Introduction
Renal cell carcinoma (RCC) is the second most
commonly diagnosed cancer type in human urological systems and
accounts for almost 3% of all adult malignancies (1). Clear cell renal cell carcinoma (ccRCC)
is the most common type of RCC and accounts for 70–80% of all
occurrences of RCC (2). At the time
of diagnosis, ~33% of patients with ccRCC have distant metastases,
which makes it difficult or occasionally impossible to perform
surgery (3). In addition, treatment
strategies for metastatic ccRCC are limited, as it is often
resistant to chemotherapy and radiotherapy (4). Therefore, biomarkers that can assist
with early diagnosis or prognosis prediction are urgently
required.
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNAs that regulate gene expression at the
post-transcriptional stage (5). It is
understood that this process is mediated via direct binding to the
3′-untranslated region (UTR) of target mRNA transcripts (6). The value of miRNAs in the diagnosis or
treatment of ccRCC has received considerable attention (7). Detection of aberrantly expressed miRNAs
can distinguish ccRCC from normal tissue and predict the prognosis
of patients with ccRCC (8,9).
miR-218 is a vertebrate-specific intronic miRNA and
the gene that encodes miR-218 is located at the introns of tumor
suppressor slit guidance ligand (SLIT)2 and SLIT3 (10). Previous studies have demonstrated that
the expression of miR-218 is significantly reduced in numerous
types of human cancer, including colorectal cancer (11), breast cancer (12), gastric cancer (10), non-small cell lung cancer (13) and ovarian cancer (14), which suggests that miR-218 can
function as a tumor suppressor. Reduced expression of miR-218 was
associated with advanced tumor stage and lymph node metastasis in
ovarian cancer, and restoration of miR-218 expression levels
inhibited cell proliferation, migration and invasion in
vitro by targeting runt-related transcription factor 2
(14). Another study demonstrated
that miR-218 serves as a tumor suppressor in lung cancer by
targeting interleukin-6/signal transducer and activator of
transcription 3, and that it negatively correlates with a poor
prognosis (15). However, to the best
of our knowledge, the expression status and the mechanism by which
miR-218 exerts its clinical significance in ccRCC remain largely
unknown.
Therefore, the present study was aimed to
investigate the expression of miR-218 in ccRCC. In addition, the
biological roles of miR-218 on proliferation and migration were
examined. In addition, the downstream target of miR-218 was
examined by bioinformation analysis tool and western blot analysis.
The present study provided a novel insight into the mechanism of
ccRCC tumor progression and identified a novel biomarker for ccRCC
treatment.
Patients and methods
Patients and tissue samples
A total of 43 patients (mean age, 56.7; range, 42–73
years old) with ccRCC treated at The First Central Hospital of
Baoding (Baoding, China) between January 2011 and October 2012 were
enrolled in the study. Paired tumor and normal tissues (≥2 cm from
the tumor) were obtained from these patients, immediately
snap-frozen in liquid nitrogen and stored at −80°C until further
use. Informed written consent was obtained from all patients who
participated in the study. The study procedure was approved by the
Ethics Committee of The First Central Hospital of Baoding.
Clinicopathological features were obtained and their associations
with the expression level of miR-218 are summarized in Table I.
 | Table I.Associations of miR-218 expression
with the clinicopathological features of patients with clear cell
renal cell carcinoma. |
Table I.
Associations of miR-218 expression
with the clinicopathological features of patients with clear cell
renal cell carcinoma.
|
|
| miR-218 expression
level |
|
|---|
|
|
|
|
|
|---|
| Variable | n | High (n=15) | Low (n=28) | P-value |
|---|
| Sex |
|
|
| 0.511 |
| Male | 22 | 7 | 15 |
|
|
Female | 21 | 8 | 13 |
|
| Age, years |
|
|
| 0.285 |
| ≥60 | 20 | 8 | 12 |
|
|
<60 | 23 | 7 | 16 |
|
| Tumor size, cm |
|
|
| 0.029 |
| ≥5 | 26 | 9 | 17 |
|
|
<5 | 17 | 6 | 11 |
|
| Tumor stage |
|
|
| 0.014 |
| I–II | 15 | 5 | 10 |
|
| III | 28 | 10 | 18 |
|
Cell culture
The human RCC Caki-1 and 786-O cell lines, and the
immortalized proximal tubule epithelial HK-2 cell line were
purchased from the American Type Culture Collection (Manassas, VA,
USA). These cell lines were maintained in Dulbecco's modified
Eagle's medium supplemented with 10% fetal bovine serum (both
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100
U/ml penicillin and 100 U/ml streptomycin. The cell lines were
incubated in a humidified incubator containing 5% CO2 at
37°C.
Cell transfection
The miR-218 mimic (5′-UGUACCAAUCUAGUUCGUGUU-3′),
miR-218 inhibitor (5′-ACAUGGUUAGAUCAAGCACAA-3′) and negative
control (NC, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were designed and
synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
Small interfering RNA (siRNA) targeting CIP2A (si-CIP2A) and NC
siRNA were also synthesized by Guangzhou RiboBio Co., Ltd.
