Introduction
Breast cancer is one of the most common cancer types
among females worldwide in 2014, which cause cancer-associated
mortalities globally (1,2). In China, breast cancer remains the most
common type of neoplasm in 2014 (3).
With the development of medical technologies over the past 20
years, including surgery, radiotherapy and chemotherapy, the
diagnosis and treatment of breast cancer have continuously improved
(4,5).
Metastasis and recurrence remain the major causes of high mortality
rates of patients with breast cancer (6); therefore, understanding the mechanisms
and investigating novel biomarkers, which are responsible for
unfavorable progression, is important. Furthermore, the
identification of novel therapeutic targets for breast cancer
treatment is essential.
Sex determining region Y-box protein 5 (SOX5) is a
member of the SOX family, which was identified based on the
conserved homology of the high-mobility group DNA-binding motif
(7). It has been reported that SOX5
is involved in the regulation of embryonic development (8), and is associated with various cancer
types, including prostate cancer (9),
glioblastoma (10), hepatocellular
carcinoma (11), osteosarcoma
(12) and nasopharyngeal carcinoma
(13). In 2014, Pei et al
(14) reported that in breast cancer,
SOX5 induces epithelial-mesenchymal transition (EMT) by
transactivation of Twist1 expression. However, the expression and
the precise regulatory mechanism underlying the biological function
of SOX5 in breast cancer remain unclear.
Patients and methods
Cell culture and reagents
The normal breast tissue cell line, MCF-10A, and the
MCF7, T47D and MDA-MB-231 breast cancer cell lines were obtained
from the American Type Culture Collection (Manassas, VA, USA).
MCF-7 and T47D cells were cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), and MDA-MB-231 cells were cultured in RPMI-1640 supplemented
with 10% fetal bovine serum (FBS; both Invitrogen; Thermo Fisher
Scientific, Inc.). MCF-10A cells were cultured in DMEM-F12
supplemented with 5% horse serum (Invitrogen; Thermo Fisher
Scientific, Inc.). The cells were incubated in an atmosphere
containing 5% CO2 at 37°C. Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection to transfected into MCF-7 or MDA-MB-231 cells. The
relative small interfering (si)RNAs targeting SOX5 (si-SOX5-1 and
si-SOX5-2) or enhancer of zeste 2 polycomb repressive complex 2
subunit (EZH2) or negative control and G418 were purchased from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). SOX5 and vector
plasmid were purchased from Genepharma (Shanghai, China). The 70%
confluence of MCF-7 or MDA-MB-231 cells were achieved overnight
prior to transfection. In each group, 2 µg oligonucleotide were
used for transfection. At 48 h following transfection, the cells
were harvested for experimentation.
Patients
The present study was approved by the Research
Ethics Committee of Weifang People's Hospital (Weifang, China). All
patients provided written informed consent. A total of 58 pairs of
breast cancer tissues from female patients aged from 40–55 years
old and relative adjacent healthy mammary tissues were collected
between May 2010 and January 2013. The fresh specimens were frozen
immediately at −80°C in liquid nitrogen for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) use.
Patients who received tumor-specific therapy prior to diagnosis
were excluded. The pathological information was retrieved by the
Pathology Department of Weifang People's Hospital. Overall survival
times were calculated as the duration between the date of diagnosis
and date of cancer-associated mortality in the follow-up
period.
RNA extraction and RT-qPCR
analysis
Total RNA was extracted from cells or tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. First-strand
complementary DNA was synthesized using SuperScript II Reverse
Transcriptase kit (Invitrogen; Thermo Fisher Scientific, Inc.).
qPCR was performed using the Fast SYBR® Green Master mix
system (Roche Applied Science, Penzberg, Germany) on an ABI 7500
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
qPCR reaction was subsequently performed according to the following
conditions: Initial step, 95°C for 5 min; second step, 95°C for 10
sec, 60°C for 30 sec and 72°C for 10 sec for a total of 35 cycles.
The primers used were as follows: EZH2 forward,
5′-TTTCCAACACAAGTCATCCC-3′, and reverse,
5′-ATAAACCCACATTCTCTATCCC-3′; GAPDH forward,
5′-CCGTCTAGAAAAACCTGCC-3′, and reverse, 5′-GCCAAATTCGTTGTCATACC-3′.
The relative mRNA level was calculated using the 2−∆∆Cq
method and normalized to GAPDH (15).
The experiment was performed in triplicate.
Western blot analysis
MCF-7 or MDA-MB-231 cells were harvested and protein
was extracted using radioimmunoprecipitation buffer (50 mM tris-HCl
pH 7.4, 1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl and
0.1% SDS). The Bradford assay reagent (Thermo Fisher Scientific,
Inc.) was then used to determine the protein concentration in the
lysates. Equal amounts of protein (30 µg) were separated by 10%
SDS-PAGE gel, and then transferred to a polyvinylidene fluoride
membrane (EMD Millipore, Billerica, MA, USA). The membranes were
blocked in 5% non-fat milk in PBS containing 0.5% Tween-20 at room
temperature for 1 h and incubated with the primary antibodies
overnight at 4°C, and then washed three times with washing buffer
Tris-buffered saline Tween-20 (Sigma-Aldrich, Merck KGaA).
Horseradish peroxidase-conjugated anti-rabbit (sc-2357; 1:3,000) or
anti-mouse (sc-2789, 1:3,000; both Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) antibodies were used as the secondary antibodies
at room temperature for 1 h. The signal was visualized using
enhanced chemiluminescence reagents (Pierce; Thermo Fisher
Scientific, Inc.). The primary antibodies used were as follows:
SOX5 (ab94396; 1:1,000), EZH2 (ab186006; 1:1,000; both Abcam).
β-actin (sc-47778; 1:1,000; Santa Cruz Biotechnology, Inc.) was
used as the control.
