Introduction
Lung cancer is the leading cause of
cancer-associated mortality in males and females in China. The
majority of patients present with advanced non-small cell lung
cancer (NSCLC) at the time of diagnosis. NSCLC may be categorized
by the driving mutations identified in certain subtypes, including
epidermal growth factor receptor (EGFR), Kirsten rat sarcoma
viral oncogene (KRAS) and anaplastic lymphoma kinase
(ALK) mutations (1–4). Oncogenic ROS1 rearrangements have
become one of the established molecular targets in lung cancer;
however, they have only been identified in 1–2% of NSCLC cases
(5,6).
A total of 15 ROS1 fusion partner genes have been reported,
including H-2 class II histocompatibility antigen gamma chain
(CD74), sodium-dependent phosphate transport protein 2B
(SLC34A2), golgi-associated PDZ and coiled-coil
motif-containing protein (GOPC), coiled-coil
domain-containing protein 6 (CCDC6), syndecan-4
(SDC4), tropomyosin alpha-3 chain (TPM3), ezrin
(EZR), leucine-rich repeats and immunoglobulin-like domains
protein 3 (LRIG3), ER lumen protein-retaining receptor 2
(KDELR2), LIM domain and actin-binding protein 1
(LIMA1), zinc finger protein MSN2 (MSN), clathrin
heavy chain 1 (CLTC), tumor protein D53 (TPD52L1),
L-amino-acid oxidase (FIG) and transmembrane protein 106B
(TMEM106B) (7–9).
For patients with advanced NSCLC, chemotherapy and
radiation provide only palliative relief, however, prognosis is
poor for these patients. Molecular targeted therapy is effective
for patients with advanced NSCLC with associated gene mutations.
The EGFR tyrosine kinase inhibitors (TKIs), including geftinib,
have been widely used as first-line treatments and have higher a
sensitivity compared with platinum-based chemotherapy in advanced
NSCLC with EGFR mutations (10). Crizotinib, an ALK inhibitor, was the
first targeted agent approved by the US food and drug
administration for the treatment of advanced ROS1-rearranged
NSCLC, based on a series of trials (11,12). These
trials revealed that the objective response rate was 72%, with a
median progression-free survival (mPFS) time of 19.2 months. In
addition, crizotinib demonstrated higher overall response and
disease control rates and a longer progression-free survival (PFS)
time in patients with NSCLC with ROS1 rearrangements, when
compared with pemetrexed.
However, the majority of the aforementioned studies
were performed among Caucasian populations. Therefore, the present
study analyzed the clinicopathological features and clinical
efficacy of crizotinib in Chinese patients with NSCLC and
ROS1 rearrangement.
Patients and methods
Patients
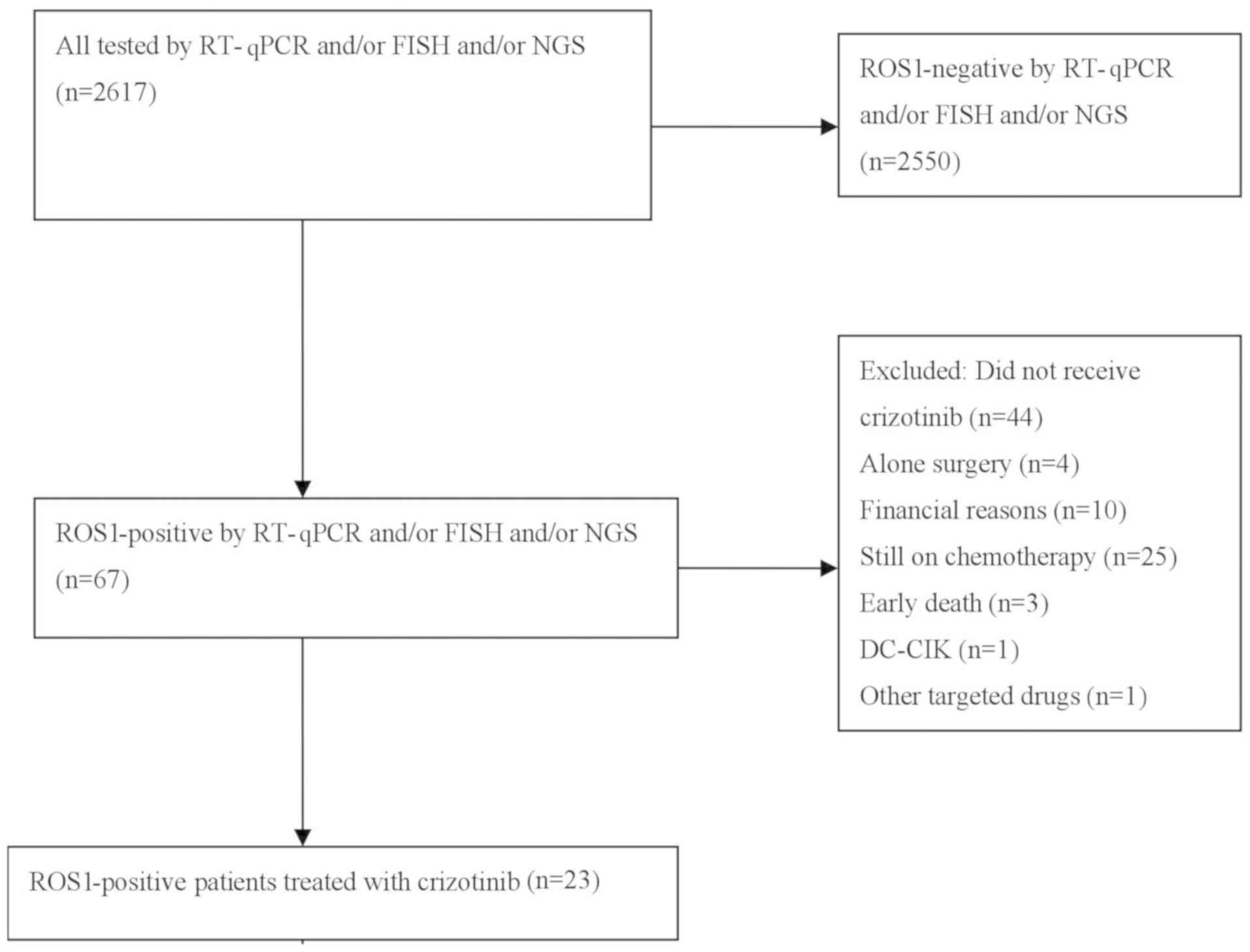
A total of 2,617 patients diagnosed with NSCLC at
Zhejiang Rongjun Hospital (Jiaxing, China), Zhejiang Cancer
Hospital (Hangzhou, China) and Fujian Cancer Hospital (Fuzhou,
China) between January 2013 and December 2016 were included in the
current study (Fig. 1). The
clinicopathological features of the patients are presented in
Table I. The median age of all
patients was 52 years (range, 22–92 years), including 1,415 males
and 1,202 females. The inclusion criteria were as follows: i)
Pathologically confirmed NSCLC with at least one measurable lesion;
and ii) ROS1-positive cancer, as assessed by reverse
transcription quantitative-polymerase chain reaction (RT-qPCR),
fluorescence in situ hybridization (FISH) or next-generation
sequencing (NGS) techniques. The exclusion criteria were as
follows: i) ROS1-negetive cancer, and ii) patients who could
not tolerate crizotinib therapy. The specimen types included
fine-needle aspirate or bronchoscopic biopsy specimens (1,293
cases), surgical specimens (758 cases) and cytology specimens (566
cases). The patients' medical records were reviewed to evaluate
clinicopathological features and treatment regimens. All clinical
data included the patient's age, sex, smoking status, tumor
histological type, performance status (PS), Tumor-Node-Metastasis
(TNM) staging (13), EGFR gene
status and previous treatment regimens. Non-smokers were defined as
patients with a smoking dose of <100 cigarettes in their
lifetime. The study was approved by the Ethics Committee of
Zhejiang Rongjun Hospital (Jiaxing, China) and written informed
consent was obtained from each participant.