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection of the miRNAs (100 nM)
or siRNAs into Caki-1 and 786-O cell lines, according to the
manufacturer's protocols. Cells were harvested for the following
experiments following transfection for 48 h.
Cell proliferation assay
An MTT assay was conducted to investigate the cell
proliferation ability. Briefly, Caki-1 and 786-O cells transfected
with or without miRNAs or siRNAs were seeded at a density of
3×103 cells/well in a 96-well plate and incubated for 0,
24, 48 or 72 h at 37°C. Subsequently, 20 µl MTT (5 mg/ml) was added
to each well and further incubated for 4 h. Following the removal
of supernatant, 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was added to stop the reaction. The
optical density was measured with a microplate reader at a
wavelength of 595 nm.
Cell migration assay
A wound-healing assay was conducted to measure cell
migration ability. Briefly, Caki-1 and 786-O cells transfected with
or without miRNAs or siRNAs were seeded at a density of
2×105 cells/well in a 6-well plate and incubated for 24
h at 37°C. A wound was then created using a sterile 200-µl pipette
tip at the cell surface of each well. Subsequently, the cells were
observed and the wound closure was quantified at 0 and 48 h
following generation of the wound using Image J 1.42 software
(National Institutes of Health, Bethesda, MA, USA).
3′-UTR luciferase reporter assay
The online prediction algorithm TargetScan
(http://www.targetscan.org/) was used to
predict the direct targets of miR-218. Among these potential
targets, CIP2A was selected as it is recognized as an oncogene in
human cancer (16). For the
luciferase reporter assay, the wild-type and mutant 3′-UTR of CIP2A
was cloned into psiCHECK-2 luciferase reporter vector (Promega
Cooperation, Madison, WI, USA) and termed CIP2A WT and CIP2A MUT,
respectively. Briefly, Caki-1 and 786-O cells were seeded at a
density of 2×105cells/well in a 24-well plate. The cells
were co-transfected with CIP2A WT or CIP2A MUT together with the
miR-218 mimic or NC miRNA using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Luciferase activities were
measured using the Dual-Luciferase Reporter Assay kit (Promega
Cooperation) with Renilla luciferase activity as the
internal control, following co-transfection for 48 h, according to
the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissue or Caki-1 and 786-O cells
transfected with or without miRNAs or siRNAs was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. RNA quality was
measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher
Scientific, Inc.). M-MLV Reverse Transcriptase (Promega
Cooperation) was used to synthesize complementary DNA at the
prepared buffer of 250 mM Tris-HCl (pH 8.3, 25°C), 375 mM KCl, 15
mM MgCl2, 50 mM DTT. The expression levels of miR-19a-3p were
determined using the miScript SYBR Green PCR kit (Qiagen GmbH,
Hilden, Germany) using an ABI7500 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions were
as follows: Initial denaturation at 95°C for 10 min, followed by 40
cycles of denaturation at 95°C for 1 min and annealing/extension at
56°C for 1 min. The following primer sequences were used: miR-218
forward, 5′-ACACTCCAGCTGGGTTGTGCTTGATCTAA-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACATGGTT-3′; and U6 small
nuclear RNA (snRNA) forward, 5′-GCGCGTCGTGAAGCGTTC-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′. The relative gene expression levels were
determined using the 2−ΔΔCq method (17). The U6 snRNA was used for normalizing
the expression of miR-218.
Western blot analysis
Total protein from tissues and Caki-1 and 786-O
cells transfected with or without miRNAs or siRNAs was isolated
using radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China). The concentration of
protein was measured using a BCA protein assay kit (Beyotime
Institute of Biotechnology). Subsequently, protein samples (50 µg)
were separated by 10% SDS-PAGE and then transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billercia, MA,
USA). The membranes were blocked with 5% skimmed milk in
Tris-buffered saline with 0.1% Tween-20 (TBS-T) at room temperature
for 1 h and incubated with mouse primary antibodies against CIP2A
(1:1,000; catalog no. ab128179; Abcam, Cambridge, UK) or β-actin
(1:1,000; catalog no. ab8226; Abcam) at 4°C for overnight.
Following washing with TBS plus Tween-20, the membranes were
incubated with goat anti-mouse horseradish peroxidase-conjugated
secondary antibodies (1:2,000; catalog no. ab6789; Abcam) at room
temperature for 4 h. The protein band signals were developed using
an enhanced chemiluminescence detection kit (Beyotime Institute of
Biotechnology) and analyzed using Image J 1.42 software (National
Institutes of Health).