Chromatin immunoprecipitation assay
(ChIP)
The ChIP assay was performed using Chip-IT Express
kit (Active Motif; Carlsbad, CA, USA), according to manufacturer's
protocols. The PCR products were resolved using a ABI 7500 system.
PCR was performed with 5 µl of the immunoprecipitated target DNA, 1
µl primers and 9 µl mixture (1 µl enzyme, 2 µl dNTP and 6 µl SYBR
green solution buffer all were included in the ChIP-IT kit.
ChIP sequencing
For ChIP sequencing, the DNA was purified with the
Qiagen PCR purification kit. In-depth whole genome DNA sequencing
was performed by the CapitalBio Corporation. The raw sequencing
image data were examined by the Illumina analysis pipeline, aligned
to the unmasked human reference genome (NCBI v36, hg18) using ELAND
(Illumina), and further analyzed by MACS (Model-based Analysis for
ChIP-Seq).
Luciferase report assay
A luciferase report assay was performed using a dual
luciferase assay kit according to the manufacturer's protocol
(Promega Corporation, Madison, WI, USA). The EZH2 promoter was
cloned into the luciferase reporter pGL3-basic vector plasmid,
which was part of the kit. A total of 5×104 cells-well
were cultured in DMEM at 37°C in 24-well plates for 48 h. The
report plasmid was transfected into the cells with the relative
plasmid SOX5 or shRNA targeting SOX5 or a negative control plasmid.
After 24 h transfection, the luciferase activities were measured
according to the aforementioned kit. The result was normalized to
Renilla. The transfections were performed in triplicate.
Cell proliferation assay
For the MTT assay, 5×103 cells were
seeded into 96-well plates with 100 µl culture medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) and cultured at 37°C for different
periods of time at 0, 24, 48 and 72 h. A total of 10 µl 5 mg-ml MTT
reagent (Beyotime Institute of Biotechnology, Shanghai, China) was
added into each well and the culture was continued for 4 h.
Subsequently, 100 µl dimethyl sulfoxide was used to replace the
medium. After 30 min of incubation, the absorbance at 570 nm
wavelength was measured on a SpectraMax 190 microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) and cell growth
curves were determined. Experiments were performed in triplicate
independently.
Colony formation assay
For the colony formation assay, the cells were
seeded into 6-well plates with 1×103 cells-well. Fresh
culture medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) was
replaced every 3 days and cultured at 37°C in an atmosphere
containing 5% CO2 for 2 weeks, to form colonies.
Subsequently, the cells were fixed with 70% methanol at room
temperature for 30 mins and stained with 5% crystal violet at room
temperature for 10 mins. The colonies containing >50 cells were
counted under a Leica DMI 3000B inverted microscope (Leica
Microsystems, Inc., Buffalo Grove, IL, USA) at magnification,
×40.
Scratch assay
Wound healing was used to observe the migration
ability of breast cancer cells. A total of 5×104 cells
were plated in 6-well plates and cultured until 95% confluency. A
plastic 20 µl pipette tip was used to scratch a vertical wound.
Detached cells were removed and phase contrast images of the
scratched fields were captured at 0 and 24 h. In each group, at
least three scratched fields were recorded using an upright light
microscope at magnification, ×20 (Leica DM4B; Leica Microsystems,
Shanghai, China).
Invasion assay
A Matrigel assay was performed to investigate the
invasion ability. Transwell chambers (8-µm pore size) were coated
with 1 mg-ml Matrigel (both BD Biosciences, San Jose, CA, USA).
Cells were seeded into 0.2 ml serum-free medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) at a density of 1×104
cells-well and placed on the top chamber of each insert. The lower
chamber was filled with 600 µl medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS. After 24 h of incubation, cells on
the surface were wiped off by mechanical scraping, and the migrant
cells attached to the lower surface were fixed with 10% methanol
for 30 min at room temperature. Following staining with 5% crystal
violet at room temperature for 20 min, the cells were visualized
and counted under a Leica DMI 3000B inverted microscope at
magnification, ×40. A total of three different fields of view in
each group were counted.
Statistical analysis
SPSS 19.0 (IBM Corp., Armonk, NY, USA) was used for
statistical analysis. All data were expressed as the mean ±
standard deviation following ≥3 independent experiments. P<0.05
was considered to indicate a statistically significant difference.
Kaplan-Meier analysis followed by the log-rank test was used to
analyze the association between SOX5 expression and the overall
survival rate. Significant differences between two groups were
determined with a Student's t-test. One-way analysis of variance
followed by Tukey's test was used to analyze the differences
between multiple groups to compare values of test and control
samples.
