYBR® Green reagent was
purchased from Takara Biotechnology Co., Ltd. and used with the
following protocols: 94°C for 10 min followed by 35 cycles of 94°C
for 5 sec and 60°C for 30 sec. The primer sequences were as
follows: BX357664 forward, 5′-GGCGTGGTTTTGATGGAGTG-3′ and reverse,
5′-AGGCTGCAGAGTTGAGATCG-3′; and GAPDH forward,
5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCCTGTTGCTGTA-3′.
RT-qPCR amplification was performed in triplicate reactions. The
transcription level of BX357664 was normalized to that of GAPDH
using the 2−ΔΔCq method (14).
Colony formation assay
A549 and 95D cells (1×104/well) were
seeded into 12-well plates 24 h prior to the transfection.
Subsequently, A549 and 95D cells were treated with
BX357664-expressing plasmid. A total of 500 cells were seeded into
a 6-well plate in each treatment group. The plates were incubated
at 37°C for two weeks without changing the culture medium. Finally,
the colonies were stained with 1% crystal violet, and images were
captured of five randomly selected fields of view. The whole plates
were counted manually and statically analyzed.
Cell proliferation determination
MTT assays (Promega Corporation, Madison, WI, USA)
were performed to determine cell proliferation abilities according
to the manufacturer's protocol. In brief, A549 and 95D cell lines
were seeded into 96-well plates at an initial concentration of
5×103 cells/well in the DMEM supplemented with 10% FBS.
A549 and 95D cells were transfected with the BX357664 plasmid, as
aforementioned. Each experimental group of cells was seeded in
sextuplicate and the culture medium was replaced every other day.
The purple formazan was dissolved in dimethyl sulfoxide. Cell
proliferation was detected for 5 consecutive days. For each
checking point, the cell proliferation rate was detected using a
TECAN reader (Tecan Group Ltd., Männedorf, Switzerland) at an
absorbance of 490 nm.
Cell cycle analysis
Prior to experimentation, A549 and 95D cells were
transfected with pcDNA 3.1 vector or a BX357664-expressing plasmid
for 48 h, as aforementioned. Next, cells were collected by low
speed centrifugation (1,000 × g at 4°C for 5min) and fixed with
cold ethanol (70%) for 10 min at 4°C. The cells were then washed
and re-suspended in pre-cold PBS and incubated at 37°C for 30 min
with 10 mg/ml RNase and 1 mg/ml propidium iodide (PI;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The percentage of
cells in each cell cycle phase was determined with a flow cytometer
using the Cell Quest acquisition software (Pro version; BD
Biosciences, Franklin Lakes, NK, USA).
Transwell assay
A549 and 95D cells were transfected with
BX357664-expressing plasmid for 48 h as described earlier.
Subsequently, cells were harvested with serum-free medium and mixed
gently, and then 150 µl cell suspension (6×104 cells)
was seeded into the upper chamber (8 µm; Corning Incorporated,
Corning, NY, USA), while the lower chamber was filled with 600 µl
DMEM supplemented with 10% FBS. Following incubation for 24 h at
37°C, cells in each group were fixed with ice-cold methanol (100%)
for 5 min at room temperature and stained with 1% crystal violet
for 5 min at room temperature. Following washing with PBS, cells on
the upper surface of the chamber were gently cleaned by a cotton
swab. Images of the cells on the lower surface of the membrane were
captured and the number of cells was counted under a light
microscope (×200; Nikon Corporation, Tokyo, Japan) with five
randomly selected fields of view. For cell invasion assays, the
membrane was pre-coated with Matrigel (Corning Incorporated,
Corning, NY, USA) for 6 h at 37°C.
Wound healing assay
The A549 and 95D cell lines were seeded into 6-well
plates (3×104 cells/well) and treated with the
corresponding plasmids for 48 h at 37°C as aforementioned. After
the cells had formed a confluent monolayer, the linear wound of the
cellular monolayer was created by scratching a straight line
through the middle of the plate using a sterile plastic pipette tip
(10 µl). Cells were washed with PBS and images were captured
immediately. Following incubation for 24 h at 37°C, images of cells
were captured again (Nikon Corporation), and wound closure was
calculated in each group.
Statistical analysis
Data were calculated with SPSS 19.0 (IBM Corp.,
Armonk, NY, USA) and expressed as the mean ± standard deviation.