Statistical analysis
Associations between the miR-218 expression level
and clinicopathological parameters were evaluated using a
χ2 test. An overall survival curve was plotted using the
Kaplan-Meier method and a log-rank test. Correlations between
miR-218 and CIP2A expression levels were assessed by Pearson's
correlation coefficient method. All data are presented as the mean
± standard deviation. Two groups were analyzed by paired Student's
t-test, and three or more groups were analyzed by one-way analysis
of variance followed by Tukey's post hoc test. Statistical analysis
was performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Levels of miR-218 are frequently
downregulated in ccRCC tissues and cell lines
To identify the role of miR-218 in the progression
of ccRCC, miR-218 expression was examined in the collected tissues
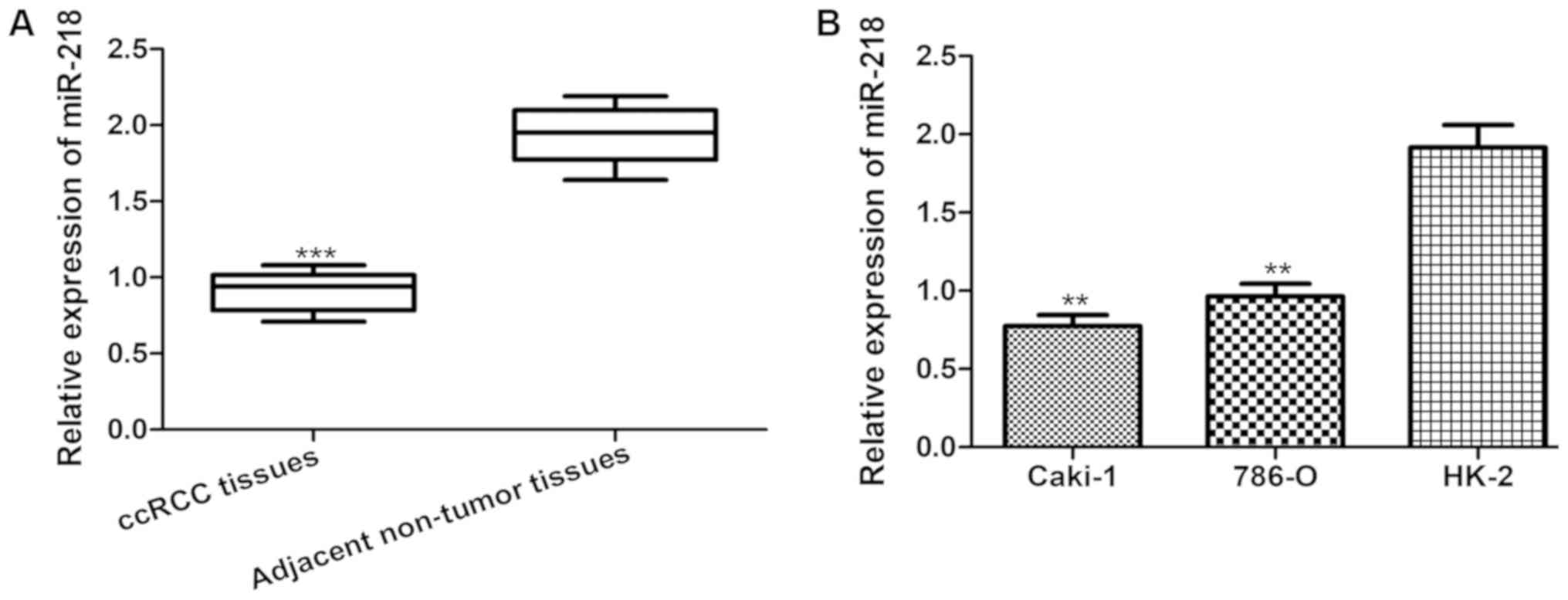
and a series of cell lines. As presented in Fig. 1A, the expression of miR-218 was
significantly lower in ccRCC tissues compared with that in the
adjacent non-tumor tissues. Furthermore, miR-218 expression was
measured in the human RCC Caki-1 and 786-O cell lines, and in the
immortalized proximal tubule epithelial HK-2 cell line. As
expected, it was identified that miR-218 expression was
significantly lower in Caki-1 and 786-O cells compared with that in
HK-2 cells (Fig. 1B).