Results
SOX5 is frequently upregulated in
breast cancer tissues and associated with a reduced overall
survival rate
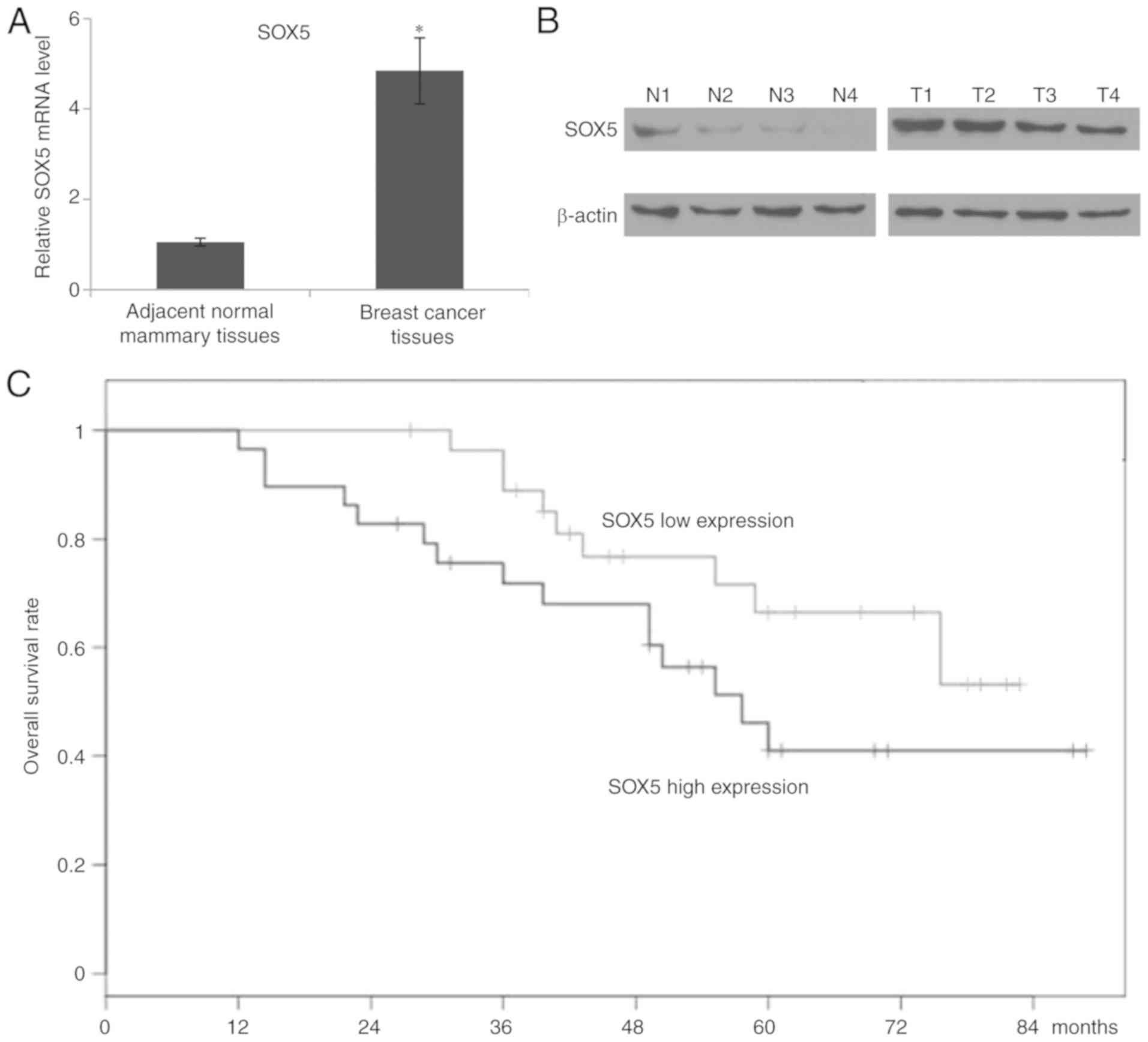
In order to identify the role of SOX5 in breast
cancer, the expression of SOX5 in 58 pairs of matched breast cancer
and adjacent healthy mammary tissues was investigated using RT-qPCR
assays. Compared with the adjacent healthy tissues, significantly
increased SOX5 mRNA expression levels were determined in breast
cancer tissues (Fig. 1A).
Furthermore, four pairs of breast cancer and relative healthy
mammary tissues were selected to detect the protein expression of
SOX5. As depicted in Fig. 1B, western
blotting demonstrated that SOX5 was notably overexpressed in breast
cancer tissues compared with healthy breast tissue. Kaplan-Meier
estimator analysis with the log-rank test was used to investigate
the prognostic significance of SOX5 in patients with breast cancer.
It was determined that an increased expression of SOX5 was
significantly associated with a reduced overall survival rate
(P=0.00213) (Fig. 1C).
SOX5 promotes breast cancer cell
proliferation in vitro
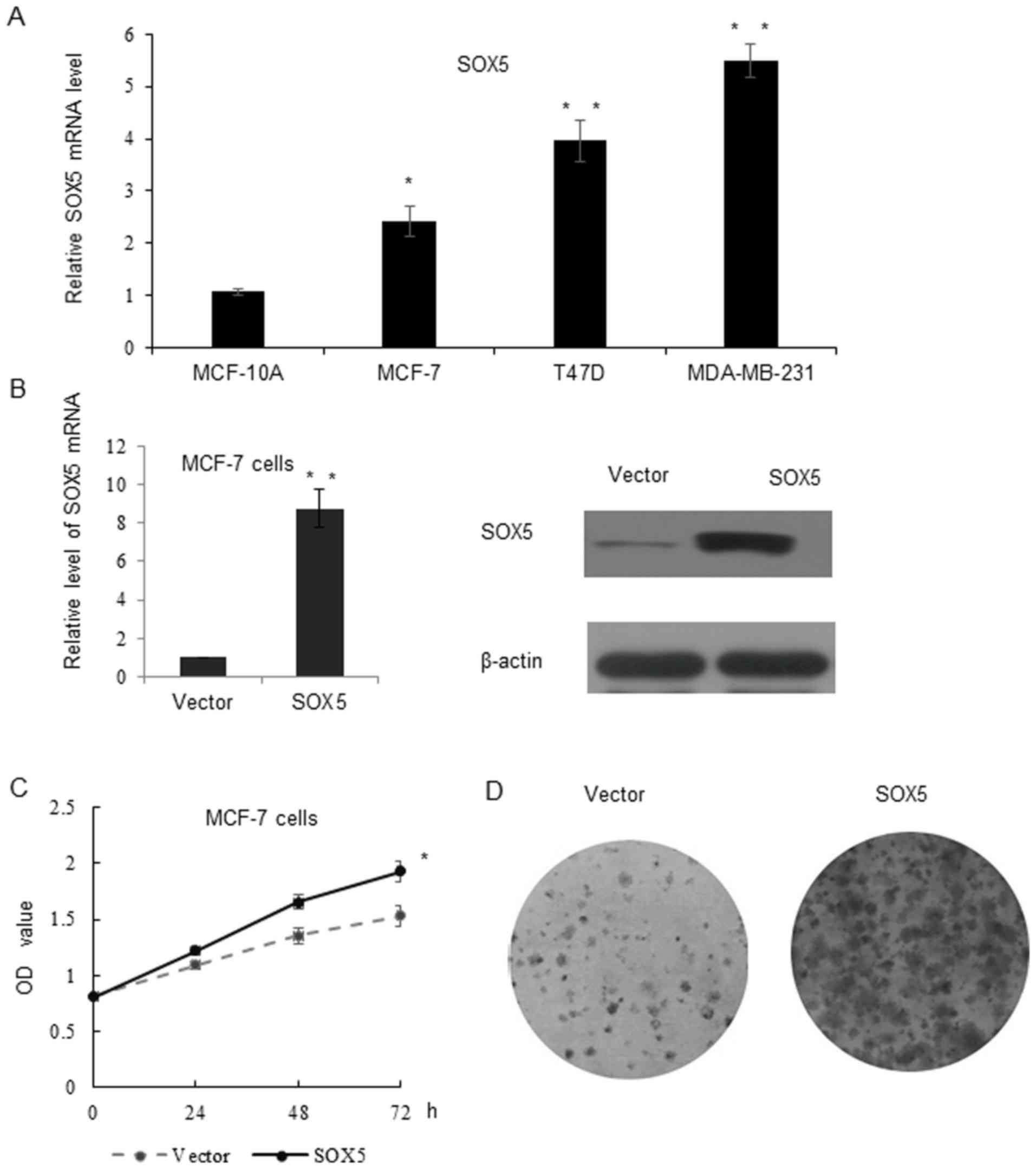
In order to examine the role of SOX5 in breast
cancer progression, RT-qPCR was performed to investigate the
expression of SOX5 in MCF-10A, MCF-7, T47D and MDA-MB-231 cells.