Differences were evaluated using paired Student's t-test or one-way
analysis of variance, followed by the Student-Newman-Keuls post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
BX357664 is downregulated in lung
cancer and differentially expressed in lung cancer cell lines
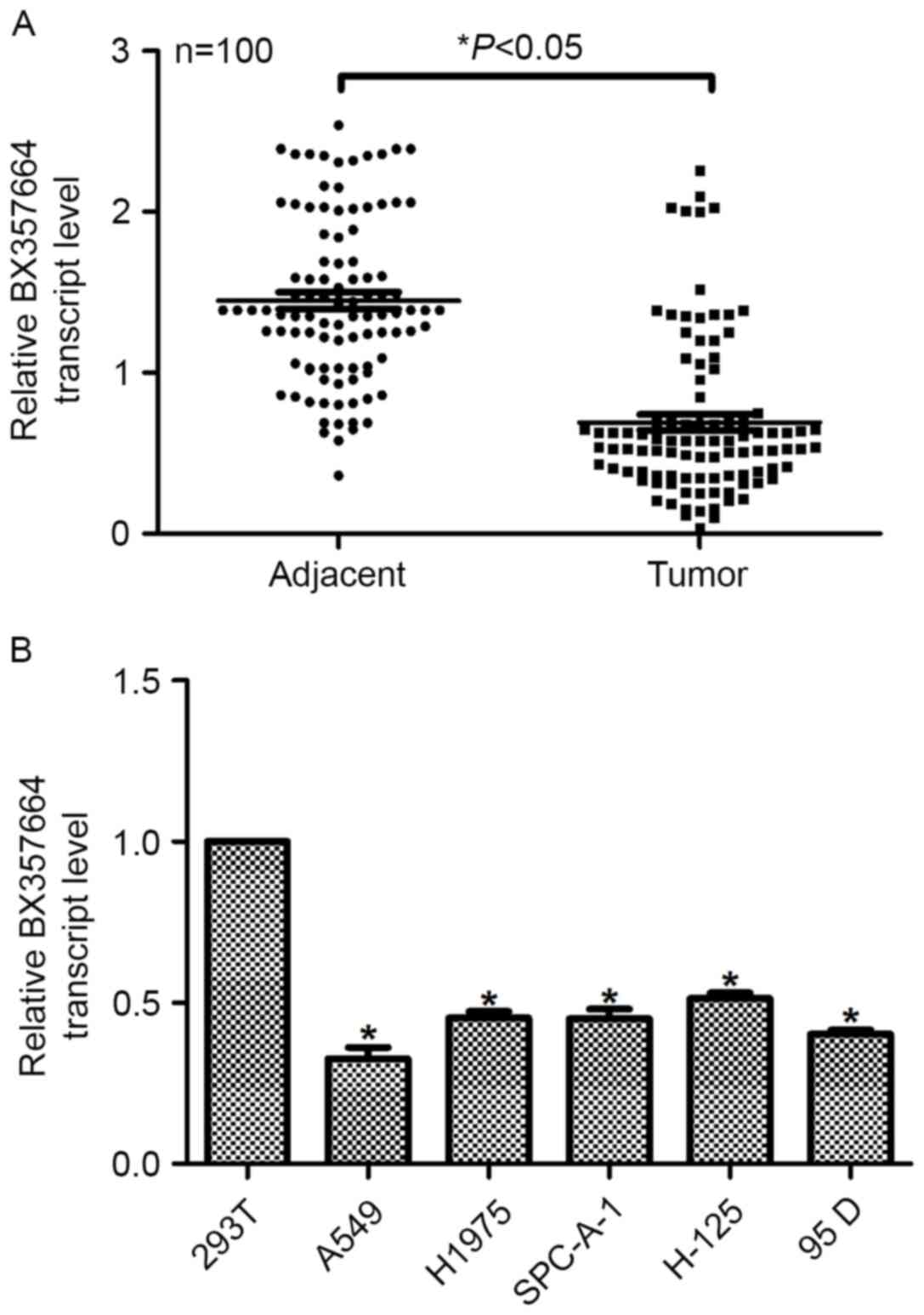
In the present study, the expression profile of
BX357664 was initially examined. In the 100 samples collected,
RT-qPCR analysis revealed that the average transcription level of
BX357664 in the tumor tissues was significantly lower, making up
approximately half of that in the adjacent non-tumor tissues
(P<0.05; Fig. 1A and B). The
expression of BX357664 in the 100 lung cancer cases was then
classified into low (less than the mean value) and high (higher
than the mean value), and was statistically analyzed using clinical
categories. It was identified that expression of BX357664 was
significantly associated with tumor size, distant metastasis and
TNM stage (P<0.001); however, BX357664 expression was not
significantly associated with other examined parameters, including
age, marriage status, symptoms and lymph node metastasis (Table I), implying that BX357664 may be
associated with the aggressive behaviors observed in lung cancer.
Following culture of a series of lung cancer cell lines and the
control 293T cells, it was further demonstrated that the relative
transcription level of BX357664 was significantly lower in the lung
cancer cell lines than in the control 293T cells. Notably, BX357664
had even lower transcription levels in the relatively more
aggressive A549 and 95D cell lines. The less aggressive H1975 and
H-125 cell lines exhibited a higher expression of BX357664 than the
A549 and 95D cell lines. Taken together, these results suggested
that BX357664 is downregulated in lung cancer and is differentially
expressed in lung cancer cell lines. The A549 and 95D cell lines
were selected for subsequent plasmid transfection.
 | Table I.Association between BX357664 and
clinical variables among 100 patients with lung cancer. |
Table I.