Low expression of miR-218 is
associated with a poorer prognosis
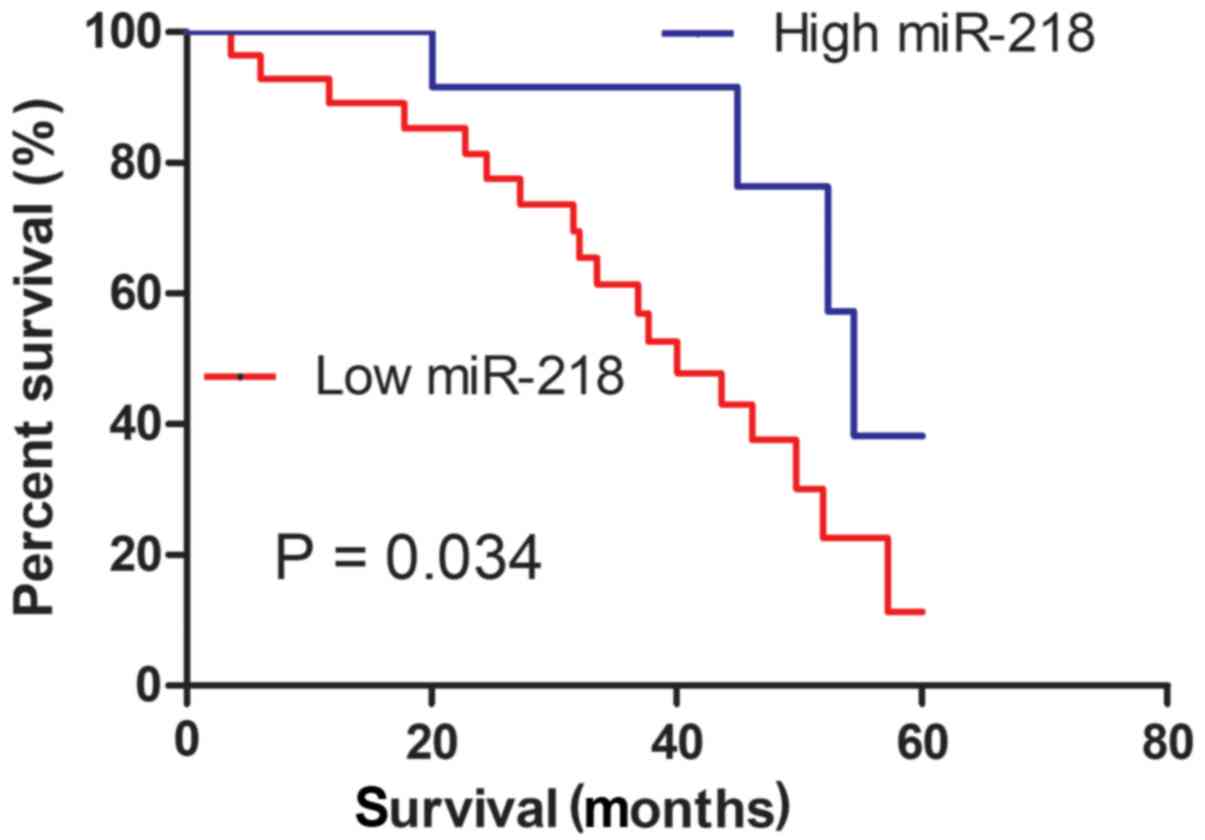
The enrolled patients were classified into two
groups, designated high and low, according to the expression level
of miR-218 obtained from RT-qPCR, with a cut-off median value of
1.03. It was revealed that the expression level of miR-218 was
significantly associated with tumor size and tumor stage (18), but not with sex and age (Table I). The association between miR-218
expression level and the prognosis of patients with ccRCC was then
evaluated. As presented in Fig. 2,
the Kaplan-Meier survival curve revealed that patients with low
miR-218 expression exhibited a significantly poorer overall
survival rate (P=0.034) compared with those with high
expression.
Overexpression of miR-218 inhibits
cell proliferation and migration in vitro
To further elucidate the biological role of miR-218
in ccRCC, gain-of and loss-of function experiments were performed