Compared with the normal breast cancer cell line MCF-10A, the SOX5
mRNA level was significantly increased in the breast cancer cell
lines. Additionally, the triple-negative cell line MDA-MB-231
exhibited the highest SOX5 expression level among all cell lines
(Fig. 2A). Therefore, the MDA-MB-231
cell line was selected to perform the loss of function assay, while
the MCF-7 cell line was selected for the gain of function assay. As
depicted in Fig. 2B, stable
transfection of SOX5 lentivirus was obtained following G418
selection and confirmed using RT-qPCR and western blotting. An MTT
assay was performed, which indicated that cells overexpressing SOX5
proliferated significantly faster, compared with vector control
cells (Fig. 2C). The colony formation
assay demonstrated that SOX5 formed larger and an increased number
of colonies, compared with the vector control group (Fig. 2D).
SOX5 enhances breast cancer cell
invasion in vitro
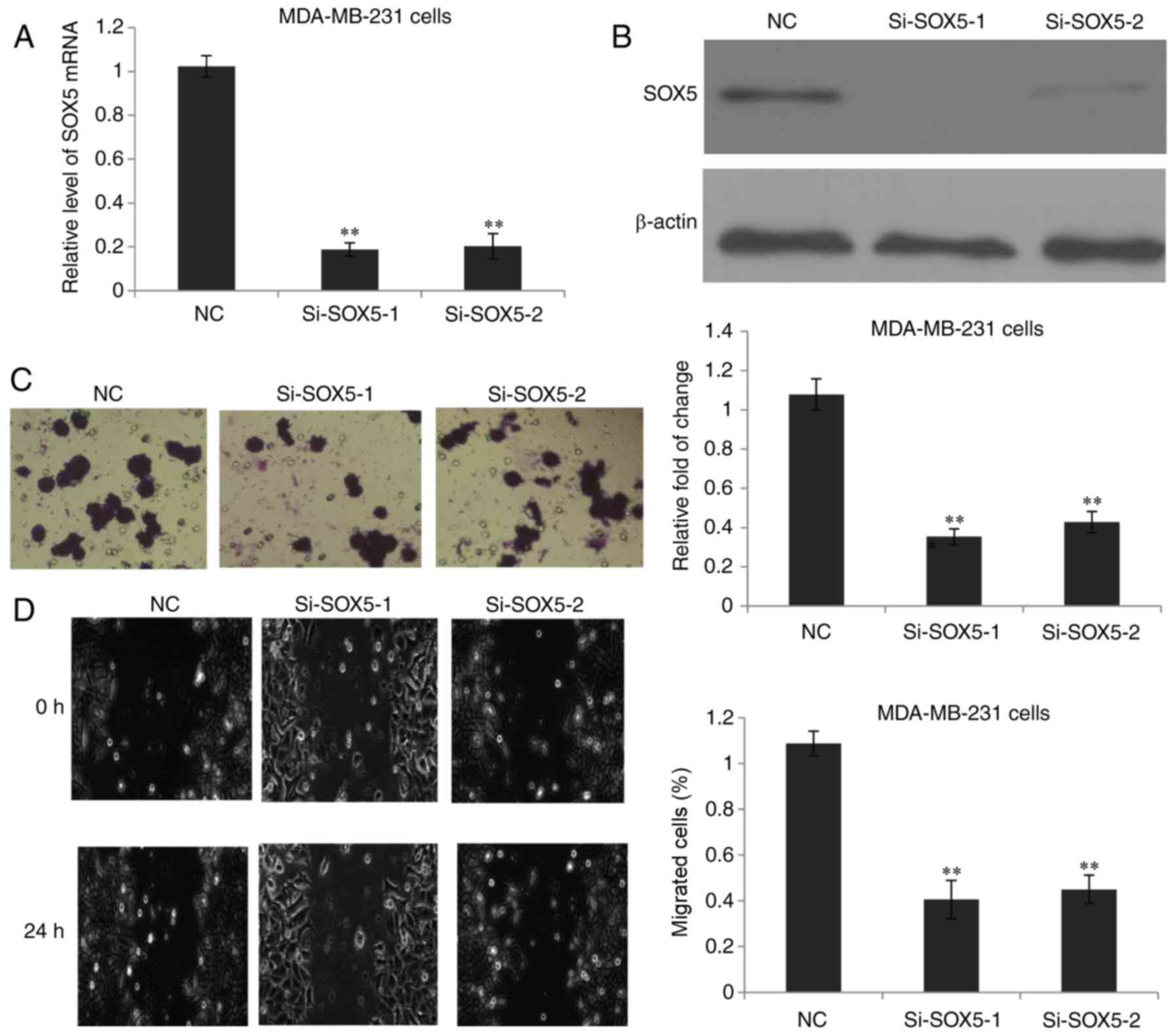
To further investigate the role of SOX5 in breast
cancer cell invasion, SOX5 expression was silenced in MDA-MB-231
cells using two different siRNAs. Successful depletion of SOX5
expression was confirmed at the mRNA (Fig. 3A) and protein levels (Fig. 3B). As expected, inhibition of SOX5
significantly impeded the MDA-MB-231 cell invasion ability compared
with the negative control group (Fig.
