Introduction
Colorectal cancer (CRC) is one of the most common
malignant tumors and is the third highest cancer for incidence and
has the second highest mortality (9.2%) of different types of
cancer worldwide (1). The principal
treatments used for CRC are surgery, radiation therapy,
chemotherapy and molecular targeted therapy. As a novel treatment
type, molecular targeted therapy has been used in a variety of
tumors including CRC (2,3). The therapeutic strategy to target the
selected epidermal growth factor receptor has been developed in
clinical trials (4). However, it
possesses drug resistance in the treatment of patients with CRC
harboring the V600E mutation of B-Raf proto-oncogene
serine/threonine kinase (BRAFV600E) mutation (5,6). BRAF is
an important oncogene and mutant BRAF has been implicated in the
pathogenesis of several types of cancer. The 1796T>A mutation
results in an amino acid substitution at position 600 in BRAF, from
valine to glutamic acid. This mutation occurs within the activation
segment of the kinase domain and leads to the continuous activation
of the MAPK/ERK signaling pathway (7,8). Although
oral BRAF inhibitors have remarkable clinical activity in
metastatic melanomas with BRAFV600E, resistance to
therapy invariably develops in patients with CRC with the same BRAF
mutation (9,10). Therefore, there is an urgent
requirement to develop a novel and effective treatment for these
patients.
Our previous study identified a novel gene monopolar
spindle protein kinase 1 (Mps1), which is a downstream target of
BRAFV600E, and continuously activated
BRAFV600E signaling may be a potential mechanism for the
deregulation of Mps1 stability and kinase activity in human
malignancies (11,12). Persistent phosphorylation of Mps1
through BRAFV600E signaling is a key event in disrupting
the control of centrosome duplication and chromosome stability that
may contribute to tumorigenesis (13,14).
Notably, phospho (p)-Ser281 Mps1 staining was
demonstrated to be positively associated with p-mitogen-activated
protein kinase (MAPK) [extracellular-signal-regulated kinase
ERK)1/2] in human melanoma tissues (13). However, to the best of our knowledge,
no previous study has investigated whether a correlation exists
between BRAF and Mps1 in CRC.
In the present study, the incidence of
BRAFV600E was determined in CRC tissues, and the
correlation of Mps1 and BRAFV600E and p-ERK in Chinese
patients with CRC was determined. The results raise the possibility
that targeting the oncogenic BRAF and Mps1, particularly when used
in combination, may potentially provide effective therapeutic
opportunities for the treatment of CRC.
Materials and methods
Patients and samples
The present study was approved by the Ethics
Committee of Shanxi Medical University (Taiyuan, China), and
patients provided written informed consent for their inclusion. A
total of 288 (156 male and 132 female) paraffin-embedded tissue
sections containing the carcinoma and its adjacent non-neoplastic
colorectal tissue were obtained from The First Hospital of Shanxi
Medical University and TaiYuan Municipal No. 2 People's Hospital
collected between January 2009 and June 2015. Among them, there
were 284 adenocarcinoma, 1 glandular squamous cell carcinoma, 1
signet-ring cell carcinoma and 2 neuroendocrine carcinoma tissues.
The age of patients at the time of diagnosis with CRC ranged
between 25 and 92 years, and the median age was 64 years. On the
basis of Tumor-Node-Metastasis classification, there were 169
patients at stage I and II, 119 patients at stage IIIb and IV
(15) 110 cases with lymph
metastasis, and 178 cases without lymph node (LN) metastasis
(Table I). All patients were
diagnosed with CRC, with no previous history of other malignant
tumor types, and had not received other treatments prior to
surgery. In 183 cases [168 cases with wild-type BRAF
(BRAFWT) and 15 cases with BRAFV600E], no
cases of squamous cell carcinoma were identified. The complete
clinical data and follow-up data were included in the survival
analysis.
 | Table I.Clinicopathological information of
the 288 patients with colorectal cancer in the present study. |
Table I.
Clinicopathological information of
the 288 patients with colorectal cancer in the present study.
| Clinicopathological
feature | n |
|---|
| Sex |
|
|
Male | 156 |
|
Female | 132 |
| Age, years |
|
|
>60 | 195 |
|
≤60 | 93 |
| Smoking status |
|
|
Yes | 47 |
| No | 241 |
| Drinking
status |
|
|
Yes | 259 |
| No | 29 |
|
Differentiation |
|
|
Well | 51 |
| Medium
and poor | 237 |
| Pathological
pattern |
|
|
Mucinous carcinoma | 24 |
|
Other | 264 |
| Ta (infiltration depth) |
|
|
>T3 | 79 |
|
≤T3 | 209 |
| Lymph node
metastasis |
|
|
Positive | 110 |
|
Negative | 178 |
| Clinical stage
(15) |
|
|
I–II | 169 |
|
III–IV | 119 |
| Location |
|
|
Rectum | 125 |
|
Colon | 163 |
DNA extraction
Surgically removed tissue was fixed in 10% buffered
formalin for 24 h at room temperature and embedded in paraffin.
Hematoxylin and eosin-stained tumor tissues (stained for 5 min and
30 sec at room temperature, respectively) were independently
reviewed by two pathologists. DNA was extracted using the FFPE DNA
kit (Omega Bio-Tek, Inc., Norcross, GA, USA), according to the
manufacturer's protocol. The DNA concentration was determined using
a NanoDrop 2000 instrument and the 260/280 nm ratio was calculated
to evaluate the quality of DNA. The sample concentration was
adjusted to 200–300 ng/µl, and DNA was stored at −80°C.
Polymerase chain reaction (PCR)
Using the DNA template above, and a 252-bp fragment
of BRAF exon 15 was obtained using PCR. The primers of BRAF were
5′-CTTGCCACAGGTCTCCCC-3′ (forward) and
5′-TCTAGTAACTCAGCAGCATCTCAGG-3′ (reverse). The PCR was carried out
in 10 µl PCR buffer, 4 µl dNTP, 2 µl DNA, 1.5 µl forward primer,
1.5 µl reverse primer, 1 µl DNA polymerase (PrimeSTAR GXL DNA
Polymerase TAKARA Japan) and double-distilled water for a total
reaction volume of 50 µl. The amplification procedure was
pre-denaturation for 3 min at 98°C, followed by 30 cycles of 30 sec
at 98°C, 30 sec at 58°C and 30 sec at 72°C. The PCR product was
examined by 2% agarose electrophoresis and stained by ethidium
bromide, then detected under UV. Sanger sequencing was performed by
Beijing Liuhe Huada Gene Technology Company (Beijing, China).