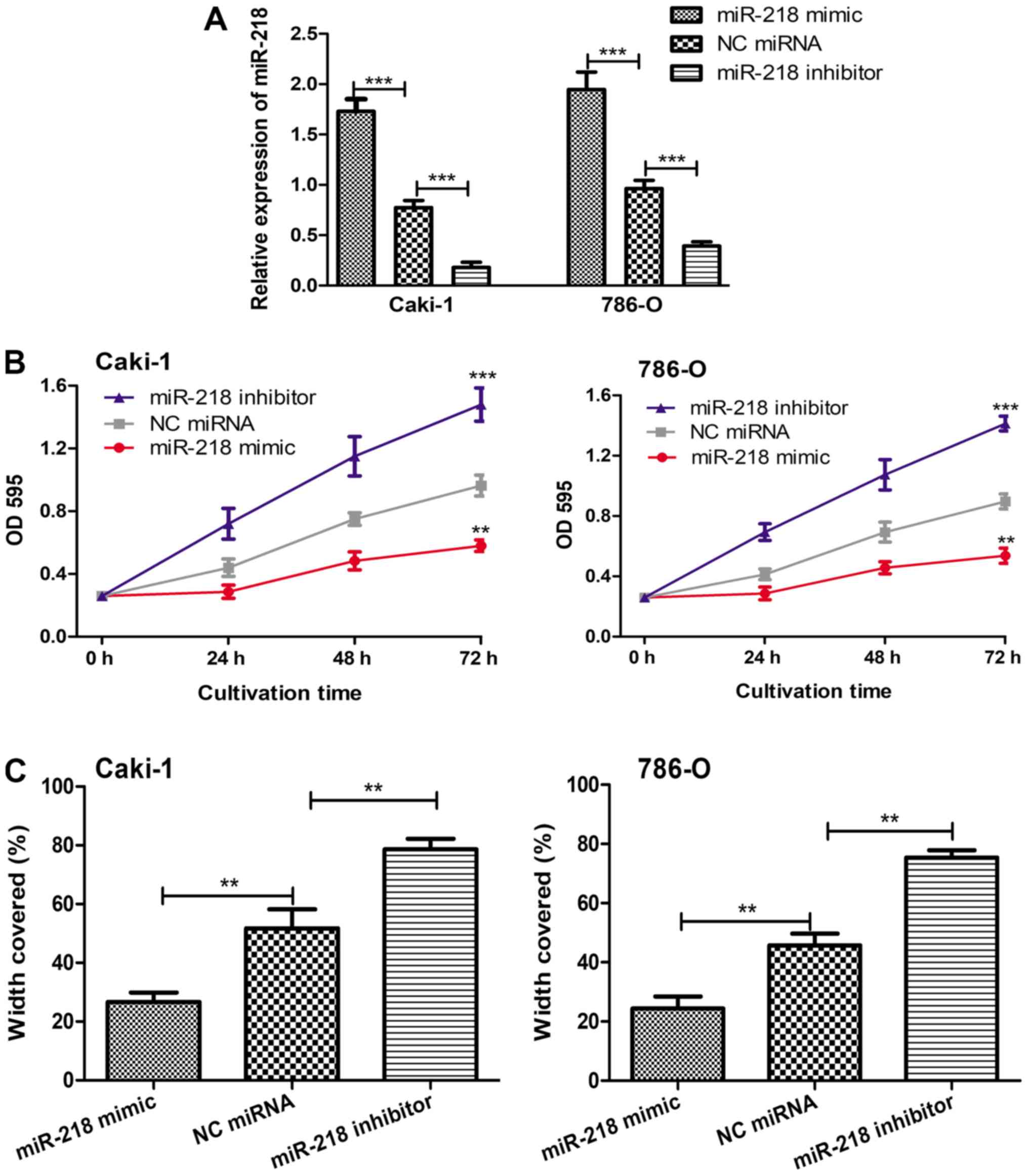
in the selected cell lines. It was confirmed that miR-218 mimic
enhanced the expression level of miR-218, while the miR-218
inhibitor reduced the expression level of miR-218 (Fig. 3A). Subsequently, the in vitro
cell proliferation and migration abilities of the cells were
examined with MTT and wound-healing assays. As demonstrated in
Fig. 3B and C, reduced expression of
miR-218 significantly enhanced cell proliferation and migration,
while induced expression of miR-218 significantly decreased cell
proliferation and migration compared with that found in the cells
transfected with NC miRNA.
miR-218 directly targets CIP2A
To elucidate how miR-218 inhibits cell proliferation
and migration, the TargetScan algorithm was used to predict targets
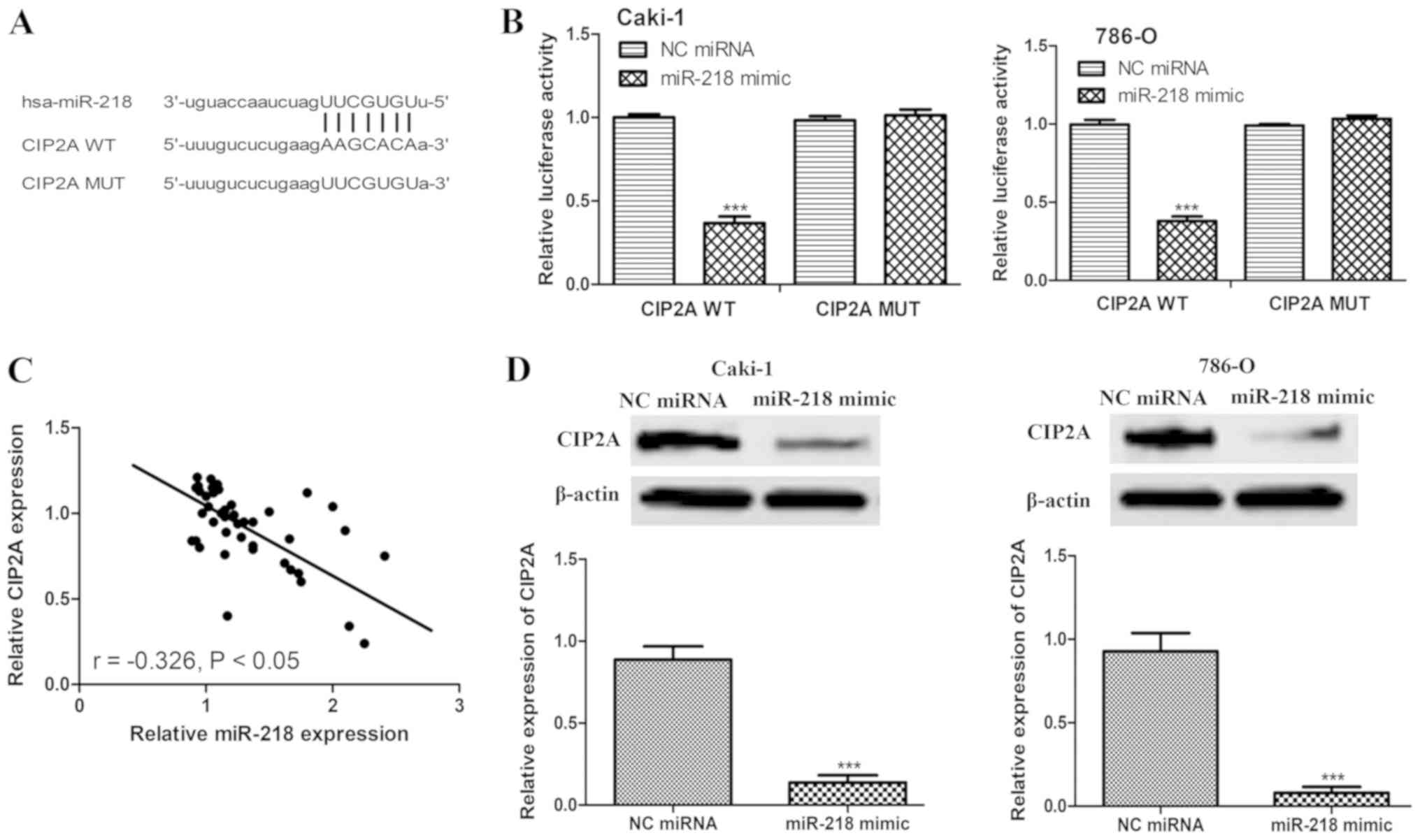
of miR-218. It was identified that CIP2A contains a putative
binding sequence for miR-218 (Fig.