3C). Subsequently, a scratch assay was performed to assess the
role of SOX5 in the migration of breast cancer cells. As depicted
in Fig. 3D, the scratch assay
revealed that SOX5 knock down significantly reduced the migratory
ability of MDA-MB-231 cells. Therefore, these data indicated that
SOX5 exhibited the ability to promote MDA-MB-231 cell invasion and
migration in vitro.
Identification of EZH2 as a downstream
target gene of SOX5
Subsequently, the potential downstream molecule
regulated by SOX5 was identified. A ChIP sequence (ChIP-seq) assay
was performed. The ChIP-seq peak distribution is depicted in
Fig. 4A, and 17.5% promoters were
identified to be targeted by SOX5. To further validate the ChIP-seq
results, a qChIP assay and the binding between SOX5 and the EZH2
promoter was demonstrated to be the most significantly enriched
among the 10 genes selected (Fig.
4B). To investigate the SOX5-regulated EZH2 promoter activity,
a luciferase report assay was performed. The EZH2 promoter reporter
or EZH2 binding site mutant promoter reporter was transiently
transfected into MCF-7 cells with pcDNA3.1-SOX5 or a vector. As
depicted in Fig. 4C, SOX5
significantly activated EZH2 wild type promoter activity, but not
the EZH2 mutant reporter activity. No significant changes in EZH2
promoter activity were observed in the vector control group.
However, in MDA-MB-231 cells transfected with the EZH2 promoter
reporter or EZH2 binding site mutant promoter reporter with si-SOX5
or si-control, it was observed that si-SOX5 significantly repressed
EZH2 promoter activity, but not the mutant promoter activity
(Fig. 4D).
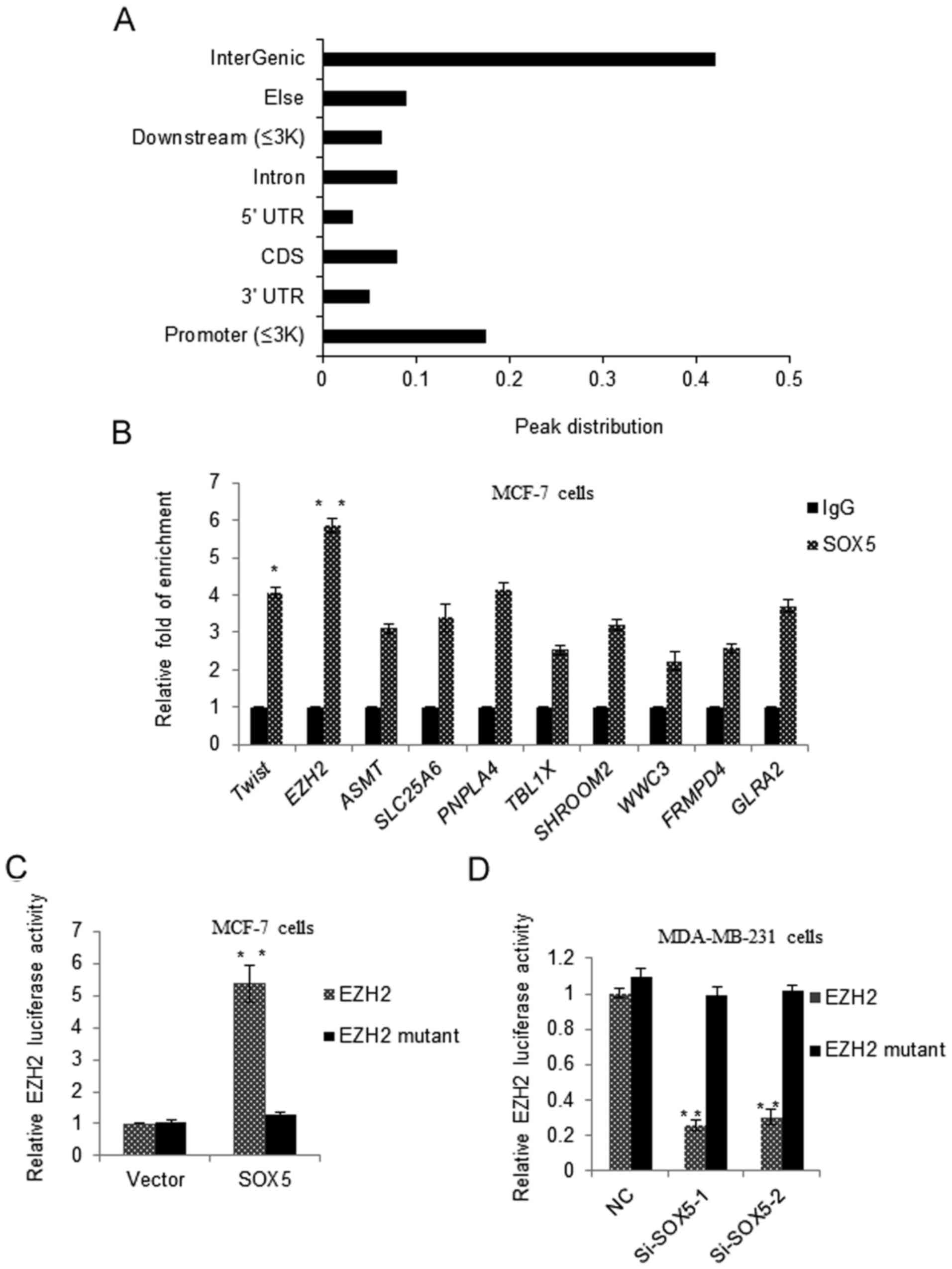 | Figure 4.Identification of EZH2 as a downstream
target gene of SOX5. (A) A ChIP-seq assay was performed in MCF-7
cells with a SOX5 antibody or a normal IgG, as a negative control,
and the peak distributions were depicted. (B) A qChIP experiment
was performed in MCF-7 or MDA-MB-231 cells, and the enrichments on
the promoter of EZH2 were detected. Each bar indicated the mean ±
standard deviation of 3 independent experiments. *P<0.05,
**P<0.01 vs. IgG. (C) MCF-7 cells were transfected with EZH2
promoter reporter or EZH2 binding site mutant promoter reporter,
and pcDNA3.1-SOX5 or vector. Luciferase activity was measured and
normalized to Renilla. Experiments were repeated 3 times.
**P<0.01 vs. Vector. (D) MDA-MB-231 cells were transfected with
EZH2 promoter reporter or EZH2 binding site mutant promoter