Immunohistochemistry (IHC)
According to the sequencing results, 15 cases
harboring BRAFV600E and the same number of patients
harboring BRAFWT were randomly selected for IHC, with
rabbit anti-human Mps1 antibody (cat. no. ab135819; Abcam,
Cambridge, UK) and rabbit anti-human p-ERK antibody (cat. no. 4376;
Cell Signaling Technology, Inc., Danvers, MA, USA) as primary
antibodies, and horseradish peroxide (HRP)-labeled goat anti-rabbit
immunoglobulin G (IgG; cat. no. A20120A0704; BioTNT, Shanghai,
China) as a secondary antibody. A known positive tissue section
served as a positive control. A negative control was established
using PBS instead of the primary antibody.
The IHC analysis was performed as follows: 4 micron
thick sections were moved to anhydrous ethanol for 2 min at room
temperature, then placed in 95% ethanol liquid cylinder for 2 min,
then moved to 85% ethanol liquid cylinder for 2 min, and finally
placed in 75% ethanol liquid cylinder for 2 min, and then incubated
in 3% hydrogen peroxide to block endogenous peroxidase for 15 min
at room temperature, followed by washing with PBS three times and
soaking in distilled water for 5 min. Antigen retrieval was
performed with citrate liquid in a microwave at 92–98°C for 15 min.
Sections were blocked with 10% goat serum (1201A Shanghai Biyun day
Biotechnology Co., Ltd China) at 37°C for 30 min and rabbit
anti-human Mps1 monoclonal antibody (1:100) or anti-p-ERK antibody
(1:400) was added, and incubated at 4°C overnight. The slides were
then washed with PBS three times and incubated with HRP-labeled
goat anti-rabbit IgG at 37°C for 20 min, followed by washing with
PBS three times and using 3,3′-diaminobenzidine as a chromogen.
Sections were counterstained with hematoxylin for 1 min at room
temperature, dehydrated in graded alcohol and sealed with resin
sealing agent.
Fully automatic digital pathological scanning
apparatus (Leica Microsystems, Inc., Buffalo Grove, IL, USA) were
used to obtain high-resolution digital images. A total of five
high-power fields of vision (×400 magnification) were randomly
selected and analyzed using Image Scope software (12.0; Leica
Microsystems, Inc.). The average value of the positive rate of five
randomly selected fields of view was obtained using standardized
cell nuclear analysis parameters. Nuclear p-ERK expression was
classified as negative for 0–35 and positive for >35. Mps1
expression in the cytoplasm was classified as negative for 0–70 and
positive for >70.
Statistical analysis
Continuous variables are presented as the mean (SD),
and categorical variables are presented as count values
(percentages). The p-ERK and Mps1 positive expression levels were
divided into four groups based on inter-quartile range
respectively, which were 35–45.71, 45.71–51.14 51.14–65.54, and
≥65.54 for p-ERK, as well as 70–88.73, 88.73–117.33, 117.33–157.12
and ≥157.12 for Mps1. The differences were analyzed by
χ2 test. χ2 test or Fisher's exact test was
also used to assess the association between BRAF mutation status
and clinical parameters. Univariate and multivariate survival
analyses were performed using a Cox proportional hazards regression
model. The contingency coefficient was used to evaluate the
correlation between p-ERK and Mps1 expression. A sensitivity
analysis was conducted using Spearman's rank analysis was performed
to determine the correlation between p-ERK and Mps1. Kaplan-Meier
survival analysis and the log-rank test were used to analyze the