4A). A dual-luciferase reporter assay was then used to validate
whether CIP2A is a direct target of miR-218. miR-218 significantly
inhibited the luciferase activity in cells transfected with CIP2A
WT, but not CIP2A MUT (Fig. 4B).
Furthermore, an inverse correlation between the expression levels
of miR-218 and CIP2A in ccRCC tissues was observed (Fig. 4C). To further confirm that CIP2A is a
target of miR-218, the expression level of CIP2A in Caki-1 and
786-O cells transfected with miR-218 mimic or NC miRNA was
measured. As expected, miR-218 mimic significantly reduced the
protein expression level of CIP2A in the two cell lines (Fig. 4D).
CIP2A functions to promote cell
proliferation and migration in vitro
To determine the biological function of CIP2A, a
loss-of function experiment was performed by transfecting Caki-1
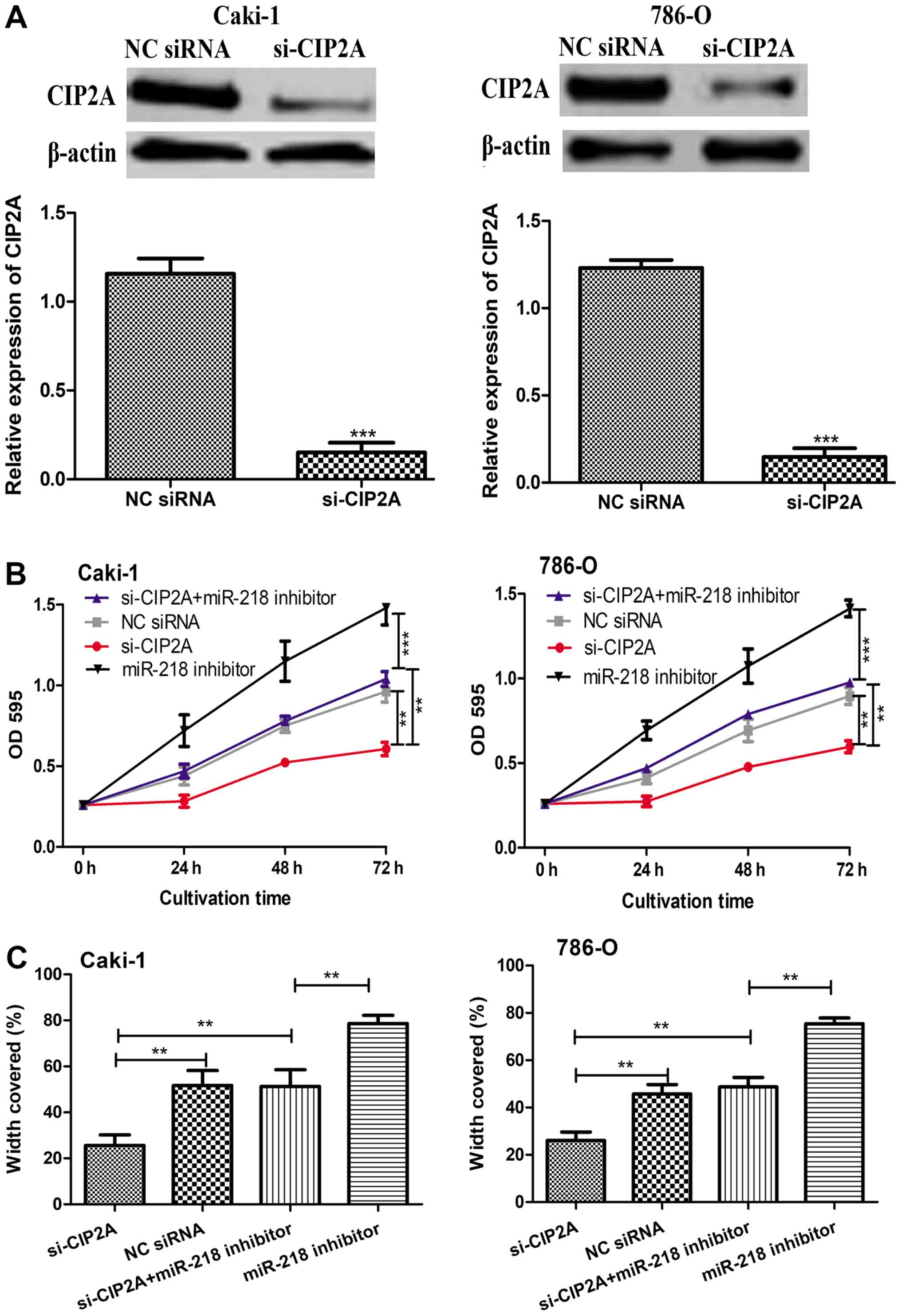
and 786-O cells with si-CIP2A. The efficiency of knockdown was
confirmed by western blot analysis (Fig.