Association between BX357664 and
clinical variables among 100 patients with lung cancer.
|
|
| Expression of
BX357664 |
|
|---|
|
|
|
|
|
|---|
| Variables | No. (%) | Low (n=60) | High (n=40) | P-value |
|---|
| Age, years |
|
|
| 0.504 |
|
<40 | 23 (23) | 8 (8) | 15 (15) |
|
|
40–55 | 34 (33) | 21 (21) | 13 (13) |
|
|
>55 | 43 (43) | 31 (31) | 13 (13) |
|
| Marriage status |
|
|
| 0.525 |
|
Single | 15 (15) | 7 (7) | 8 (8) |
|
|
Married | 64 (64) | 45 (45) | 20 (20) |
|
|
Divorced/separated | 21 (21) | 9 (9) | 12 (12) |
|
| Presenting
symptoms |
|
|
| 0.652 |
| Painless
lump | 35 (35) | 24 (24) | 11 (11) |
|
| Painful
lump | 30 (30) | 18 (18) | 12 (12) |
|
| Atypical
symptoms | 35 (35) | 18 (18) | 17 (17) |
|
| Tumor size (T) |
|
|
|
<0.001a |
| T1 (≤2
cm) | 17 (17) | 1 (1) | 16 (16) |
|
| T2 (>2
cm-<5 cm) | 23 (23) | 5 (5) | 18 (18) |
|
| T3 (≥5
cm) | 19 (19) | 15 (15) | 4 (4) |
|
| T4 (any
size with distant metastasis) | 41 (41) | 39 (39) | 2 (2) |
|
| Lymph node
metastasis |
|
|
| 0.780 |
| N0 | 45 (45) | 29 (29) | 16 (16) |
|
| N1+ | 55 (55) | 31 (31) | 24 (24) |
|
| Distant
metastasis |
|
|
|
<0.001a |
| M0 | 58 (58) | 24 (24) | 34 (34) |
|
| M1 | 42 (42) | 36 (36) | 6 (6) |
|
| TNM stage |
|
|
|
<0.001a |
| I/II | 35 (35) | 5 (5) | 30 (30) |
|
|
III/IV | 65 (65) | 55 (55) | 10 (10) |
|
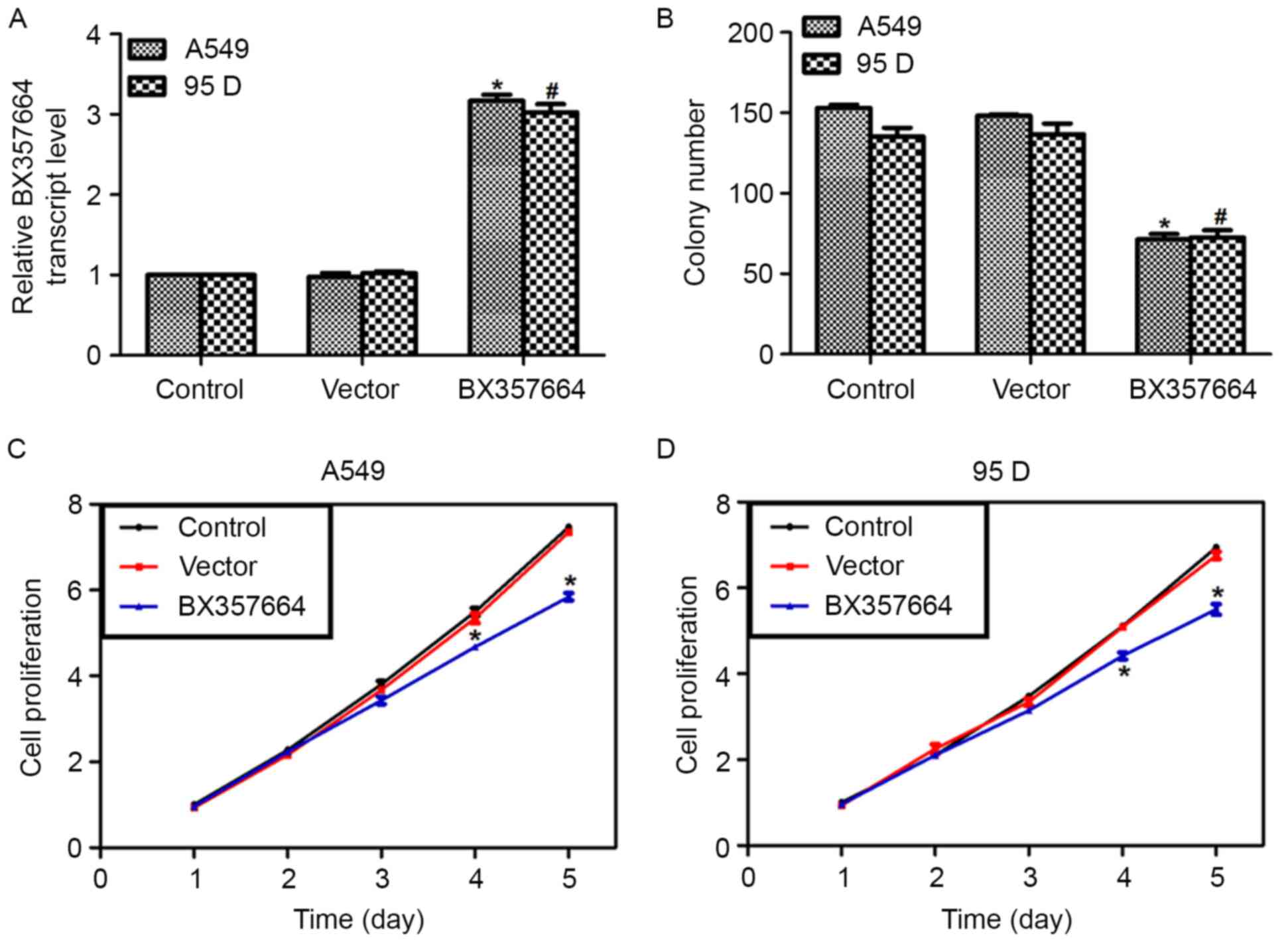
Overexpression of BX357664 in A549 and
95D cells inhibits cell proliferation in vitro
Next, the BX357664 expression plasmid was
transfected into A549 and 95D cells. Following transfection, the
vector plasmid did not cause any significant changes in the