association between BRAFV600E and prognosis. Data were
analyzed using SPSS software (version 20.0; IBM Corp., Armonk, NY,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
BRAFV600E mutation and its
association with clinical parameters in CRC
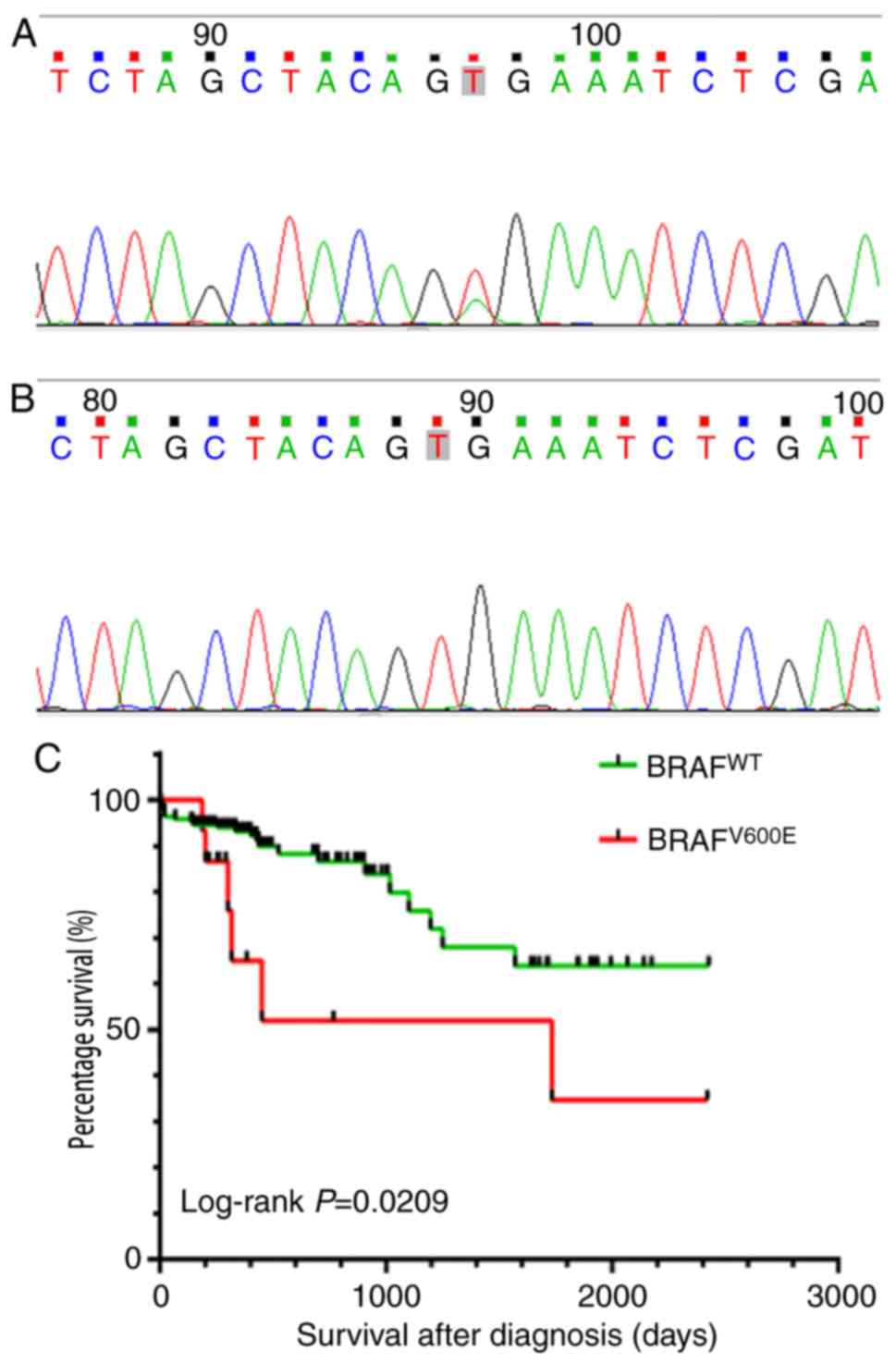
Sanger DNA sequencing was used to detect the
1796T>A (V600E) mutation, which was the most frequently observed
mutation site in BRAF (Fig. 1A and
B). In 288 cases of colorectal cancer, 15 cases of BRAF
mutation were identified. The rate of BRAFV600E was 5.2%
in CRC. A statistical analysis of BRAF mutations and clinical
parameters revealed that BRAFV600E was further
associated with the age, infiltrating depth, pathological pattern
of CRC, were more prevalent in older patients (>60 years),
infiltrating depth>T3 stage patients and the mucinous tumors
(χ2 test or Fisher's exact test P<0.05; Table II). However, no association of the
BRAF mutation with location, clinical stage, LN metastasis,
differentiation and sex were identified (χ2 test or
Fisher's exact test P>0.05; Table
II). Despite previous studies attempting to identify specific
risk factors, no dietary and lifestyle factors have been clearly
associated with the development of BRAF mutated CRC (16–18). The
results of the present study suggest that the BRAF mutation was not
associated with smoking, alcohol intake (χ2 test or
Fisher's exact test P>0.05; Table
II).
 | Table II.Association between
BRAFV600E mutation and clinicopathological parameters in
colorectal cancer. |
Table II.
Association between
BRAFV600E mutation and clinicopathological parameters in
colorectal cancer.
|
|
BRAFV600E mutation |
|---|
|
|
|
|---|
| Clinicopathological
feature | + (n=15; 5.2%) | - (n=273;
94.8%) | P-value |
|---|
| Sex |
|
| 0.739 |
|
Male | 7 | 149 |
|
|
Female | 8 | 124 |
|
| Age, years |
|
| 0.043 |
|
>60 | 14 | 181 |
|
|
≤60 | 1 | 92 |
|
| Smoking status |
|
| 0.142 |
|
Yes | 0 | 47 |
|
| No | 15 | 226 |
|
| Drinking
status |
|
| 0.378 |
|
Yes | 0 | 29 |
|
| No | 15 | 244 |
|
|
Differentiation |
|
| 0.082 |
|
Well | 0 | 51 |
|
| Medium
and poor | 15 | 222 |
|
| Pathological
pattern |
|
| 0.001 |
|
Mucinous carcinoma | 6 | 18 |
|
|
Others | 9 | 255 |
|
| Ta (infiltration depth) |
|
| <0.001 |
|
>T3 | 15 | 64 |
|
|
≤T3 | 0 | 209 |
|
| Lymph node
metastasis |
|
| 0.882 |
|
Positive | 6 | 104 |
|
|
Negative | 9 | 169 |
|
| Clinical stage
(15) |
|
| 0.871 |
|
I–II | 8 | 161 |
|
|
III–IV | 7 | 112 |
|
| Location |
|
| 0.107 |
|
Rectum | 3 | 122 |
|
|
Colon | 12 | 151 |
|
BRAFV600E mutation is
associated with a poor prognosis of patients with CRC
Kaplan-Meier survival analysis indicated that the
survival rate was significantly lower in patients with
BRAFV600E mutation compared with those with
BRAFWT (Fig. 1C). The
median survival time of patients with BRAFV600E and
BRAFWT were 300 and 429.5 days respectively.
Cox regression analysis was used to assess the
impact of BRAF mutations and clinical parameters on OS. Notably,
the results revealed that the association of BRAF mutations with OS
was statistically significant (Cox regression multivariate analysis
HR=0.32, P=0.051; Fig. 2A; Cox
regression univariate analyses, HR=0.36, P=0.03, Fig. 2B). Although the P-value of
BRAFV600E in the multivariate analysis was 0.051These
results suggested that BRAFV600E may serve an important
function in specific pathological CRC, and may function as an
independent prognostic factor and a novel oncological therapeutic
strategy.