5A). The results of MTT and wound-healing assays revealed that
the knockdown of CIP2A significantly reduced the cell proliferation
and migration in the two cell lines (Fig.
5B and C). Notably, it was also revealed that the
downregulation of CIP2A partly reversed the stimulation effect of
miR-218 inhibitor on the proliferation and migration of Caki-1 and
786-O cells (Fig. 5B and C).
Discussion
ccRCC demonstrates high aggressiveness and is
characterized with high rates of mortality and resistance to
conventional treatment options, including chemotherapy and
radiotherapy (19). In addition, the
majority of patients with ccRCC are diagnosed at a late stage and
no diagnostic or therapeutic biomarkers for ccRCC are currently
available (20). Therefore, the
prognosis of ccRCC is relatively poor (20).
A number of studies have demonstrated that miRNAs
serve crucial roles in various cellular behaviors, including cell
proliferation, apoptosis, migration, tumorigenesis and metastasis
(21,22). The value of miRNAs in predicting the
prognosis of ccRCC has been appreciated (23). In addition, it has been hypothesized
that an improved understanding of the alterations of miRNA
expression in ccRCC may lead to an improved understanding of the
mechanisms underlying ccRCC progression, which may provide
improvements in the diagnosis and treatment measures for advanced
ccRCC (7–9,23).
Downregulation of miR-218 has been reported in
multiple types of human cancer (10–15).
However, the expression pattern and significance of miR-218 in
ccRCC is poorly understood. Therefore, the current study first
examined miR-218 expression in ccRCC tissues, and it was identified
that the expression level of miR-218 was lower in ccRCC tissues
compared with that in adjacent non-tumor tissues. Low-level
expression of miR-218 exhibited a significant association with
characteristics of a more aggressive tumor phenotype, including
larger tumor size and higher tumor stage. Notably, it was revealed
that low expression of miR-218 was associated with a lower 5-year
overall survival rate for patients with ccRCC. In functional
assays, cell proliferation and migration were significantly
suppressed by miR-218 expression. These findings suggest that
miR-218 serves a tumor suppressor role in the development of
ccRCC.
It is understood that cell proliferation and
migration are important hallmarks of malignant tumors (24). Therefore, identification of the
underlying molecular mechanisms involved in regulation of ccRCC
proliferation and migration is crucial. In the present study,
miR-218 targets were predicted to investigate the mechanism
underlying the function of miR-218 in ccRCC. Using online
predication and luciferase reporter assay, it was revealed that
CIP2A may be a target of miR-218 in ccRCC. To support this, it was
identified that the expression level of CIP2A was inversely
correlated with the expression level of miR-218 in ccRCC, and that
CIP2A expression could be negatively regulated by miR-218. Further
in vitro functional assays revealed that knockdown of CIP2A
reduced cell proliferation and migration. Cell proliferation and
migration were lower in cells co-transfected with si-CIP2A and
miR-218 inhibitor compared with that in the cells only transfected
with miR-218 inhibitor. In summary, these findings indicate that
miR-218 exerts its biological function in ccRCC by regulating the
expression of CIP2A.
In conclusion, the current study investigated the
possible role of miR-218 in ccRCC progression and its underlying
mechanisms. The results suggest that miR-218 functions as a novel
tumor suppressor by regulating the expression of CIP2A in ccRCC and
that miR-218 may be used as a therapeutic target for ccRCC
treatment in the future. Although certain novel insights into the
mechanisms regarding the progression of ccRCC were provided in the
present study, the mechanisms by which miR-218 is downregulated
require elucidation in future studies.
Acknowledgements
Not applicable.