transcription level of BX357664. By contrast, the expression
plasmid increased the transcription of BX357664 by nearly 3-fold in
A549 and 95D cells (Fig. 2A). These
data indicated the high efficiency of the expression plasmid.
Colony formation assays were then performed using these gene
reconstructs. It was revealed that only ~68 colonies were formed in
A549 cells following overexpression of BX357664, causing a 58%
decrease, compared with the control A549 cells. Similarly, the
average number of colonies in BX357664-overexpressing 95D cells was
70, which was in contrast with the 138 colonies formed by control
95D cells (Fig. 2B).
In the MTT assay, the proliferative rates were
significantly decreased by overexpression of BX357664 on day 4, and
on day 5, a 28.6 and 26.3% decrease in cell numbers was observed in
A549 cells (Fig. 2C) and 95D cells
(Fig. 2D), respectively. These data
suggested that BX357664 inhibits lung cancer cell
proliferation.
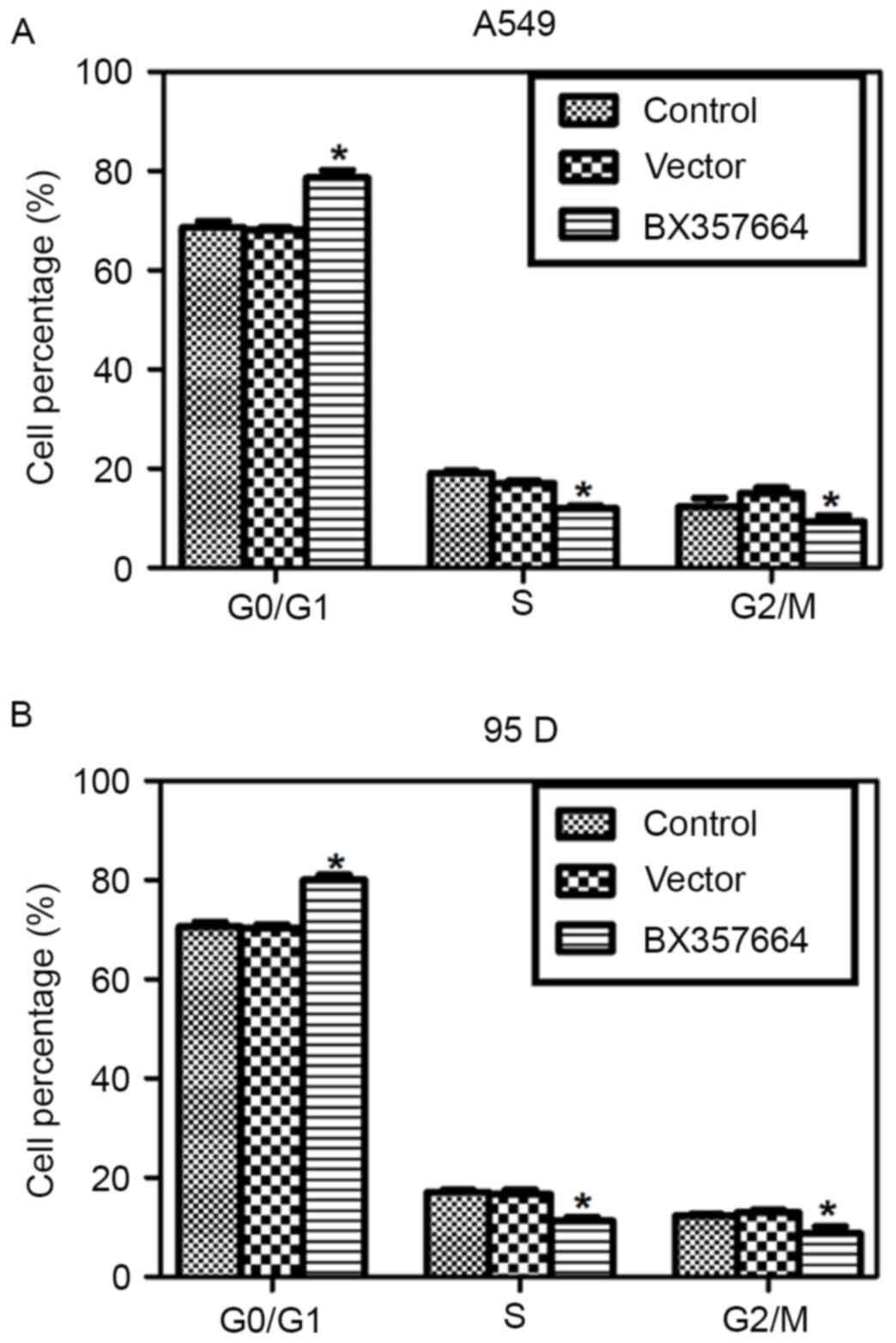
BX357664 regulates cell cycle
progression in lung cancer cells
Cell cycle analysis revealed that the cell
percentage in the G0/G1 phase was increased by ~10%, while the cell
percentages in the S and G2/M phases were decreased in
BX357664-overexpressing A549 cells (Fig.
3A). Similarly, Overexpression of BX357664 in 95D cells also
resulted in cell cycle arrest in the G0/G1 phase, as evidenced by
the increased percentage of cells in the G0/G1 phase (Fig. 3B). These observations suggested that
BX357664 regulates cell cycle progression in lung cancer cells.
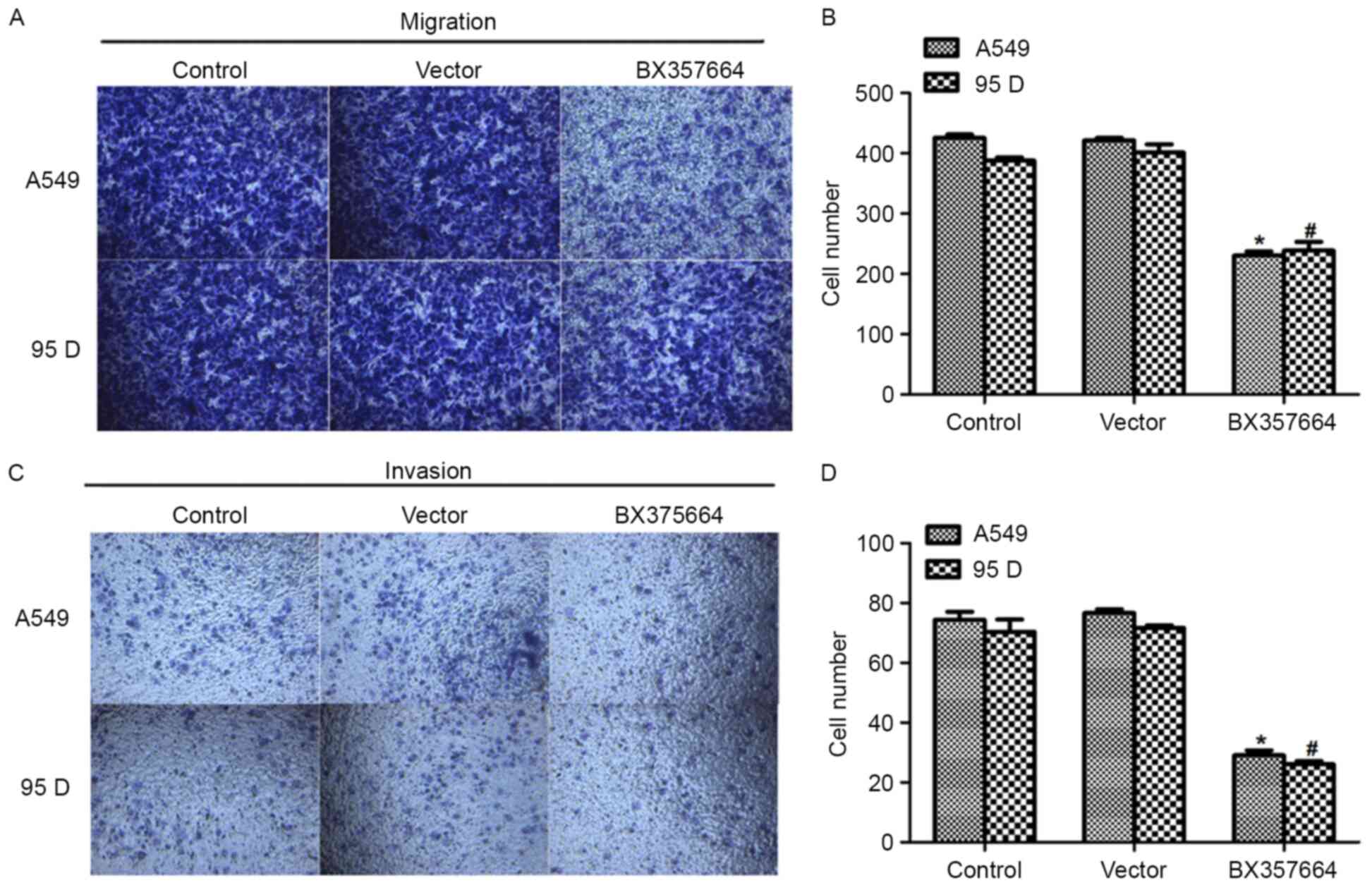
Overexpression of BX357664 in A549 and