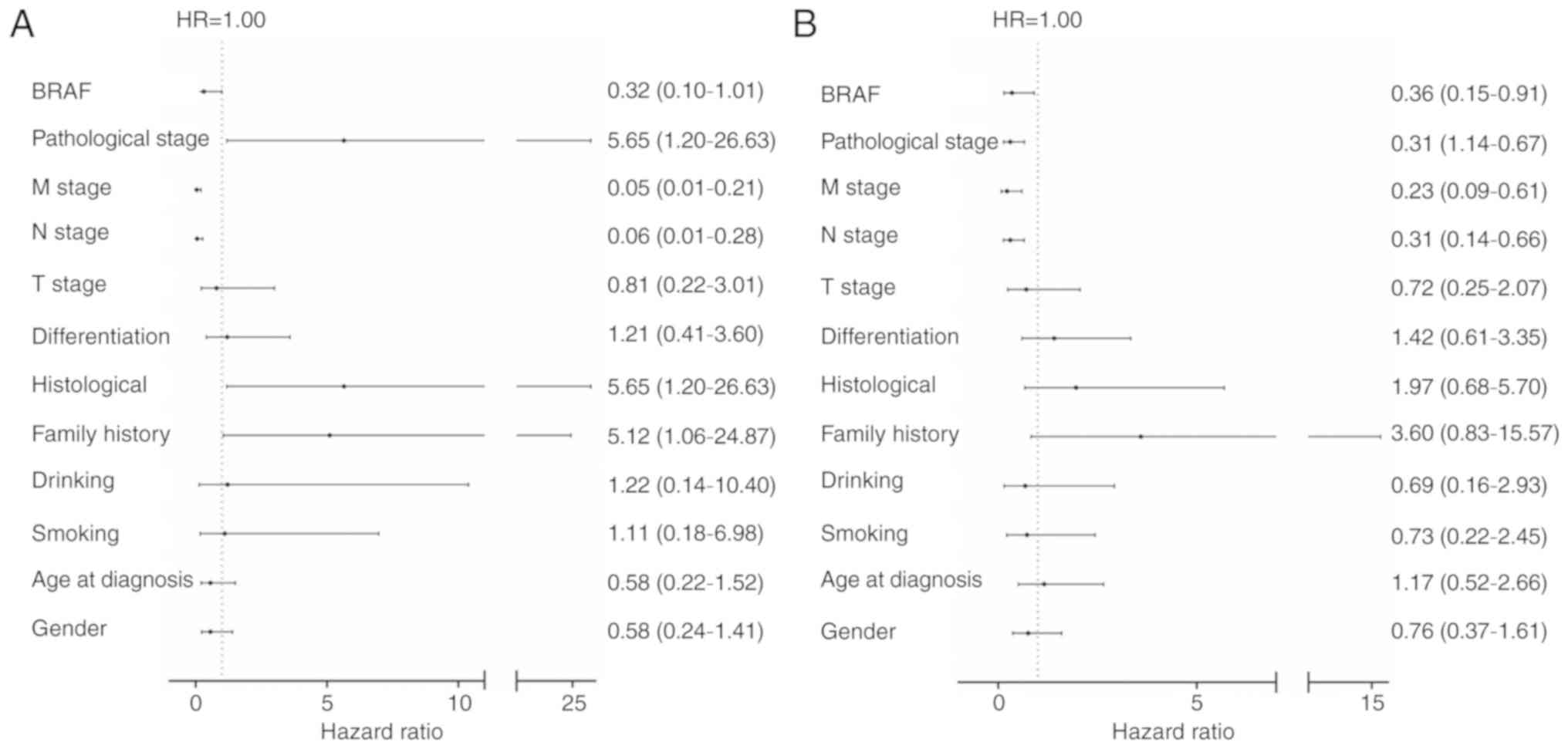 | Figure 2.Multivariate and univariate analyses
of survival prognosis in CRC. (A) Multivariate analyses of survival
prognosis in CRC (B) Univariate analysis of survival prognosis in
CRC. BRAF, WT vs. V600E; pathological stage, I–II vs. III–IV; M
stage, M0 vs. M1; N stage, N0 vs. N1; T stage, T1-2 vs. T3-4;
differentiation, poor vs. medium + well; histological, mucous
adenocarcinoma vs. others; family history, yes vs. no; drinking,
yes vs. no; smoking, yes vs. no; age at diagnosis, ≥60 years vs.
<60 years; sex, male vs. female. CRC, colorectal cancer; BRAF,
B-Raf proto-oncogene serine/threonine kinase; WT, wild-type; HR,
hazard ratio; T, tumor; N, node; M, metastasis. The bracketed
values represent 95% confidence intervals. |
IHC and evaluation of p-ERK and Mps1
in CRC
According to the sequencing results, 15 cases
harboring BRAFV600E and the same number of patients
harboring BRAFWT were randomly selected for IHC with
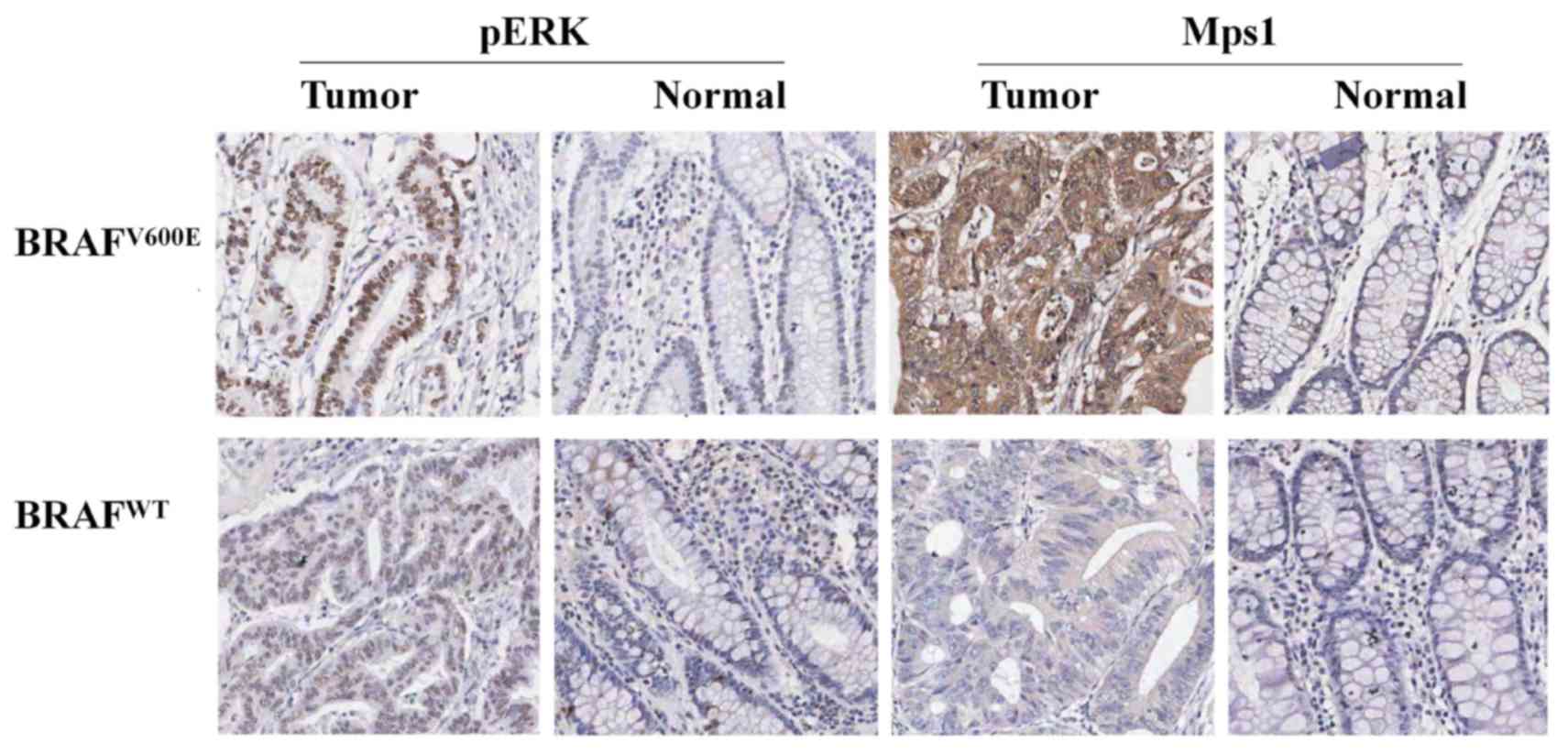
anti-Mps1 and anti-p-ERK antibodies. The positive expression of
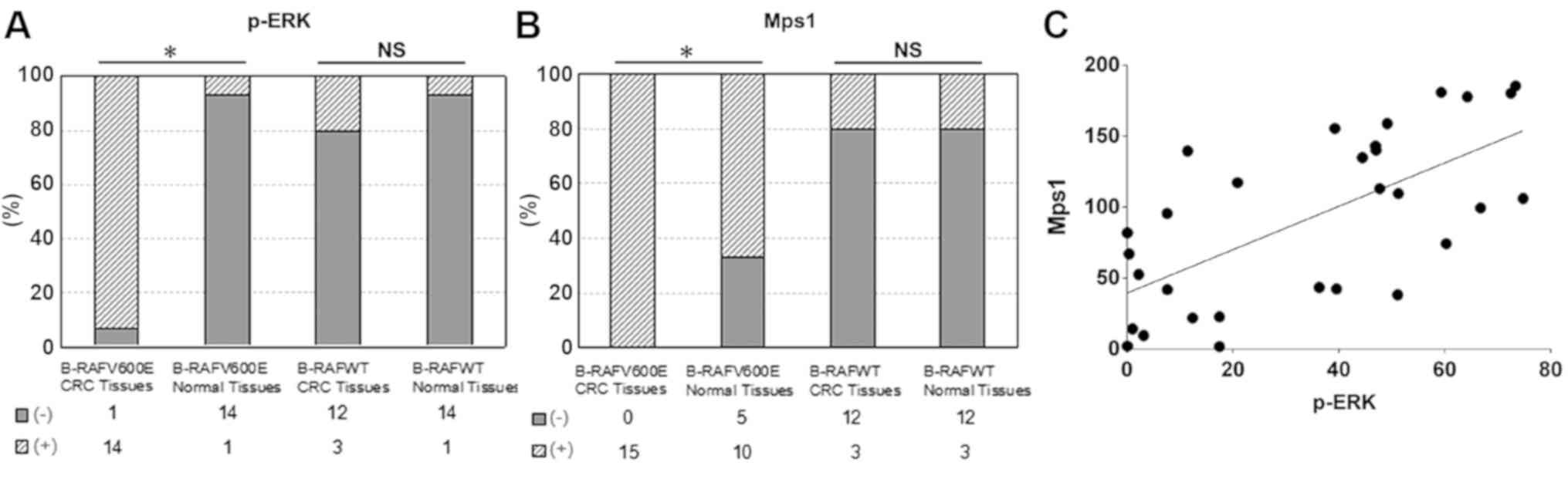
p-ERK protein was brown and localized in the nucleus (Fig. 3). And the positive rate of p-ERK
expression was 93.3% in colorectal cancer tissue with
BRAFV600E, while the positive rate of p-ERK expression
was 6.7% in paired normal tissues, which the difference was
statistically significant in BRAFV600E mutation cases
(Fig. 4A, χ2 test,
P<0.05). However, In BRAFWT cases, there was no