 | Table I.Clinical characteristics of
patients. |
Table I.
Clinical characteristics of
patients.
|
| Total patients
(n=2,617) |
|
|---|
|
|
|
|
|---|
| Clinical
characteristic |
ROS1-positive (n=67) |
ROS1-negative (n=2,550) |
P-valuea | Crizotinib-treated
(n=23) |
|---|
| Median age, years
(range) | 68 (35–79) | 52 (22–92) | 0.126 | 64 (35–79) |
| Sex |
|
| <0.001 |
|
|
Male | 21 | 1,394 |
| 8 |
|
Female | 46 | 1,156 |
| 15 |
| Smoking status |
|
| 0.025 |
|
|
Yes | 15 |
908 |
| 2 |
| No | 52 | 1,642 |
| 21 |
| Stage |
|
| <0.001 |
|
|
I–IIIa | 4 |
809 |
| 0 |
|
IIIb–IV | 63 | 1,741 |
| 23 |
| Histology |
|
| 0.066 |
|
|
Adenocarcinoma | 59 | 2,009 |
| 23 |
|
Non-adenocarcinoma | 8 |
541 |
| 0 |
| Specimen type |
|
| >0.999 |
|
|
Fine-needle aspirate or |
|
bronchoscopy specimen | 46 | 1,247 |
| 14 |
|
Surgical specimens | 4 |
754 |
| 0 |
|
Cytology specimens | 17 |
549 |
| 9 |
| EGFR
status |
|
| <0.001 |
|
|
Wild-type | 60 |
941 |
| 17 |
|
Mutation | 1 |
485 |
| 0 |
|
Unknown | 6 | 1,124 |
| 6 |
Immunohistochemistry (IHC)
The paraffin-embedded sections (4 µm) were then
dewaxed using grade I, II and III xylene for 5 min. Subsequently,
the sections were hydrated using 95% ethanol for 1 min followed by
80% ethanol for 1 min, and washed with PBS three times for 1 min
each time. Tissue samples were subjected to antigen retrieval in a
microwave (600 W; Siemens AG, Munich, Germany) with 1 mM EDTA (pH
9.0; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) four times for
5 min. ROS1 (D4D6) rabbit monoclonal antibody (catalog no. sc-3287;
Cell Signaling Technology, Inc., Danvers, MA, USA) was applied at
1:150 in SigalStain antibody diluent (Cell Signaling Technology,
Inc.) at room temperature for 1 h. The sections were then incubated
with 500 µg/ml normal goat IgG (catalog no. sc-2004; 1:3,000; DAKO;
Agilent Technologies, Inc., Santa Clara, CA, USA) dissolved in 1%
BSA in PBS (pH 7.4) for 1 h at room temperature. The sections were
washed with PBS and then incubated with horseradish peroxidase
(HRP)-conjugated goat anti-rabbit antibody (catalog no. 414162;
Nichirei Corporation, Tokyo, Japan) in 1% BSA in PBS for 1 h at
room temperature. Following washing with PBS, the sites of HRP were
visualized with diaminobenzidine (DAB). As a negative control,
certain sections were reacted with goat IgG. DAB or
3-amino-9-ethylcarbazole was used as the chromogen and slides were
counterstained with haematoxylin for 2 min at room temperature
prior to mounting. The color development was observed with a light
microscope) (magnification, ×200). ROS1 IHC was scored using the
following scoring system: 0, no staining; 1+, faint cytoplasmic
reactivity without any background staining; 2+, moderate
cytoplasmic reactivity; and 3+, granular cytoplasmic reactivity of
strong intensity in 10% of tumor cells.
RT-qPCR
Total RNA was extracted from three to four sections
of 3-µm thick formalin-fixed paraffin-embedded (FFPE) tissues using
an RNeasy FFPE kit (Qiagen GmbH, Hilden, Germany). The ROS1
fusion was readily detected by PCR using a ROS1 fusion gene
detection kit (Amoy Diagnostics Co., Ltd, Xiamen, China), according
to the manufacturer's protocol. Briefly, total RNA was subjected to
reverse transcription with a RT-PCR kit (catalog no. M1701, Promega
Corperation, Madison, WI, USA) under the following conditions: 42°C
for 1 h and 95°C for 5 min. The resulting complementary DNA
solutions were used in multiplex RT-qPCR to detect ROS1
fusion gene mRNA. For each case, four reactions were performed to
amplify SLC34A2-ROS1, SDC4-ROS1, CD74-ROS1, EZR-ROS1, TPM3-ROS1,
LRIG3-ROS1 and GOPC-ROS1, and the reference gene
HPRT1. All primers were included in the AmoyDx®
ROS1 Gene Fusions Detection kit (AmonyDiagnostics Co., Ltd.,
Xiamen, China). All the assays were performed on an Agilent Mx3000P
QPCR instrument (Agilent Technologies, Inc.). The following PCR
procedure was used: An initial denaturation at 95°C for 5 min,
followed by 95°C for 25 sec, 64°C for 20 sec and 72°C for 20 sec to
ensure the specificity, and 31 cycles of 93°C for 25 sec, 60°C for
35 sec and 72°C for 20 sec to perform the data collection.