95D cells inhibits cell metastasis in vitro
In light of the association of BX357664 expression
with TNM staging and distant metastasis (Table I), the effects of BX357664 on cell
migration and invasion capacities were further evaluated. In the
Transwell migration assay, a significant number of A459 and 95D
cells migrated to the lower surface of the membrane, which was
evidenced by the crystal violet staining (Fig. 4A and B). In the
BX357664-overexpressing cells, only an average of 208 A549 cells
migrated, which was in contrast to the 408 control A549 cells. A
similar result was also observed in the invasion assay in
BX357664-overexpressing 95D cells (Fig.
4C). The potential of cells to invade through the membrane was
inhibited by >65% when 95D cells were transfected with the
BX357664-expressing plasmid (Fig.
4D). The multi-cell lines-based observations suggested that
BX357664 may function as a negative regulator of cell metastasis in
lung cancer.
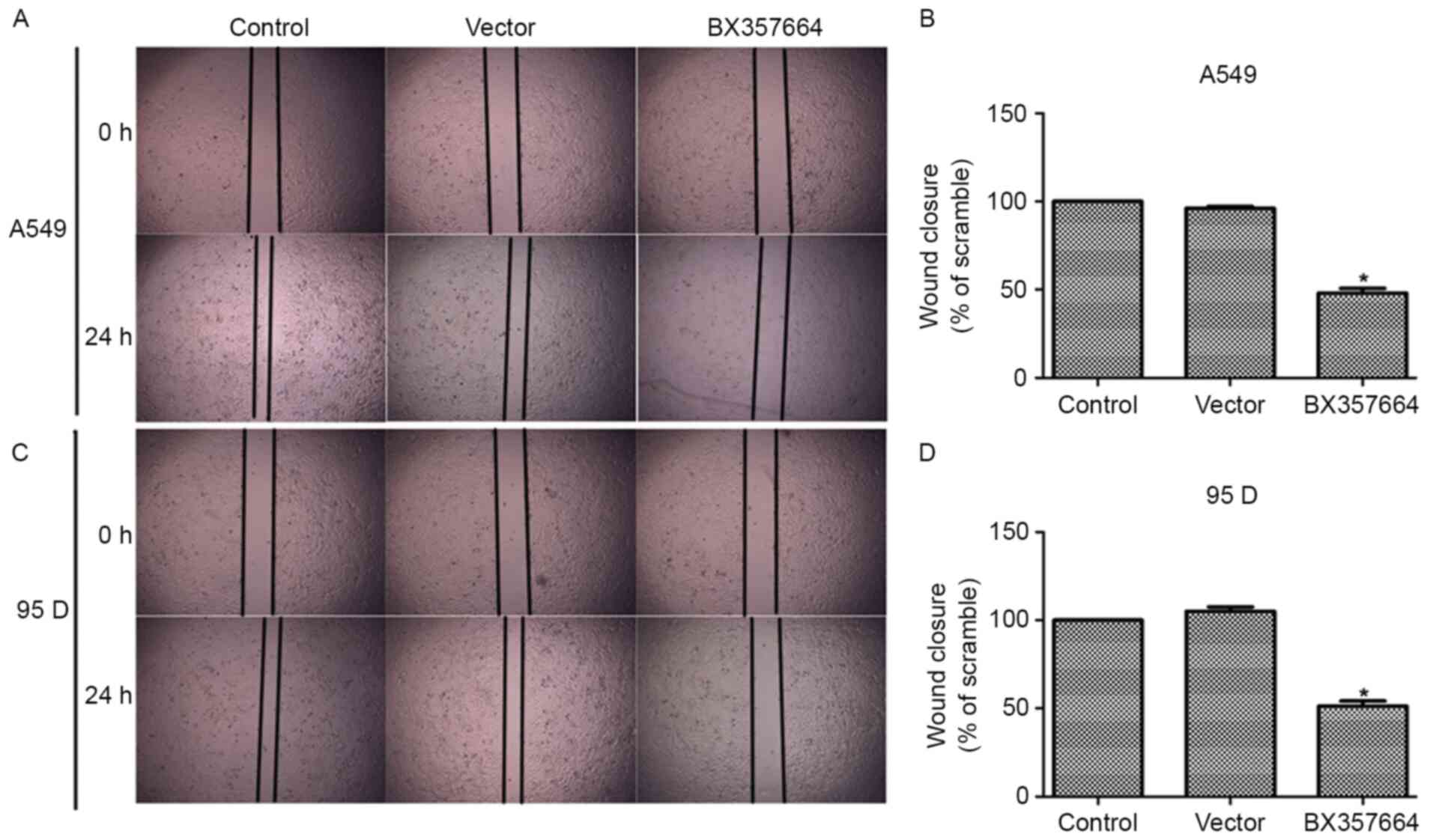
In the wound healing assay, when BX357664 was
overexpressed, the wound recovery was significantly decreased in
A549 (Fig. 5A and B) and 95D
(Fig. 5C and D) cells. In particular,
the overexpression of BX357664 in A549 cells inhibited cell
migration by 52% since the area of wound closure was decreased
(Fig. 5B). Likewise, the
overexpression of BX357664 in 95D cells resulted in similar effects
(Fig. 5D). The wound recovery rates
corroborated the Transwell migration and invasion results.
Discussion
LncRNAs are a class of non-coding RNAs with a length
of >200 nucleotides (15). It has
been demonstrated that the aberrant expression of lncRNAs may