Funding
This study was supported by the Hebei Province
Natural Science Foundation of China (grant no. H2017201052).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RW, XY, YZ, NJ and TL conceived and designed the
experiments. RW, XY, YZ, NJ, TL, WL, HW, GZ and LS performed the
experiments. RW and YZ analyzed the data. RW, XY, NJ and TL
contributed reagents/materials/analysis tools. RW wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The First Central Hospital of Baoding (Baoding, China). Informed
written consent was obtained from all patients who participated in
the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ljungberg B, Hanbury DC, Kuczyk MA,
Merseburger AS, Mulders PF, Patard JJ and Sinescu IC; European
Association of Urology Guideline Group for renal cell carcinoma, :
Renal cell carcinoma guideline. Eur Urol. 51:1502–1510. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumor-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crispen PL, Breau RH, Allmer C, Lohse CM,
Cheville JC, Leibovich BC and Blute ML: Lymph node dissection at
the time of radical nephrectomy for high-risk clear cell renal cell
carcinoma: Indications and recommendations for surgical templates.
Eur Urol. 59:18–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al-Ali BM, Ress AL, Gerger A and Pichler
M: MicroRNAs in renal cell carcinoma: implications for
pathogenesis, diagnosis, prognosis and therapy. Anticancer Res.
32:3727–3732. 2012.PubMed/NCBI
|
|
5
|
D'Angelo B, Benedetti E, Cimini A and
Giordano A: MicroRNAs: A puzzling tool in cancer diagnostics and
therapy. Anticancer Res. 36:5571–5575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li M, Wang Y, Song Y, Bu R, Yin B, Fei X,
Guo Q and Wu B: MicroRNAs in renal cell carcinoma: A systematic
review of clinical implications (Review). Oncol Rep. 33:1571–1578.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu
X, Xiao P, Shi H, Wang R, Chen L, et al: MiR-141 Is a key regulator
of renal cell carcinoma proliferation and metastasis by controlling
EphA2 expression. Clin Cancer Res. 20:2617–2630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nofech-Mozes R, Khella HW, Scorilas A,
Youssef L, Krylov SN, Lianidou E, Sidiropoulos KG, Gabril M, Evans
A and Yousef GM: MicroRNA-194 is a marker for good prognosis in
clear cell renal cell carcinoma. Cancer Med. 5:656–664. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S,
Guo X, Wang B, Gang Y, Zhang Y, et al: mIr-218 inhibits invasion
and metastasis of gastric cancer by targeting the Robo1 receptor.
PLos Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao
X, Jia W and Huang J: Decreased expression of miR-218 is associated
with poor prognosis in patients with colorectal cancer. Int J Clin
Exp Pathol. 6:2904–2911. 2013.PubMed/NCBI
|
|
12
|
Liu B, Tian Y, Li F, Zhao Z, Jiang X, Zhai
C, Han X and Zhang L: Tumor-suppressing roles of miR-214 and
miR-218 in breast cancer. Oncol Rep. 35:3178–3184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi ZM, Wang L, Shen H, Jiang CF, Ge X, Li
DM, Wen YY, Sun HR, Pan MH, Li W, et al: Downregulation of miR-218
contributes to epithelial mesenchymal transition and tumor
metastasis in lung cancer by targeting Slug/ZEB2 signaling.
Oncogene. 36:2577–2588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li N, Wang LF, Tan GY, Guo ZH, Liu L, Yang
M and He J: MicroRNA-218 inhibits proliferation and invasion in
ovarian cancer by targeting Runx2. Oncotarget. 8:91530–91541.
2017.PubMed/NCBI
|
|
15
|
Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang
X, Xiong H, Gurbani D, Li L, Liu Y and Liu A: MicroRNA-218
functions as a tumor suppressor in lung cancer by targeting
IL-6/STAT3 and negatively correlates with poor prognosis. Mol
Cancer. 16:1412017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu N, Zhang T, Zhao D, Cao Z, Du J and
Zhang Q: CIP2A is overexpressed in human endometrioid
adenocarcinoma and regulates cell proliferation, invasion and
apoptosis. Pathol Res Pract. 214:233–239. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
International Agency for Research on
Cancer. Tumours of the kidney. In: Pathology and Genetics: Tumors
of the Urinary System and Male Genital Organs. Moch H, Humphrey PA,
Ulbright T and Reuter VE: 4th. WHO Classification of Tumors. IARC
Press Zurich; Switzerland: pp. pp7–10. 2015
|
|
19
|
De Meerleer G, Khoo V, Escudier B, Joniau
S, Bossi A, Ost P, Briganti A, Fonteyne V, Van Vulpen M, Lumen N,
et al: Radiotherapy for renal-cell carcinoma. Lancet Oncol.
15:e170–e177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang QB, Ma X, Zhang X, Liu SW, Ai Q, Shi
TP, Zhang Y, Gao Y, Fan Y, Ni D, et al: Down-Regulated miR-30a in
clear cell renal cell carcinoma correlated with tumor hematogenous
metastasis by targeting angiogenesis-Specific DLL4. PLoS One.
8:e672942013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Esquela-Kerscher A and Slack FJ: Oncomirs-
microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ran LJ, Liang J, Deng X and Wu JY: miRNAs
in Prediction of Prognosis in Clear Cell Renal Cell Carcinoma.
Biomed Res Int. 2017:48329312017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|