reporter, and si-SOX5 or si-control. Luciferase activity was
measured and normalized to Renilla. Experiments were
repeated 3 times. **P<0.01 vs. NC. SOX5, sex determining region
Y-box protein 5; NC, negative control; siRNA, small interfering
RNA; EZH2, enhancer of zeste 2 polycomb repressive complex 2
subunit; ChIP, chromatin immunoprecipitation; UTR, untranslated
region; CDS, coding sequence; ASMT, acetylserotonin
O-methyltransferase; SLC25A6, solute carrier family 25 member 6;
PNPLA4, patatin like phospholipase domain containing 4; TBL1X,
transducing β like 1 X-linked; FRMPD4, FERM and PDZ domain
containing 4; GLRA2, glycine receptor α 2. |
SOX5 induces breast cancer cell
proliferation and invasion by modulating EZH2
EZH2 is the catalytic subunit of polycomb repressive
complex 2, and EZH2 had been demonstrated to serve a role in breast
tumor initiation and progression (16–18).
Therefore, we hypothesized that the modulation of EZH2 was involved
with SOX5, increasing breast cancer cell proliferation and
invasion. As depicted in Fig. 5A, the
mRNA expression of EZH2 was significantly upregulated in
SOX5-transfected MCF-7 cells, compared with vector-transfected
cells, as demonstrated by RT-qPCR. This was further confirmed
through western blotting. While in MDA-MB-231 cells, the knockdown
of SOX5 resulted in significantly decreased EZH2 mRNA expression
and markedly reduced EZH2 protein levels (Fig. 5B). These results indicated that SOX5
transactivated EZH2 expression. Notably, the knockdown of EZH2 was
able to overcome the SOX5 promoter effect on the proliferation of
MCF-7 cells (Fig. 5C). Additionally,
the invasion rate of MDA-MB-231 cells was significantly increased
following treatment with EZH2, compared with the SOX5 knockdown
group (Fig. 5D). These results
indicated that SOX5 may regulate breast cancer cell proliferation
and invasion through targeting EZH2 expression.
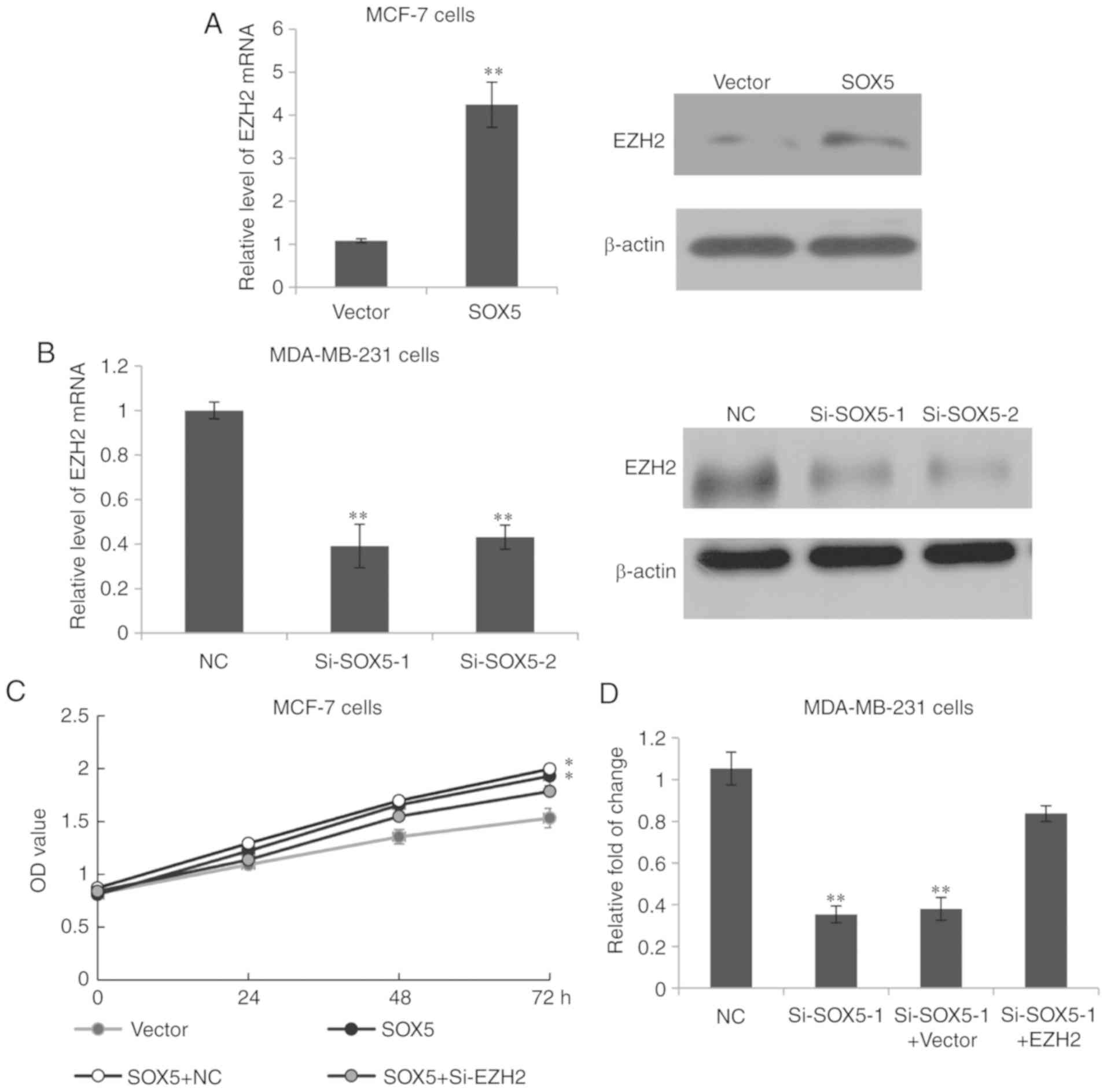 | Figure 5.SOX5 induces breast cancer cell
proliferation and invasion by modulation of EZH2. (A) RT-qPCR and
western blotting was used to analyze EZH2 expression in SOX5- and
vector-transfected MCF-7 cells. **P<0.01 vs. Vector. (B) RT-qPCR
and western blotting was used to analyze EZH2 expression in
MDA-MB-231 cells transfected with SOX5 siRNA (si-SOX5-1 and
si-SOX5-2) or si-control. **P<0.01 vs. NC. (C) MCF-7 cells were