Quantification was achieved using the 2−ΔΔCq
method (14) according to the fusion
fluorescence signal. Assay reactions achieving Cq values of <30
cycles were considered positive. The housekeeping gene HPRT1
was used to control the integrity of the RNA.
FISH
FISH analysis was performed on 3-µm thick tissue
microarrays with a break-apart probe specific to the ROS1
locus (ZytoLight® SPEC ROS1 Dual Color Break Apart
Probe; ZytoVision GmbH, Bremerhaven, Germany), according to the
manufacturer's protocol. The slides were observed using a
fluorescent microscope equipped with a digital camera (Shanghai
Xinsheng Photoelectric Technology Co., Ltd., Shanghai, China).
Tumor cells whose nuclei exhibited ≥1 FISH signals of each color
were enumerated. Cells positive for the rearrangement were defined
as those with split signals or isolated green signals. A specimen
was considered as ROS1-rearranged if the
rearrangement-positive cells constituted 15% of the enumerated
tumor cells.
NGS
Genomic DNA sequencing libraries were prepared using
the protocols recommended by the Illumina TruSeq DNA Library
Preparation kit (Illumina, Inc., San Diego, CA, USA). For samples
close to the minimum input requirement, additional pre-capture PCR
cycles were performed to generate sufficient PCR product for
hybridization. The libraries were hybridized to custom-designed
probes (Integrated DNA Technologies, Inc., Coralville, IA, USA),
including all exons of 170 genes and selected introns of ALK,
RET and ROS1, for the detection of genomic
rearrangements. DNA sequencing was performed on a HiSeq3000
sequencing system (Illumina, Inc.) with 2×75 base pair paired-end
reads. The reads were aligned to the human genome build GRCh37
using Burrows-Wheeler aligner (https://sourceforge.net/projects/bio-bwa/files/bwa-0.7.15.tar.bz2/download).
Somatic single nucleotide variant and indel calls were generated
using MuTect (https://software.broadinstitute.org/gatk/documentation/version-history.php?id=7712&page=4)
and GATK version 3.0 (https://software.broadinstitute.org/gatk/documentation/version-history.php?id=7712&page=4),
respectively. Somatic copy number alterations were identified with
CONTRA version 2.0.8 (https://sourceforge.net/projects/contra-cnv/files/).
Genomic rearrangements were identified by software developed
in-house for analyzing chimeric read pairs.
Evaluation of response
Patients received 250 mg crizotinib twice daily and
continued to receive therapy as long as they did not have
progressive disease (PD) or intolerable side effects. The patients
were evaluated for safety at least once every 2 weeks for the first
two cycles and then at least every 4 weeks thereafter. Radiologic
assessments were performed at baseline and generally following
every two cycles of treatment. Additional assessment could be
performed at any time when symptoms or signs suggested that the
disease may be progressive. Clinical responses were evaluated
according to the response evaluation criteria in solid tumors
version 1.1 (15). PFS time was
measured from the first day of treatment until either tumor
progression or mortality from any cause. The last follow-up was on
December 30, 2016.
Statistical analysis
Categorical variables were compared using the
χ2 test or Fisher's exact test when necessary. Survival
curves were estimated using the Kaplan-Meier method and the
log-rank test, with Bonferroni's correction applied for >2
groups. Statistical analysis was performed using SPSS version 19.0
software (IBM Corp., Armonk, NY, USA). All P-values were two-sided
and P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics
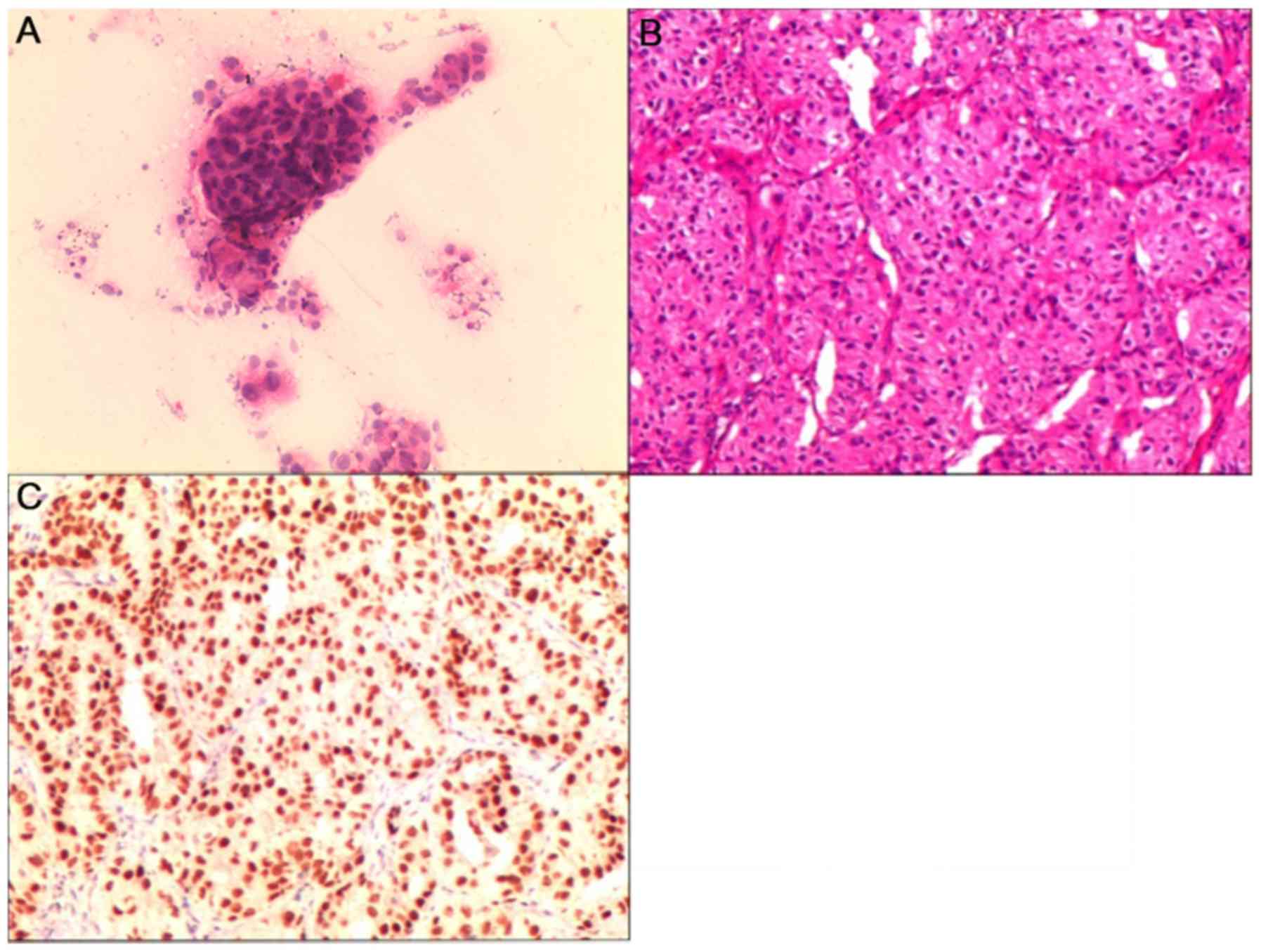
The histopathological findings from different
specimen types are demonstrated in Fig.