significant difference in the expression of p-ERK between
colorectal cancer and paired normal tissues (Fig. 4A, χ2 test, P>0.05).
Further evaluations were made concerning the
expression of Mps1 in colorectal cancer. the positive expression of
Mps1 protein was brown and localized in the cytoplasm (Fig. 3). The positive expression of Mps1 in
CRC with BRAFV600E was significantly higher compared
with that in paired normal tissues (Fig.
4B, χ2 test, P<0.05). While there was no
significant difference in the expression of Mps1 between colorectal
cancer and paired normal tissues in BRAFWT cases.
(Fig. 4B, χ2 test,
P>0.05).
Association between p-ERK or Mps1
expression and clinical parameters
Subsequently, the association of expression of p-ERK
and Mps1 with clinical pathological features was analyzed (Table III). p-ERK expression was associated
with LN metastasis, pathology type and degree of differentiation
(χ2 test, P<0.05; Table
III). The expression of p-ERK was significantly higher in
poorly differentiated adenocarcinoma and mucinous adenocarcinoma
compared with that in highly differentiated adenocarcinoma and
non-mucinous adenocarcinoma, as well as in the group with LN
metastasis compared with without LN metastasis. (χ2
test, P<0.05; Table III);
however, p-ERK expression was not associated with age, sex, smoking
status, drinking status, location or T stage in CRC (χ2
test, P>0.05; Table III).
 | Table III.Association between p-ERK/Mps1 and
clinicopathological parameters in colorectal cancer. |
Table III.