transfected with empty vector or the SOX5 overexpression construct,
SOX5 overexpression construct plus control siRNA or SOX5
overexpression construct plus si-EZH2, and the MTT assay was
performed with the results being detected at 0, 24, 48 and 72 h.
*P<0.05 vs. Vector. (D) MDA-MB-231 cells were transfected with
si-control, si-SOX5-1, si-SOX5-1 plus a vector or si-SOX5-1 plus
EZH2, and a Matrigel assay was performed. The data are presented as
the fold of change. **P<0.01 vs. NC. SOX5, sex determining
region Y-box protein 5; NC, negative control; siRNA, small
interfering RNA; EZH2, enhancer of zeste 2 polycomb repressive
complex 2 subunit; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; OD, optical density. |
Discussion
Previously, a number of members of the SOX family,
including SOX2 and SOX4, have been reported to be involved in tumor
progression. SOX2 is a well-established stem cell regulator that is
highly expressed in multiple tissue stem cells and sustain the
infiltrative behavior in ≥25 different cancer types, including
cancers of the ovary, lung, skin, brain, breast, prostate and
pancreas (19–21). Increased expression of SOX4 serves as
an important role in human tumor development such as through
regulating cell growth, invasion, EMT and apoptosis (22–25).
However, the research regarding SOX5 remains limited.
To the best of our knowledge, the present study is
the first to demonstrate that SOX5 directly regulated EZH2
expression by transactivation, and thus promotes the proliferation
and invasion of human breast cancer cells. Using ChIP-seq, qChIP
and luciferase reporter assays, EZH2 was identified as a downstream
target gene of SOX5. Using RT-qPCR and western blotting analysis,
it was demonstrated that SOX5 regulates the expression of EZH2. The
present data added to accumulating evidence regarding SOX family
members being involved in breast cancer progression. As reported by
Pei et al (14), SOX5 was
overexpressed in highly invasive breast cancer cell lines,
including MDA-MB-435 and MDA-MB-231 cells, and suppression of SOX5
expression inhibited the proliferation and migration of MDA-MB-231
cells. These data were consistent with the present study. In the
present study, SOX5 was demonstrated to be frequently upregulated
in breast cancer tissues compared with healthy breast tissue, and
associated with a reduced overall survival rate, indicating that
SOX5 may serve as a poor prognostic biomarker in breast cancer.