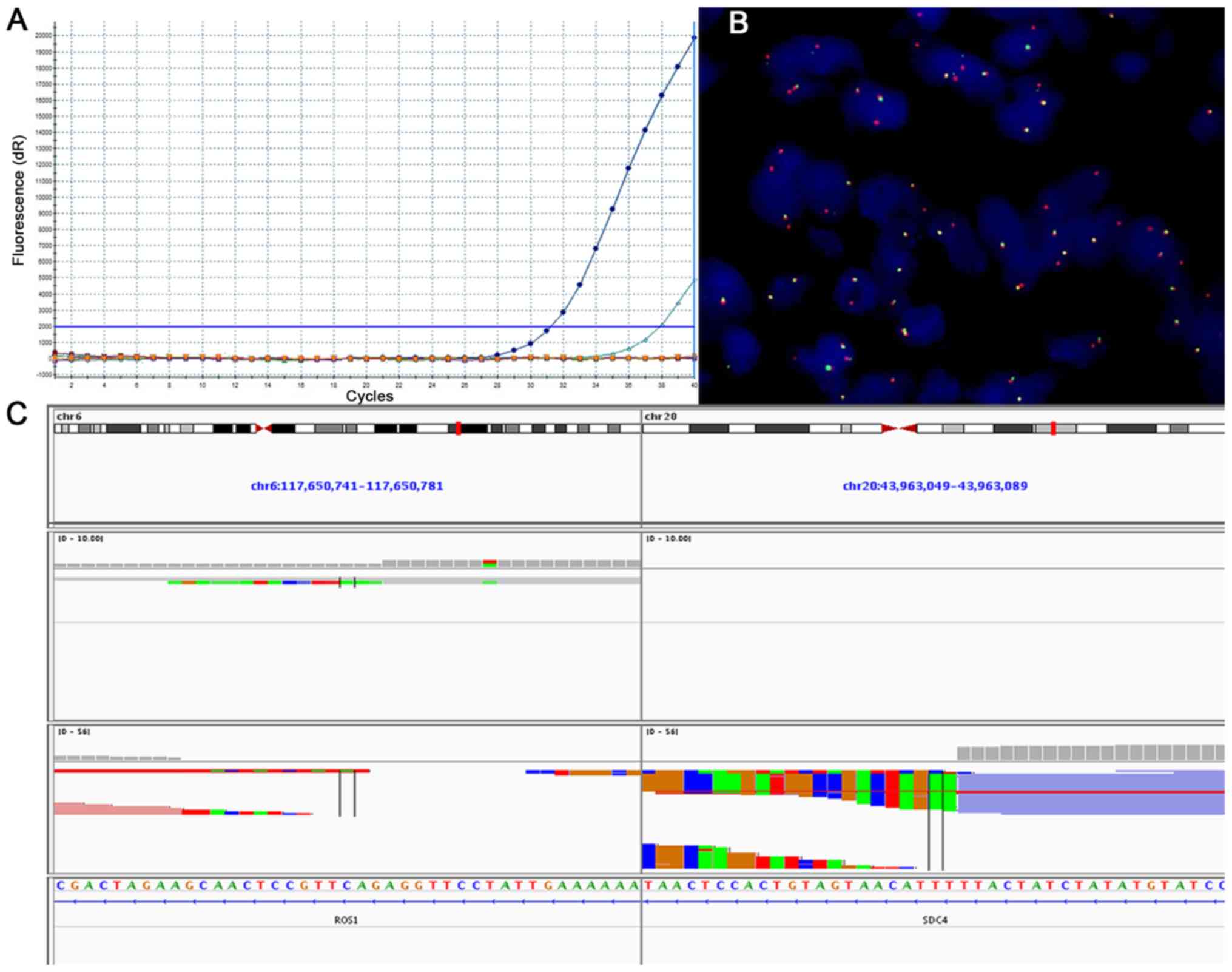
2. Results from tumors revealed to be ROS1-positive with
the use of RT-qPCR, FISH or NGS are presented in Fig. 3. A total of 67 patients (2.56%) were
identified with ROS1 rearrangements, including 21 males and
46 females. The median age was 68 years. Among these cases, 59
(88.05%) were adenocarcinoma and 8 were non-adenocarcinoma.
According to the TNM staging, 4 cases were stage I–IIIa and 63
(94.03%) were stage IIIb–IV. The EGFR gene status included
60 cases of wild-type, 1 case of co-mutation and 6 cases of unknown
status. Statistically significant differences were identified with
regard to sex, smoking history, TNM staging and EGFR gene
status between ROS1 fusion gene-positive and -negative
patients (P<0.05). No statistical significance was observed for
age (P=0.126), histopathological type (P=0.066) or specimen type
(P>0.999) between ROS1 fusion-positive and ROS1
fusion-negative patients. The clinicopathological features of the
patients are outlined in Table I. A
total of 23 patients with advanced NSCLC and a median age of 64
years (range, 35–79 years) received crizotinib, including 4 as
first-line, 5, as second-line and 14 as third-line or later
therapy.