Association between p-ERK/Mps1 and
clinicopathological parameters in colorectal cancer.
|
| p-ERK in tumor | Mps1 in tumor |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | + (n=17;
56.7%) | - (n=13;
13.3%) | P-value | + (n=18; 60%) | - (n=12; 40%) | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
Male | 10 | 9 |
| 9 | 10 |
|
|
Female | 7 | 4 | 0.558 | 9 | 2 | 0.121 |
| Age, years |
|
|
|
|
|
|
|
>60 | 15 | 11 |
| 17 | 9 |
|
|
≤60 | 2 | 2 | 0.773 | 1 | 3 | 0.274 |
| Smoking status |
|
|
|
|
|
|
|
Yes | 1 | 1 |
| 0 | 2 |
|
| No | 16 | 12 | 0.844 | 18 | 10 | 0.152 |
| Drinking
status |
|
|
|
|
|
|
|
Yes | 1 | 0 |
| 0 | 1 |
|
| No | 16 | 13 | 1.000 | 18 | 11 | 0.400 |
|
Differentiation |
|
|
|
|
|
|
|
Well | 1 | 6 |
| 1 | 6 |
|
| Medium
and poor | 16 | 7 | 0.01 | 17 | 6 | 0.009 |
| Pathological
pattern |
|
|
|
|
|
|
|
Mucinous carcinoma | 6 | 0 |
| 6 | 0 |
|
|
Others | 11 | 13 | 0.024 | 12 | 12 | 0.057 |
| Ta (infiltration depth) |
|
|
|
|
|
|
|
>T3 | 4 | 1 |
| 5 | 0 |
|
|
≤T3 | 13 | 12 | 0.355 | 13 | 12 | 0.066 |
| Lymph node
metastasis |
|
|
|
|
|
|
|
Positive | 8 | 1 |
| 6 | 3 |
|
|
Negative | 9 | 12 | 0.02 | 12 | 9 | 0.626 |
| Clinical stage
(15) |
|
|
|
|
|
|
|
I–II | 8 | 9 |
| 10 | 7 |
|
|
III–IV | 9 | 4 | 0.225 | 8 | 5 | 0.88 |
| Location |
|
|
|
|
|
|
|
Rectum | 13 | 9 |
| 14 | 8 |
|
|
Colon | 4 | 4 | 0.698 | 4 | 4 | 0.679 |
Positive Mps1 expression was significantly greater
in poorly differentiated carcinoma compared with in
well-differentiated adenocarcinoma in CRC (χ2 test,
P<0.05, Table III). There were
no significant associations between positive expression of Mps1 and
age, sex, smoking status, drinking status, location, pathological
type, LN metastasis or T stage in CRC (χ2 test,
P>0.05; Table III).
In addition, the expression of p-ERK and Mps1
between the BRAFV600E and BRAFWT groups were
then compared, and the expression of p-ERK and Mps1 in the
BRAFV600E group was higher compared with those in the
BRAFWT group (Fisher's exact test, P<0.05).
Expression of p-ERK was correlated positively with the Mps1
expression, with a contingency coefficient of 0.679 (P=0.002;
Table IV). In the sensitivity
analysis, it was also identified that p-ERK expression was
positively correlated with the expression of Mps1 (Spearman's rank
correlation analysis correlation coefficient 0.623; P<0.001;
Fig. 4C).
 | Table IV.Association between the p-ERK or Mps1
expression and BRAF mutation in colorectal cancer. |
Table IV.
Association between the p-ERK or Mps1
expression and BRAF mutation in colorectal cancer.
|
| p-ERK | Mps1 |
|---|
|
|
|
|
|---|
| CRC | + | − | P-value | + | − | P-value |
|---|
|
BRAFV600E | 14 (93.3%) | 1 (6.7%) |
| 15 (100%) | 0 (0%) |
|
|
BRAFWT | 3 (20%) | 12 (80%) | <0.001 | 3 (20%) | 12 (80%) | 0.002 |
Discussion
The results of the present study demonstrated that
the incidence of BRAFV600E was 5.2% in CRC, which was
consistent with previously published rates, between 5 and 15%
(19–24). The difference mentioned above may be
due to the complicated genetic background of different ethnicities.
In the study by Yoon et al (25), BRAF mutation frequency in CRC from
Caucasians (13.9%) was twice that of tumors from Asians (5.6%) or
individuals of African (6.4%) descent.
Malignant tumors with the BRAFV600E
mutation have been demonstrated to be associated with mortality in
patients with colorectal cancer (26). Numerous studies have demonstrated that
the malignant tumor with BRAFV600E is insensitive to the
traditional treatments and patients have a poor prognosis (27–29). With
the success of BRAF inhibitors in malignant melanoma, there is
concern about the efficacy of BRAF inhibitors in other tumors with
BRAFV600E mutations (9,30).
However, BRAF inhibitors exhibited severe adverse effects in the
treatment of patients with CRC harboring the BRAFV600E mutation
(31). A previous clinical study
compared the expression of p-ERK between pre- and post-treatment
with BRAF inhibitors, but the results showed that the
downregulation of p-ERK only occurred in 47% of patients with CRC
harboring the BRAFV600E mutation (32). This indicates that the inhibition of
the MAPK signaling pathway by this BRAF inhibitor is insufficient,
which may be a principal reason for the low response rate of BRAF
inhibitors. Thus, identifying novel strategies for the full and
sustained inhibition of the MAPK pathway in patients with CRC with
the BRAF mutant is of marked clinical importance.
Mps1, a member of the spindle-monitoring complex, is
involved in centrosome duplication and spindle checkpoint (33) and cell cycle regulation, and has
maximum kinase activity in the M phase of the cell cycle (34). Typically, Mps1 is an unstable protein,
which is degraded by the ubiquitin-proteasome pathway when
centrosome duplication is completed, and cells enter anaphase
(35). It has been reported that
either too high or too low Mps1 kinase activity results in
aberrations in centrosome duplication (36), lead to aneuploidy formation and result
in malignant tumor formation (37).
Currently, Mps1 has been reported in the breast, colon and other
malignant tumors with high expression, and may facilitate tumor
cell evasion from apoptosis, culminating in carcinogenesis
(38–40). Our previous study identified that Mps1
is a downstream target of BRAFV600E (12). Persistent phosphorylation of Mps1
through BRAFV600E signaling is a key event in disrupting
the control of centrosome duplication and chromosome stability that
may contribute to tumorigenesis (13). Thus, Mps1 may serve as a novel
therapeutic target for patients with CRC harboring the
BRAFV600E mutation.