Additionally, the function of SOX5 was investigated in different
cell lines, including MCF-7, the promotion of breast cancer cells
proliferation and invasion indicated that SOX5 may be a potential
oncogene. Notably, as reported by Tiwari et al (26), SOX4 directly regulated the expression
of EZH2, and thus serves an indispensable role in EMT and cell
survival in breast cancer (26). In
patients with pancreatic cancer, the SOX4-EZH2 axis was
demonstrated to be associated with the clinical outcome (27). Thus, we hypothesized that SOX4 and
SOX5 may have a coordinated function on the EZH2 promoter to
transactivate its expression. In the future, studies regarding the
mechanistic association between SOX5 and EZH2 may be used for
development of potential specifically-targeted therapies, and may
benefit patients with breast cancer metastasis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and-or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CS conceived and designed the study. YB provided
technical assistance and performed the Transwell assay. YS and ZZ
analyzed the data, KW performed the cell culture and wrote the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Weifang People's Hospital. All patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bombonati A and Sgroi DC: The molecular
pathology of breast cancer progression. J Pathol. 223:307–317.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hong W and Dong E: The past, present and
future of breast cancer research in China. Cancer Lett. 351:1–5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu J, Steeg PS, Price JE, Krishnamurthy S,
Mani SA, Reuben J, Cristofanilli M, Dontu G, Bidaut L, Valero V, et
al: Breast cancer metastasis: Challenges and opportunities. Cancer
Res. 69:4951–4953. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steeg PS: Targeting metastasis. Nat Rev
Cancer. 16:201–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Martinez-Morales PL, Quiroga AC, Barbas JA
and Morales AV: SOX5 controls cell cycle progression in neural
progenitors by interfering with the WNT-beta-catenin pathway. EMBO
Rep. 11:466–472. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rescan PY and Ralliere C: A Sox5 gene is
expressed in the myogenic lineage during trout embryonic
development. Int J Dev Biol. 54:913–918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma S, Chan YP, Woolcock B, Hu L, Wong KY,
Ling MT, Bainbridge T, Webber D, Chan TH, Guan XY, et al: DNA
fingerprinting tags novel altered chromosomal regions and
identifies the involvement of SOX5 in the progression of prostate
cancer. Int J Cancer. 124:2323–2332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Y, Liu W, Chao T, Zhang Y, Yan X,
Gong Y, Qiang B, Yuan J, Sun M and Peng X: MicroRNA-21
down-regulates the expression of tumor suppressor PDCD4 in human
glioblastoma cell T98G. Cancer Lett. 272:197–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang D, Han S, Wang X, Peng R and Li X:
SOX5 promotes epithelial-mesenchymal transition and cell invasion
via regulation of twist1 in hepatocellular carcinoma. Med Oncol.
32:4612015.PubMed/NCBI
|
|
12
|
Zhang D and Liu S: SOX5 promotes
epithelial-mesenchymal transition in osteosarcoma via regulation of
Snail. J BUON. 22:258–264. 2017.PubMed/NCBI
|
|
13
|
Huang DY, Lin YT, Jan PS, Hwang YC, Liang
ST, Peng Y, Huang CY, Wu HC and Lin CT: Transcription factor SOX-5
enhances nasopharyngeal carcinoma progression by down-regulating
SPARC gene expression. J Pathol. 214:445–455. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pei XH, Lv XQ and Li HX: Sox5 induces
epithelial to mesenchymal transition by transactivation of Twist1.
Biochem Biophys Res Commun. 446:322–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gonzalez ME, Moore HM, Li X, Toy KA, Huang
W, Sabel MS, Kidwell KM and Kleer CG: EZH2 expands breast stem
cells through activation of NOTCH1 signaling. Proc Natl Acad Sci
USA. 111:3098–3103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Truax AD, Thakkar M and Greer SF:
Dysregulated recruitment of the histone methyltransferase EZH2 to
the class II transactivator (CIITA) promoter IV in breast cancer
cells. PLoS One. 7:e360132012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wuebben EL and Rizzino A: The dark side of
SOX2: Cancer-a comprehensive overview. Oncotarget. 8:44917–44943.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bulstrode H, Johnstone E, Marques-Torrejon
MA, Ferguson KM, Bressan RB, Blin C, Grant V, Gogolok S, Gangoso E,
Gagrica S, et al: Elevated FOXG1 and SOX2 in glioblastoma enforces
neural stem cell identity through transcriptional control of cell
cycle and epigenetic regulators. Genes Dev. 31:757–773. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu K, Xie F, Gao A, Zhang R, Zhang L,
Xiao Z, Hu Q, Huang W, Huang Q, Lin B, et al: SOX2 regulates
multiple malignant processes of breast cancer development through
the SOX2-miR-181a-5p, miR-30e-5p-TUSC3 axis. Mol Cancer. 16:622017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Inoue H, Takahashi H, Hashimura M, Eshima
K, Akiya M, Matsumoto T and Saegusa M: Cooperation of Sox4 with
β-catenin-p300 complex in transcriptional regulation of the slug
gene during divergent sarcomatous differentiation in uterine
carcinosarcoma. BMC Cancer. 16:532016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bilir B, Osunkoya AO, Wiles WG VI,
Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD and
Moreno CS: SOX4 is essential for prostate tumorigenesis initiated
by PTEN ablation. Cancer Res. 76:1112–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun R, Jiang B, Qi H, Zhang X, Yang J,
Duan J, Li Y and Li G: SOX4 contributes to the progression of
cervical cancer and the resistance to the chemotherapeutic drug
through ABCG2. Cell Death Dis. 6:e19902015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoon TM, Kim SA, Cho WS, Lee DH, Lee JK,
Park YL, Lee KH, Lee JH, Kweon SS, Chung IJ, et al: SOX4 expression
is associated with treatment failure and chemoradioresistance in
oral squamous cell carcinoma. BMC Cancer. 15:8882015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tiwari N, Tiwari VK, Waldmeier L, Balwierz
PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schübeler D, van
Nimwegen E and Christofori G: Sox4 is a master regulator of
epithelial-mesenchymal transition by controlling Ezh2 expression
and epigenetic reprogramming. Cancer Cell. 23:768–783. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hasegawa S, Nagano H, Konno M, Eguchi H,
Tomokuni A, Tomimaru Y, Asaoka T, Wada H, Hama N, Kawamoto K, et
al: A crucial epithelial to mesenchymal transition regulator,
Sox4-Ezh2 axis is closely related to the clinical outcome in
pancreatic cancer patients. Int J Oncol. 48:145–152. 2016.
View Article : Google Scholar : PubMed/NCBI
|