Treatment response to crizotinib in
patients with ROS1 rearrangement
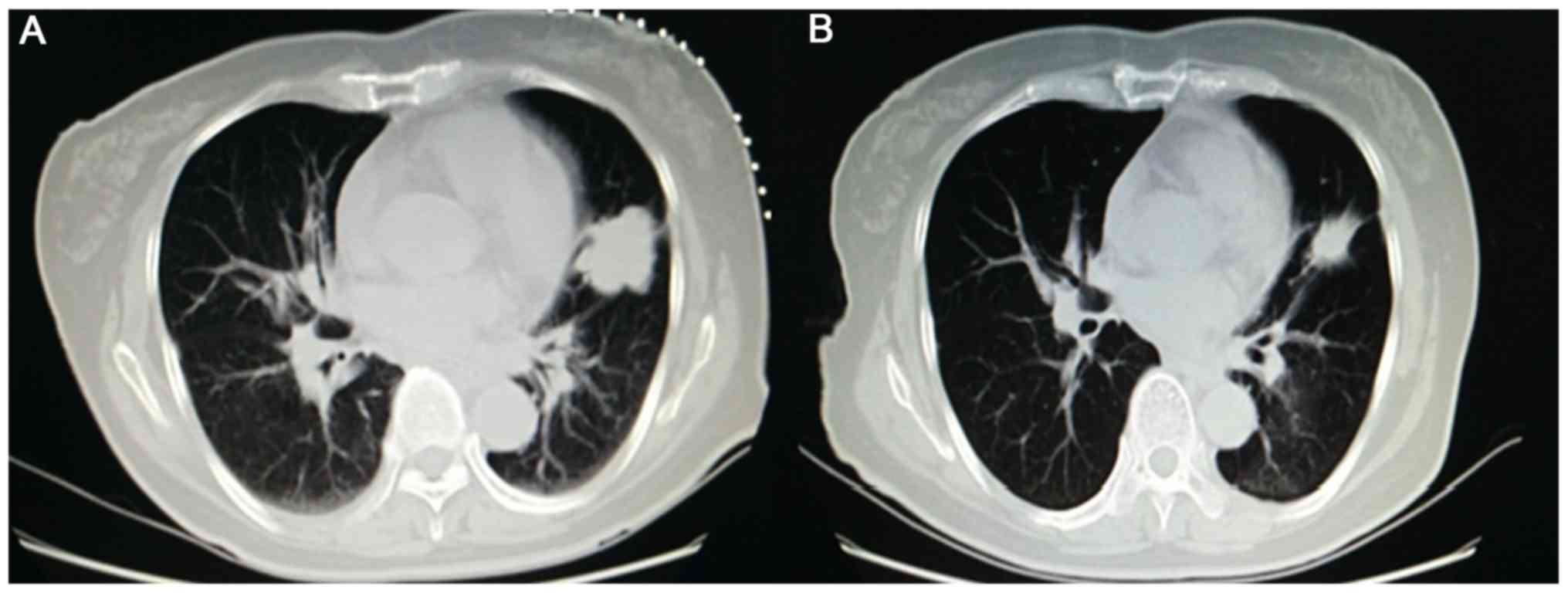
The tumor responses of patients who received
crizotinib are presented in Table
II. No patient exhibited a complete response, 13 achieved a
partial response (Fig. 4), 5
exhibited stable disease and 5 presented with PD. The objective
response rate (ORR) was 56.52% and the disease control rate (DCR)
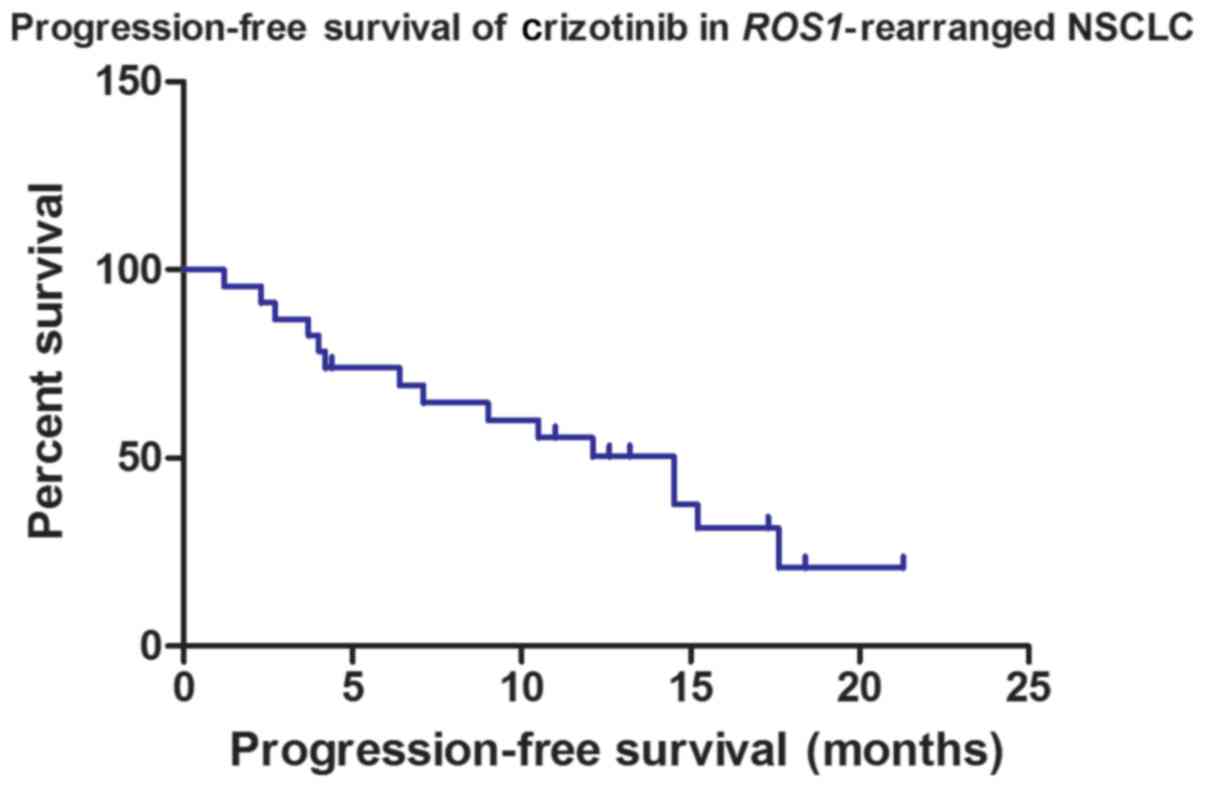
was 78.26%. Among all cases, the median PFS time was 14.5 months
(Fig. 5). To assess whether different
factors could be used as predictive biomarkers of the median PFS
time of patients treated with crizotinib, single and multiple
factor analyses were performed, including for age, sex, smoking
history, PS score, pathology type, TNM staging, tumor protein p53
gene status, EGFR gene status and treatment line of
crizotinib. Although EGFR status was significantly associated with
median PFS time (P=0.038) in signal factor analysis (Table III), the results revealed no
significant differences in median PFS time associated with these
factors following multiple factor analysis (Tables III and IV).
 | Table II.Efficacy of crizotinib in 23 cases of
C-ros oncogene 1 receptor tyrosine kinase-positive non-small cell
lung cancer. |
Table II.