The effect of Mps1 kinase inhibitors have been
investigated in a variety of malignant tumors, with promising
results (41,42). The present study initially identified
that Mps1 was significantly associated with
BRAFV600E/p-ERK in CRC. It was indicated that Mps1, the
downstream target of the BRAFV600E/MAPK/ERK kinase/ERK
signaling pathway, may serve a significant function in the
development of CRC. The use of a BRAF inhibitor combined with an
Mps1 inhibitor may provide a novel therapeutic approach for
treating patients with CRC harboring the BRAFV600E
mutation, who were previously resistant or insensitive to the BRAF
inhibitor.
However, there were some limitations to the present
study. For example, the data pertaining to the 5-year survival rate
are still being collected, and the sample size of
BRAFV600E is not large enough, which leads to the lack
of representativeness. However, even if the sample size of
BRAFV600E is small, the data still conform to the normal
distribution, ensuring the accuracy and integrity of the results.
We will analyze the 5-year survival data in further study. More
samples of BRAFV600E will be selected in statistical
analysis in further research.
In conclusion, the present study demonstrated that
Mps1 was significantly associated with BRAFV600E/p-ERK
and may serve a crucial function in the development of CRC.
Targeting the oncogenic BRAFV600E and Mps1, particularly
when used in combination, could potentially provide therapeutic
opportunities for the treatment of cancer.
Acknowledgements
The authors would like to thank Professor Yongping
Cui (Translational Medicine Research Center; Key Laboratory of
Cellular Physiology, Ministry of Education of Shanxi Medical
University, Taiyuan, China) for providing the experimental platform
and guidance with writing the paper, and Dr Heyang Cui's
(Translational Medicine Research Center; Key Laboratory of Cellular
Physiology, Ministry of Education of Shanxi Medical University,
Taiyuan, China) help in data analysis.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81201956 and
81602176), Natural Science Foundation of Shanxi (grant no.
2013011043-1), the Science and Technology Innovation Fund of Shanxi
Medical University (grant no. 01201309) and the Youth Research Fund
of Shanxi Medical University (grant no. Q02201202).
Availability of data and materials
The datasets used or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and JG analyzed and interpreted the data of the
study. YZ, JD, RS and YL are responsible for specific experimental
work including DNA extraction of colorectal cancer tissues and
polymerase chain reaction. CC, BS, YB and HH took charge for
collection of case samples and IHC examination. LF, PK and LZ
performed the statistical analysis. YZ was also a major contributor
in writing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shanxi Medical University and patients provided
written informed consent.
Patient consent for publication
There is no disclosure of any personal,
identifiable, non-anonymized patient information in the manuscript.
The patients in the study provided their consent for the
publication of this data and any associated images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stintzing S: Recent advances in
understanding colorectal cancer. F1000Res. 7:F1000 Faculty
Rev–1528. 2018. View Article : Google Scholar
|
|
4
|
Tímár J, Hegedüs B and Rásó E: KRAS
mutation testing of colorectal cancer for anti-EGFR therapy: Dogmas
versus evidence. Curr Cancer Drug Targets. 10:813–823. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Prahallad A, Sun C, Huang S, Di
Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A
and Bernards R: Unresponsiveness of colon cancer to BRAF(V600E)
inhibition through feedback activation of EGFR. Nature.
483:100–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Königsberg R, Hulla W, Klimpfinger M,
Reiner-Concin A, Steininger T, Büchler W, Terkola R and Dittrich C:
Clinical and economic aspects of KRAS mutational status as
predictor for epidermal growth factor receptor inhibitor therapy in
metastatic colorectal cancer patients. Oncology. 81:359–364. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dizdar L, Werner TA, Drusenheimer JC,
Möhlendick B, Raba K, Boeck I, Anlauf M, Schott M, Göring W,
Esposito I, et al: BRAFV600E mutation: A promising target in
colorectal neuroendocrine carcinoma. Int J Cancer. 2018:
|
|
9
|
Kopetz S, Desai J, Chan E, Hecht JR,
O'Dwyer PJ, Maru D, Morris V, Janku F, Dasari A, Chung W, et al:
Phase II pilot study of vemurafenib in patients with metastatic
BRAF-mutated colorectal cancer. J Clin Oncol. 33:4032–4038. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su F, Viros A, Milagre C, Trunzer K,
Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT,
et al: RAS mutations in cutaneous squamous-cell carcinomas in
patients treated with BRAF inhibitors. N Engl J Med. 366:207–215.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui Y and Guadagno TM: B-Raf(V600E)
signaling deregulates the mitotic spindle checkpoint through
stabilizing Mps1 levels in melanoma cells. Oncogene. 27:3122–3133.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui Y, Borysova MK, Johnson JO and
Guadagno TM: Oncogenic B-Raf(V600E) induces spindle abnormalities,
supernumerary centrosomes, an aneuploidy in human melanocytic
cells. Cancer Res. 70:675–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Cheng X, Zhang Y, Li S, Cui H,
Zhang L, Shi R, Zhao Z, He C, Wang C, et al: Phosphorylation of
Mps1 by BRAFV600E prevents Mps1 degradation and contributes to
chromosome instability in melanoma. Oncogene. 32:713–723. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Shi R, He C, Cheng C, Song B, Cui
H, Zhang Y, Zhao Z, Bi Y, Yang X, et al: Oncogenic B-Raf(V600E)
abrogates the AKT/B-Raf/Mps1 interaction in melanoma cells. Cancer
Lett. 337:125–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
NCCN Guidelines version 2.2015 Staging
Colon Cancer. https://www.nccn.org/professionals/physician_gls/default.aspx#site
|
|
16
|
Fu X, Huang Y, Fan X, Deng Y, Liu H, Zou
H, Wu P, Chen Z, Huang J, Wang J, et al: Demographic trends and
KRAS/BRAFV600E mutations in colorectal cancer patients
of South China: A single-site report. Int J Cancer. 2018 Nov
10;Doi: 10.1002/ijc.31973. View Article : Google Scholar
|
|
17
|
Sawada K, Nakamura Y, Yamanaka T, Kuboki
Y, Yamaguchi D, Yuki S, Yoshino T, Komatsu Y, Sakamoto N, Okamoto W
and Fujii S: Prognostic and predictive value of HER2 amplification
in patients with metastatic colorectal cancer. Clin Colorectal
Cancer. 17:198–205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bläker H, Alwers E, Arnold A, Herpel E,
Tagscherer KE, Roth W, Jansen L, Walter V, Kloor M, Chang-Claude J,
et al: The association between mutations in BRAF and colorectal
cancer-specific survival depends on microsatellite status and tumor
stage. Clin Gastroenterol Hepatol. S1542-3565(18)30371-9. 2018.