contribute toward carcinogenesis and specific lncRNAs have been
identified as oncogenes or tumor suppressor genes (16–18). The
present study investigated the role of a novel lncRNA, BX357664, in
the behaviors of lung cancer. BX357664 was implicated to serve
functional roles in human cancer (12), including renal cell carcinoma (RCC)
(9). The upregulation of BX357664 was
demonstrated to reduce the migration, invasion and proliferation
capabilities of RCC cells (13).
These data suggested that lncRNAs serve a functional role in solid
tumors. The study of BX357664 in human lung cancer would enhance
the present understanding of lung carcinogenesis and provide novel
evidence for the diagnosis and treatment of lung cancer.
In the present study, it was initially demonstrated
that BX357664 was downregulated in clinical lung cancer tissues and
in a series of lung cancer cell lines, which was consistent with
previous reports that BX357664 was significantly deregulated by a
microarray analysis in lung cancer (9) and in RCC tissues (13). BX357664 expression was also
demonstrated to be significantly associated with clinical
parameters, including tumor size, distant metastasis and TNM
staging. This was corroborated by the results of the present study,
where the expression of BX357664 was observed to be higher in less
aggressive lung cancer A549 and 95D cell lines, whereas BX357664
expression was lower in the more aggressive H1975 and H-125 cell
lines, implying that BX357664 may be a determining factor in the
aggressiveness of lung cancer cells. Therefore, the A549 and 95D
cell lines were utilized for the subsequent gain-of-function