Efficacy of crizotinib in 23 cases of
C-ros oncogene 1 receptor tyrosine kinase-positive non-small cell
lung cancer.
| Response | CR, n | PR, n | SD, n | PD, n | ORR, % | DCR, % | mPFS, months |
|---|
| Efficacy | 0 | 13 | 5 | 5 | 56.52 | 78.26 | 14.5 |
 | Table III.Single-factor analysis of the mPFS
time of the patients. |
Table III.
Single-factor analysis of the mPFS
time of the patients.
| Clinical
features | n | mPFS, months | 95% CI | P-value |
|---|
| Sex |
|
|
| 0.386 |
|
Male | 8 | 14.5 | 8.3–20.7 |
|
|
Female | 15 | 10.5 | 0.1–20.9 |
|
| PS |
|
|
| 0.489 |
|
0–1 | 21 | 14.5 | 7.3–21.7 |
|
| 2 | 2 |
6.4 | − |
|
| Age, years |
|
|
| 0.426 |
|
<60 | 14 | 15.2 | 7.6–22.8 |
|
|
≥60 | 9 | 10.5 | 2.5–18.6 |
|
| Smoking
history |
|
|
| 0.191 |
|
Yes | 2 |
6.4 | − |
|
| No | 21 | 14.5 | 9.5–19.5 |
|
| Pathology |
|
|
|
|
|
Adenocarcinoma | 23 | 14.5 | 9.5–19.5 |
|
|
Non-adenocarcinoma | 0 | − | − |
|
| Crizotinib |
|
|
| 0.106 |
|
First-line | 5 | 14.5 | 9.5–19.5 |
|
|
Second-line | 4 | 10.5 | 2.7–18.3 |
|
| Third-
or further-line | 14 |
3.7 | 0.0–9.9 |
|
| TNM staging |
|
|
|
|
|
I–IIIA | 0 | − | − |
|
|
IIIB–IV | 23 | 14.5 | 9.5–19.5 |
|
| EGFR
status |
|
|
| 0.038 |
|
Mutation | 0 | − | − |
|
|
Wild-type | 19 |
9.0 | 3.7–14.3 |
|
|
Unknown | 4 | − | − |
|
| TP53
status |
|
|
| 0.254 |
|
Mutation | 2 |
4.2 | 1.0–7.4 |
|
|
Wild-type | 2 | 14.5 | − |
|
|
Unknown | 19 | 14.5 | 9.5–19.5 |
|
 | Table IV.EGFR status analysis of the
median progression-free survival time of the patients. |
Table IV.
EGFR status analysis of the
median progression-free survival time of the patients.
| Index | B | SE | Wald | P-value | RR | 95% CI |
|---|
| EGFR
status | −0.927 | 0.519 | 3.192 | 0.074 | 0.396 | 0.143–1.094 |
The most common treatment-associated adverse events
were of grades 1 and 2, including nausea, abdominal pain, diarrhea,
visual disturbances, increased transaminase concentrations and skin
rash. However, grade 3/4 gastrointestinal disturbance and increased
levels of hepatic transaminases were observed in 4 (17.39%) and 2
(8.69%) patients who received crizotinib, respectively. Two
patients stopped crizotinib treatment for 3 days due to
gastrointestinal disturbance. Patients with elevated transaminase
levels were treated at a lower dose without recurrence of dose
limiting toxic effects.
Discussion
In 2007, it was first identified that ROS1
fusion genes are potential oncogenic drivers in NSCLC, as they were
revealed in a rare subset of lung adenocarcinomas (16). ROS1 gene rearrangements were
detected in 0.9–1.7% of unselected patients with NSCLC (17,18).
However, the frequency of ROS1 fusions increased to 3% in
lung adenocarcinoma and to 3.9–7.4% in patients with lung
adenocarcinoma who possessed wild-type
EGFR/KRAS/ALK (5,19–21). The frequency of ROS1
rearrangement was 2.56% among unselected patients with NSCLC in the
current study, which was slightly higher compared with the results
of previous studies. This may have been associated with the higher
incidence of lung adenocarcinoma (79.02%) in the present study.
Commonly, studies have demonstrated that ROS1
fusions are mutually exclusive with respect to EGFR, KRAS
mutations or ALK fusions (19,21).
However, co-mutation of ROS1 with other driver genes has
been reported in the literature (22–25). In
these previous studies, 2 advanced NSCLC patients with concomitant
ROS1 rearrangements and deletion of EGFR exon 19
presented with a partial response following first-line treatment
with gefitinib. In addition, 1 patient with ALK/ROS1
coexistence was treated with crizotinib for 3 months and responded
to this inhibitor (26). However,
KRAS oncogene point mutations may interfere with ROS1
signaling that is otherwise intact, resulting in a lack of response
to crizotinib, and these are therefore associated with a poor
ROS1-targeted therapy response (27). For patients with co-mutation
ROS1-positive NSCLC, further studies are required to
summarize the clinical presentation, different therapy regimens and
prognostic factors.
Patients with EGFR mutations in Eastern Asia
tend to be female, non-smokers and exhibit adenocarcinoma
histology, on the basis of subgroup analysis (28). In addition, certain studies have
demonstrated that patients with ROS1 rearrangements are
often younger, non-smokers and exhibit adenocarcinoma histology
(20,29). However, other studies have reported
contradictory findings (19,30). The current results demonstrated that
patients with ROS1 rearrangement were more frequently female
with advanced-stage disease. Similar to EGFR-mutated NSCLC,
differences have been revealed in clinicopathological features
between Eastern Asian and Caucasian patients (31). For patients with ROS1-positive
NSCLC, the biological features between different ethnicities
require further research.