|
|
19
|
Tie J, Gibbs P, Lipton L, Christie M,
Jorissen RN, Burgess AW, Croxford M, Jones I, Langland R, Kosmider
S, et al: Optimizing targeted therapeutic development: Analysis of
a colorectal cancer patient population with the BRAF(V600E)
mutation. Int J Cancer. 128:2075–2084. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pinheiro M, Ahlquist T, Danielsen SA, Lind
GE, Veiga I, Pinto C, Costa V, Afonso L, Sousa O, Fragoso M, et al:
Colorectal carcinomas with microsatellite instability display a
different pattern of target gene mutations according to large bowel
site of origin. BMC Cancer. 10:5872010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shaukat A, Arain M, Thaygarajan B, Bond JH
and Sawhney M: Is BRAF mutation associated with interval colorectal
cancers? Dig Dis Sci. 55:2352–2356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogino S, Nosho K, Kirkner GJ, Kawasaki T,
Meyerhardt JA, Loda M, Giovannucci EL and Fuchs CS: CpG island
methylator phenotype, microsatellite instability, BRAF mutation and
clinical outcome in colon cancer. Gut. 58:90–96. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rozek LS, Herron CM, Greenson JK, Moreno
V, Capella G, Rennert G and Gruber SB: Smoking, gender, and
ethnicity predict somatic BRAF mutations in colorectal cancer.
Cancer Epidemiol Biomarkers Prev. 19:838–843. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fariña-Sarasqueta A, van Lijnschoten G,
Moerland E, Creemers GJ, Lemmens VE, Rutten HJ and van den Brule
AJ: The BRAFV600E mutation is an independent prognostic
factor for survival in stage II and stage III colon cancer
patients. Ann Oncol. 21:2396–2402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoon HH, Shi Q, Alberts SR, Goldberg RM,
Thibodeau SN, Sargent DJ and Sinicrope FA; Alliance for Clinical
Trials in Oncology, : Racial differences in BRAF/KRAS mutation
rates and survival in stage III colon cancer patients. J Natl
Cancer Inst. 107:djv1862015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barras D, Missiaglia E, Wirapati P, Sieber
OM, Jorissen RN, Love C, Molloy PL, Jones IT, McLaughlin S, Gibbs
P, et al: BRAFV600E mutant colorectal cancer subtypes based on gene
expression. Clin Cancer Res. 23:104–115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bokemeyer C, Bondarenko I, Hartmann JT, de
Braud F, Schuch G, Zubel A, Celik I, Schlichting M and Koralewski
P: Efficacy according to biomarker status of cetuximab plus
FOLFOX-4 as first line treatment for metastatic colorectal cancer:
The OPUS study. Ann Oncol. 22:1535–1546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Strub T, Ghiraldini FG, Carcamo S, Li M,
Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ,
et al: SIRT6 haploinsufficiency induces BRAFV600E
melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat
Commun. 9:34402018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang G, Frederick DT, Wu L, Wei Z,
Krepler C, Srinivasan S, Chae YC, Xu X, Choi H, Dimwamwa E, et al:
Targeting mitochondrial biogenesis to overcome drug resistance to
MAPK inhibitors. J Clin Invest. 126:1834–1856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Knauf JA, Luckett KA, Chen KY, Voza F,
Socci ND, Ghossein R and Fagin JA: Hgf/Met activation mediates
resistance to BRAF inhibition in murine anaplastic thyroid cancers.
J Clin Invest. 128:4086–4097. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Korphaisarn K and Kopetz S: BRAF-directed
therapy in metastatic colorectal cancer. Cancer J. 22:175–178.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Corcoran RB, Atreya CE, Falchook GS, Kwak
EL, Ryan DP, Bendell JC, Hamid O, Messersmith WA, Daud A, Kurzrock
R, et al: Combined BRAF and MEK inhibition with dabrafenib and
trametinib in BRAFV600-mutant colorectal cancer. J Clin Oncol.
33:4023–4031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kasbek C, Yang CH and Fisk HA: Mps1 as a
link between centrosomes and genomic instability. Environ Mol
Mutagen. 50:654–665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Restuccia A, Yang F, Chen C, Lu L and Dai
W: Mps1 is SUMO-modified during the cell cycle. Oncotarget.
7:3158–3170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gorbsky GJ: The spindle checkpoint and
chromosome segregation in meiosis. FEBS J. 282:2471–2487. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Edelmann MJ, Nicholson B and Kessler BM:
Pharmacological targets in the ubiquitin system offer new ways of
treating cancer, neurodegenerative disorders and infectious
diseases. Expert Rev Mol Med. 13:e352011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hiruma Y, Sacristan C, Pachis ST,
Adamopoulos A, Kuijt T, Ubbink M, von Castelmur E, Perrakis A and
Kops GJ: CELL DIVISION CYCLE. Competition between MPS1 and
microtubules at kinetochores regulates spindle checkpoint
signaling. Science. 348:1264–1267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Daniel J, Coulter J, Woo JH, Wilsbach K
and Gabrielson E: High levels of the Mps1 checkpoint protein are
protective of aneuploidy in breast cancer cells. Proc Natl Acad Sci
USA. 108:5384–5389. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shankavaram U, Maachani UB, Zhao S,
Camphausen K and Tandle A: Molecular profiling of MPS1 gene
silencing in U251 glioma cell line. Genom Data. 6:36–39. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ling Y, Zhang X, Bai Y, Li P, Wei C, Song
T, Zheng Z, Guan K, Zhang Y, Zhang B, et al: Overexpression of Mps1
in colon cancer cells attenuates the spindle assembly checkpoint
and increases aneuploidy. Biochem Biophys Res Commun.
450:1690–1695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dominguez-Brauer C, Thu KL, Mason JM,
Blaser H, Bray MR and Mak TW: Targeting mitosis in cancer: Emerging
strategies. Mol Cell. 60:524–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martinez R, Blasina A, Hallin JF, Hu W,
Rymer I, Fan J, Hoffman RL, Murphy S, Marx M, Yanochko G, et al:
Mitotic checkpoint kinase Mps1 has a role in normal physiology
which impacts clinical utility. PLoS One. 10:e01386162015.
View Article : Google Scholar : PubMed/NCBI
|