experiments. It was demonstrated that the overexpression of
BX357664 inhibited the cell clonogenic potential and proliferative
abilities.
Cell cycle arrest is a hallmark of carcinogenesis
(19). Based on the cell viability
assays, cell cycle progression was assessed. As a consequence of
BX357664 overexpression in A549 and 95D cells, cell percentages in
the G0/G phase were significantly increased. These results
suggested that BX357664 inhibits cell proliferation in lung cancer
cells. In addition, it was further demonstrated that overexpression
of BX357664 inhibited cell migration and invasion abilities in A549
and 95D cells. Wound recovery abilities were accordingly impaired.
Together with cell viability assays, the multi-cell lines-based
observations suggested that BX357664 serves a crucial role in cell
proliferation and metastasis in lung cancer. Overexpression of
BX357664 may be a novel therapeutic strategy for the treatment of
lung cancer.
It would be of scientific value to investigate the
mechanisms underlying BX357664-mediated biological behaviors in
lung cancer. However, a limitation of the present study is that the
expression of lncRNA BX357664 within each pathological subtype of
lung cancer was not analyzed (20,21).
Whether lncRNA BX357664 has any distinct functional roles in
different pathological subtypes remains to be elucidated. This
limitation requires the collection of substantial cases categorized
as different pathological subtypes. Our future studies will address
this, as well as attempting to investigate the mechanisms
underlying the functions of BX357664 in human cancer.