Although FISH has been approved for the detection of
gene rearrangement in NSCLC, the high cost and complex operation
limits its use (5,32). Based on experience with ALK,
assays for the detection of ROS1 have included FISH, RT-qPCR
and NGS (33). These methods of
detection each have their own advantages and disadvantages. In the
current study, RT-qPCR was used for ROS1 fusion detection
due to its high sensitivity, requirement for small samples and
shorter detection time. NGS is a novel method for detecting large
numbers of gene fusions with known and unknown genes, and gene
mutations, at the same time (34).
Crizotinib has been demonstrated to be an effective
drug in improving the prognosis of patients with NSCLC and
ROS1 rearrangements. The objective response rate was
previously identified to be 72% with an mPFS time of 19.2 months
(11). A study reported that East
Asian patients with ROS1 fusion who were treated with
crizotinib demonstrated an ORR of 69.3% and an mPFS time of 13.4
months (35). In Chinese NSCLC
patients with ROS1 rearrangements, crizotinib demonstrated a
higher ORR (80.0%) and DCR (90.0%), and a longer PFS time (294
days) when compared with pemetrexed (30).
The current results demonstrated that crizotinib is
effective against advanced NSCLC carrying activated ROS1
kinase. In 23 patients, the majority of whom had received multiple
previous therapies, an ORR of 56.52% and DCR of 78.26% were
observed. Among all cases, the mPFS time was 14.5 months. This
response rate is high when compared with the response rate of ~10%
in such cancer cases treated with second-line chemotherapy. In
addition, no associations were revealed between biological features
and mPFS time.
However, similar to EGFR TKIs and ALK
TKIs, there is inevitable acquisition of resistance to targeted
therapies. The acquired resistance mechanism for crizotinib in
patients with NSCLC and ROS1 rearrangements is as yet
unidentified. Molecular changes associated with this type of
resistance in NSCLCs with ROS1 rearrangement are
heterogeneous and include ROS1 tyrosine kinase mutations,
EGFR activation and epithelial-to-mesenchymal transition
(36).
In conclusion, the present study suggests that the
rate of ROS1 fusion in Chinese patients with NSCLC is low.
As an effective and safe drug, crizotinib may be use for treating
patients with ROS1-positive advanced NSCLC. However, for
this subtype, further studies are required to summarize the
biological features and optimal treatments. Additionally, the
current study had certain limitations. Firstly, it was a
retrospective study, which may have allowed selection bias, and
secondly, the number of patients treated with crizotinib was
relatively small. Further large-scale studies are therefore
required.
Acknowledgements
This abstract was presented at the IASLC 19th World
Conference on Lung Cancer, 23–26 September 2018, in Toronto,
Canada, and was published as P1.01-113 in J Thorac Oncol 13 (S508),
2018.
Funding
This study was supported by the Science and
Technology Planning Project of Zhejiang Province (grant no.
2015C33194), the Medical Scientific Research Foundation of Zhejiang
Province (grant no. 2019RC027), the Technology Bureau of Jiaxing
City (grant nos. 2016AY23087, 2017BY18050 and 2018AD32163) and the
National Clinical Key Specialty Construction Program (grant no.
2013).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YCZ analysed and interpretated the data, drafted the
manuscript, gave final approval of the version and agreed to be
accountable for all aspects of the work. XGZ and WXW made
substantial contributions to conception and design of the study.
CWX and KQD obtained and analyzed the data. XPL, XFL, LXW and HFC
performed the experiments. All authors read and approved the final
manuscript.
Ethical approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhejiang Rongjun Hospital (Jiaxing, China) and all
participants provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
ROS1
|
C-ros oncogene 1 receptor tyrosine
kinase
|
|
EGFR
|
epidermal growth factor receptor
|
|
KRAS
|
Kirsten rat sarcoma viral oncogene
|
|
ALK
|
anaplastic lymphoma kinase
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
FISH
|
fluorescent in situ
hybridization
|
|
NGS
|
next-generation sequencing
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
ORR
|
objective response rate
|
|
DCR
|
disease control rate
|
References
|
1
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shi Y, Au JS, Thongprasert S, Srinivasan
S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G and Yang PC: A
prospective, molecular epidemiology study of EGFR mutations in
Asian patients with advanced non-small-cell lung cancer of
adenocarcinoma histology (PIONEER). J Thorac Oncol. 9:154–162.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: Molecular testing guideline for selection of lung
cancer patients for EGFR and ALK tyrosine kinase inhibitors:
Guideline from the College of American pathologists, international
association for the study of lung cancer, and association for
molecular pathology. J Thorac Oncol. 8:823–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wood K, Hensing T, Malik R and Salgia R:
Prognostic and predictive value in KRAS in non-small-cell lung
cancer: A review. JAMA Oncol. 2:805–812. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bergethon K, Shaw AT, Ou SH, Katayama R,
Lovly CM, McDonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang
R, et al: ROS1 rearrangements define a unique molecular class of
lung cancers. J Clin Oncol. 30:863–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Davies KD, Le AT, Theodoro MF, Skokan MC,
Aisner DL, Berge EM, Terracciano LM, Cappuzzo F, Incarbone M,
Roncalli M, et al: Identifying and targeting ROS1 gene fusions in
non-small cell lung cancer. Clin Cancer Res. 18:4570–4579. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ou SH, Chalmers ZR, Azada MC, Ross JS,
Stephens PJ, Ali SM and Miller VA: Identification of a novel
TMEM106B-ROS1 fusion variant in lung adenocarcinoma by
comprehensive genomic profiling. Lung Cancer. 88:352–354. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu VW, Upadhyay D, Schrock AB, Gowen K,
Ali SM and Ou SH: TPD52L1-ROS1, a new ROS1 fusion variant in lung
adenosquamous cell carcinoma identified by comprehensive genomic
profiling. Lung Cancer. 97:48–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Geftinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaw AT, Ou SH, Bang YJ, Camidge DR,
Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa
DB, et al: Crizotinib in ROS1-rearranged non-small-cell lung
cancer. N Engl J Med. 371:1963–1971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mazières J, Zalcman G, Crinò L, Biondani
P, Barlesi F, Filleron T, Dingemans AM, Léna H, Monnet I,
Rothschild SI, et al: Crizotinib therapy for advanced lung
adenocarcinoma and a ROS1 rearrangement: Results from the EUROS1
cohort. J Clin Oncol. 33:992–999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chansky K, Detterbeck FC, Nicholson AG,
Rusch VW, Vallières E, Groome P, Kennedy C, Krasnik M, Peake M,
Shemanski L, et al: The IASLC lung cancer staging project: External
validation of the revision of the TNM stage groupings in the eighth
edition of the TNM classification of lung cancer. J Thorac Oncol.