In conclusion, the results of the present study
suggested that the lncRNA BX357664 is a critical inhibitor of cell
proliferation and metastasis, and an inducer of cell apoptosis in
lung cancer. The results may also provide novel insight into the
clinical diagnosis of lung cancer and suggested that overexpression
of BX357664 may be a promising therapeutic strategy for the
treatment of lung cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
SHX and GXL performed the majority of the
experiments. LQ analyzed the data and revised the manuscript. YF
designed the project, analyzed the data and wrote the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Central Hospital of Zhuzhou City (Zhuzhou, China)
and written informed consent was obtained from all
participants.
Patient consent for publication
All patients showed their full intention for
publication and provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P,
Lu B, Liu G and Wang Z: The long noncoding RNA MEG3 contributes to
cisplatin resistance of human lung adenocarcinoma. PLoS One.
10:e01145862015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang L, He T, Yan Y, Zhang Y, Zhou X,
Huang P, Kong Y, Xie M, Zhang L, Sun Q, et al: Expression and
clinical significance of the novel long noncoding RNA ZNF674-AS1 in
human hepatocellular carcinoma. Biomed Res Int. 2016:6089142016.
View Article : Google Scholar
|
|
5
|
Yu J, Fang Q and Meng S: Knockdown of long
noncoding RNA ENST457720 inhibits proliferation of non-small cell
lung cancer cells in vitro and in vivo. Oncol Res. Mar 1–2018.(Epub
ahead of print). View Article : Google Scholar
|
|
6
|
Yu W, Peng W, Jiang H, Sha H and Li J:
LncRNA HOXA11-AS promotes proliferation and invasion by targeting
miR-124 in human non-small cell lung cancer cells. Tumour Biol.
39:10104283177214402017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chansky K, Detterbeck FC, Nicholson AG,
Rusch VW, Vallières E, Groome P, Kennedy C, Krasnik M, Peake M,
Shemanski L, et al: The IASLC lung cancer staging project: External
validation of the revision of the TNM stage groupings in the eighth
edition of the TNM classification of lung cancer. J Thorac Oncol.
12:1109–1121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172.
2015.PubMed/NCBI
|
|
9
|
Qin C, Han Z, Qian J, Bao M, Li P, Ju X,
Zhang S, Zhang L, Li S, Cao Q, et al: Expression pattern of long
non-coding RNAs in renal cell carcinoma revealed by microarray.
PLoS One. 9:e993722014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Y, Zhao L, Lei L, Lau WB, Lau B, Yang
Q, Le X, Yang H, Wang C, Luo Z, et al: LncRNAs: The bridge linking
RNA and colorectal cancer. Oncotarget. 8:12517–12532.
2017.PubMed/NCBI
|
|
11
|
Liu T, Zhang X, Gao S, Jing F, Yang Y, Du
L, Zheng G, Li P, Li C and Wang C: Exosomal long noncoding RNA
CRNDE-h as a novel serum-based biomarker for diagnosis and
prognosis of colorectal cancer. Oncotarget. 7:85551–85563. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang L, Park HJ, Dasari S, Wang S, Kocher
JP and Li W: CPAT: Coding-Potential Assessment Tool using an
alignment-free logistic regression model. Nucleic Acids Res.
41:e742013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Qian J, Li X, Chen W, Xu A, Zhao K,
Hua Y, Huang Z, Zhang J, Liang C, et al: Long noncoding RNA
BX357664 regulates cell proliferation and epithelial-to-mesenchymal
transition via inhibition of TGF-β1/p38/HSP27 signaling in renal
cell carcinoma. Oncotarget. 7:81410–81422. 2016.PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rinn JL: lncRNAs: Linking RNA to
chromatin. Cold Spring Harb Perspect Biol. 6(pii): a0186142014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo X, Xia J and Deng K: Long non-coding
RNAs: Emerging players in gastric cancer. Tumour Biol.
35:10591–10600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Guo Y, Song Y and Shang C: Long
noncoding RNA GAS5 inhibits malignant proliferation and
chemotherapy resistance to doxorubicin in bladder transitional cell
carcinoma. Cancer Chemother Pharmacol. 79:49–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee J, Jung JH, Chae YS, Park HY, Kim WW,
Lee SJ, Jeong JH and Kang SH: Long noncoding RNA snaR regulates
proliferation, migration and invasion of triple-negative breast
cancer cells. Anticancer Res. 36:6289–6295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seki N, Eguchi K, Kaneko M, Ohmatsu H,
Kakinuma R, Matsui E, Kusumoto M, Tsuchida T, Nishiyama H and
Moriyama N: Stage-size relationship in long-term repeated CT
screening for lung cancer: Anti-lung cancer association project. J
Clin Oncol. 27:S15402009.
|
|
21
|
Nitsche U, Stangel D, Pan Z, Schlitter AM,
Esposito I, Regel I, Raulefs S, Friess H, Kleeff J and Erkan M:
Periostin and tumor-stroma interactions in non-small cell lung
cancer. Oncol Lett. 12:3804–3810. 2016. View Article : Google Scholar : PubMed/NCBI
|