12:1109–1121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rikova K, Guo A, Zeng Q, Possemato A, Yu
J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al: Global survey
of phosphotyrosine signaling identifies oncogenic kinases in lung
cancer. Cell. 131:1190–1203. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rimkunas VM, Crosby KE, Li D, Hu Y, Kelly
ME, Gu TL, Mack JS, Silver MR, Zhou X and Haack H: Analysis of
receptor tyrosine kinase ROS1-positive tumors in non-small cell
lung cancer: Identification of a FIG-ROS1 fusion. Clin Cancer Res.
18:4449–4457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen YF, Hsieh MS, Wu SG, Chang YL, Shih
JY, Liu YN, Tsai MF, Tsai TH, Yu CJ, Yang JC and Yang PC: Clinical
and the prognostic characteristics of lung adenocarcinoma patients
with ROS1 fusion in comparison with other driver mutations in East
Asian populations. J Thorac Oncol. 9:1171–1179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu S, Wang J, Zhou L, Su D, Liu Y, Liang
X, Zhang S and Zeng X: Clinicopathological characteristics and
outcomes of ROS1-rearranged patients with lung adenocarcinoma
without EGFR, KRAS mutations and ALK rearrangements. Thorac Cancer.
6:413–420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takeuchi K, Soda M, Togashi Y, Suzuki R,
Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, et
al: RET, ROS1 and ALK fusions in lung cancer. Nat Med. 18:378–381.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mescam-Mancini L, Lantuéjoul S,
Moro-Sibilot D, Rouquette I, Souquet PJ, Audigier-Valette C,
Sabourin JC, Decroisette C, Sakhri L, Brambilla E and McLeer-Florin
A: On the relevance of a testing algorithm for the detection of
ROS1-rearranged lung adenocarcinomas. Lung Cancer. 83:168–173.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song Z, Zheng Y, Wang X, Su H, Zhang Y and
Song Y: ALK and ROS1 rearrangements, coexistence and treatment in
epidermal growth factor receptor-wild type lung adenocarcinoma: A
multicenter study of 732 cases. J Thorac Dis. 9:3919–3926. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu YC, Liao XH, Wang WX, Xu CW, Zhuang W,
Wei JG and Du KQ: Dual drive coexistence of EML4-ALK and TPM3-ROS1
fusion in advanced lung adenocarcinoma. Thorac Cancer. 9:324–327.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu YC, Lin XP, Li XF, Wu LX, Chen HF,
Wang WX, Xu CW, Shen JF, Wei JG and Du KQ: Concurrent ROS1 gene
rearrangement and KRAS mutation in lung adenocarcinoma: A case
report and literature review. Thorac Cancer. 9:159–163. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu YC, Xu CW, Ye XQ, Yin MX, Zhang JX, Du
KQ, Zhang ZH and Hu J: Lung cancer with concurrent EGFR mutation
and ROS1 rearrangement: A case report and review of the literature.
Onco Targets Ther. 9:4301–4305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Uguen A, Schick U and Quéré G: A rare case
of ROS1 and ALK double rearranged non-small cell lung cancer. J
Thorac Oncol. 12:e71–e72. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan W, Yang Y, Zhu H, Zhang Y, Zhou R and
Sun X: KRAS mutation is a weak, but valid predictor for poor
prognosis and treatment outcomes in NSCLC: A meta-analysis of 41
studies. Oncotarget. 7:8373–8388. 2016.PubMed/NCBI
|
|
28
|
Zhou J, Mo W, Zhao J, Zheng J, Ding W and
Zhou J: Clinicopathological features associated with EGFR gene
mutation in non-small cell lung cancer patients. Zhonghua Yi Xue Za
Zhi. 94:2332–2336. 2014.(In Chinese). PubMed/NCBI
|
|
29
|
Cai W, Li X, Su C, Fan L, Zheng L, Fei K,
Zhou C, Manegold C and Schmid-Bindert G: ROS1 fusions in Chinese
patients with non-small-cell lung cancer. Ann Oncol. 24:1822–1827.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Jiang T, Zhao C, Li W, Li X, Zhao
S, Liu X, Jia Y, Yang H, Ren S and Zhou C: Efficacy of crizotinib
and pemetrexed-based chemotherapy in Chinese NSCLC patients with
ROS1 rearrangement. Oncotarget. 7:75145–75154. 2016.PubMed/NCBI
|
|
31
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han JY, Kim SH, Lee YS, Lee SY, Hwang JA,
Kim JY, Yoon SJ and Lee GK: Comparison of targeted next-generation
sequencing with conventional sequencing for predicting the
responsiveness to epidermal growth factor receptor-tyrosine kinase
inhibitor (EGFR-TKI) therapy in never-smokers with lung
adenocarcinoma. Lung Cancer. 85:161–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin JJ and Shaw AT: Recent advances in
targeting ROS1 in lung cancer. J Thorac Oncol. 12:1611–1625. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pekar-Zlotin M, Hirsch FR, Soussan-Gutman
L, Ilouze M, Dvir A, Boyle T, Wynes M, Miller VA, Lipson D, Palmer
GA, et al: Fluorescence in situ hybridization immunohistochemistry,
next-generation sequencing for detection of EML4-ALK rearrangement
in lung cancer. Oncologist. 20:316–322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu YL, Yang JC, Kim DW, Lu S, Zhou J, Seto
T, Yang JJ, Yamamoto N, Ahn MJ, Takahashi T, et al: Phase II study
of crizotinib in East asian patients with ROS1-positive advanced
non-small-cell lung cancer. J Clin Oncol. 36:1405–1411. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song A, Kim TM, Kim DW, Kim S, Keam B, Lee
SH and Heo DS: Molecular changes associated with acquired
resistance to crizotinib in ROS1-rearranged non-small cell lung
cancer. Clin Cancer Res. 21:2379–2387. 2015. View Article : Google Scholar : PubMed/NCBI
|