Introduction
Glioma is the most malignant and incurable brain
tumor with a poor outcome globally (1). Despite advances in standard therapy,
including surgical resection, radiotherapy and chemotherapy, the
5-year survival rate remains dismal (2,3).
Therefore, identifying novel diagnostic and prognostic makers is
essential.
Pseudogenes were initially regarded as
non-functional genomic fossils resulting from inactivating gene
mutations during evolution (4).
Pseudogene-derived RNAs serve multifaceted roles, including
post-transcriptional regulation identified as antisense RNAs,
endogenous small-interference RNAs and competing endogenous RNAs
(5,6).
Previous studies revealed that peudogenes exhibit multilayered
biological functions in multiple cellular processes, including
proliferation, migration and invasion, in numerous tumor types
(5,7).
For instance, small ubiquitin-like modifier 1 pseudogene 3
(SUMO1P3) had a significantly increased expression in gastric
cancer, and its expression was significantly associated with tumor
size, differentiation, lymphatic metastasis and invasion (8). Increased expression of SUMO1P3 predicts
poor prognosis and promotes tumor growth and metastasis in bladder
cancer (9). Zinc finger protein 91
pseudogene promotes the migration of BXPC-3-H cells and may be a
novel marker for early diagnosis for pancreatic cancer (10). DUXAP8 is identified to act as an
oncogene in non-small cell lung cancer (NSCLC) and promotes NSCLC
progression (11). In gastric cancer,
DUXAP8 could epigenetically suppress the expression of pleckstrin
homology domain containing O1 and enhance proliferation and
migration (12). However, the role of
pseudogene DUXAP8 in glioma progression remains unknown.
In the present study, it was revealed that
pseudogene DUXAP8 is significantly upregulated in glioma tissues.
Patients with increased DUXAP8 expression levels demonstrated poor
survival rate, implying that DUXAP8 was a prognostic marker for
patients with glioma. It was further demonstrated that knockdown of
DUXAP8 suppressed proliferation. Therefore, the results of the
present study indicated that pseudogene DUXAP8 may be a potential
prognostic biomarker and target of glioma treatment.
Materials and methods
Patient tissue samples
A total of 58 paired of human glioma tissues and
adjacent normal tissue sample were collected from patients
including 34 males and 23 females (age range, 31–72 years; median,
52.22 years), who were undergoing surgical resection at the
Department of Neurosurgery, The Second Hospital of Shandong
University (Jinan, China) between January 2011 and December 2014.
The adjacent normal brain tissue was defined as 1 cm away from the
lesions. None of the patients receive treatment, including
radiation or chemotherapy, prior to surgery. The patients with
glioma were classified as World Health Organization (WHO) I, II,
III and IV stage, according to a previous report (13). The tissue samples were snap-frozen in
liquid nitrogen immediately following resection and stored at
−80°C. The follow-up date was between March 2012 and January 2017.
The follow-up date was between the date of the primary surgery and
relapse, patient mortality or the late follow-up date prior to
mortality. The study was approved by the Ethics Committee of The
Second Hospital of Shandong University (Jinan, China). Written
informed consent was obtained from all patients in the study.
Cell lines culture
Human glioma U87 (U-87MG Uppsala), U251 (U-251 MG)
and H4 cell lines, and normal human astrocyte NHA cells, were
purchased from The Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences (Shanghai, China). All of cells were
cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in a
humidified incubator at 37°C containing 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples using
TRIzol® reagent (Takara Biotechnology Co., Ltd., Dalian,
China). RNA integrity was analyzed by using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). RNA was reversed transcribed to cDNA with a
PrimeScript® RT reagent kit (Takara Biotechnology Co.,
Ltd.), according to the manufacturer's protocols. The RT-qPCR
reaction was performed on a CFX-96 Real-Time PCR system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) with SYBR® Premix
Ex Taq™ II (Takara Biotechnology Co., Ltd.), according to the
manufacturer's protocols. The thermocycling conditions were as
follows: Denaturation at 95°C for 5 min followed by 35 cycles of
denaturation at 95°C for 15 sec, and annealing/elongation at 60°C
for 30 sec. The results were normalized to the expression of GAPDH.
The primer sequences for DUXAP8 (Access number: NR_122113.1) are as
follows: DUXAP8-forward: 5′-GAGAAGCAGTGGTGGGTTCC-3′, and
DUXAP8-reverse: 5′-GAGCAACACAGATGAACCGC-3′. GAPDH-forward:
5′-GGGAGCCAAAAGGGTCAT-3′, and GAPDH-reverse:
5′-GAGTCCTTCCACGATACCAA-3′. The mRNA expression fold changes were
calculated using the 2-ΔΔCq methods (14).
Cell transfection
A total of 2 small interfering (si)RNAs targeting
DUXAP8 (si-RNA-1, sense: 5′-UUUAGACCCAUUCUCGUAUGGAGGU-3′, and
antisense: 5′-ACCUCCAUACGAGAAUGGGUCUAAA-3′; siRNA-2, sense:
5′-CAGCAUACUUCAAAUUCACAGCAAA-3′, and antisense
5′-UUUGCUGUGAAUUUGAAGUAUGCUG-3′) and scrambled negative control
siRNA (si-NC; 5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.). The U87 and U251 cells
were transfected with si-NC (100 nM), si-RNA-1 (100 nM) or siRNA-2
(100 nM) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. Following cell transfection for 48 h, the cells were
harvested for RT-qPCR analysis of mRNA expression.
Cell Counting Kit-8 (CCK-8)
proliferation assay
The transfected cells (3×103 cells/well) were seeded
in 96-well culture plates and incubated with 10 µl CCK-٨ (Beyotime
Institute of Biotechnology, Shanghai, China) reagent per well at
37°C for 2 h. Proliferation ability was detected at the selected
time points (0, 1, 2 and 3 days following seeding). The optical
density was determined at a wavelength of 450 nm.
Cell colony formation assay
The transfected cells (1,000 cells/well) were seeded
in a 12-well plate. Following 14 days incubation at 37°C, cell
colonies were counted under a light microscope (magnification,
×200) following fixing with 100% methanol and 0.1% crystal violet
staining for 20 min at room temperature.
Statistical analysis
All of statistical analyses in the study were
performed using SPSS 19.0 software (IBM Corp., Armonk, NY, USA).
Data are presented as the mean ± standard deviation from at least
three independent experiments. Difference between two groups was
assessed by paired Student's t-test, χ2 test as appropriate and
differences between multiple groups was analyzed with one-way
analysis of variance with a post hoc Student-Newman-Keuls test. The
survival plots were calculated by the Kaplan-Meier method and
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
DUXAP8 expression is upregulated in
human glioma tissues
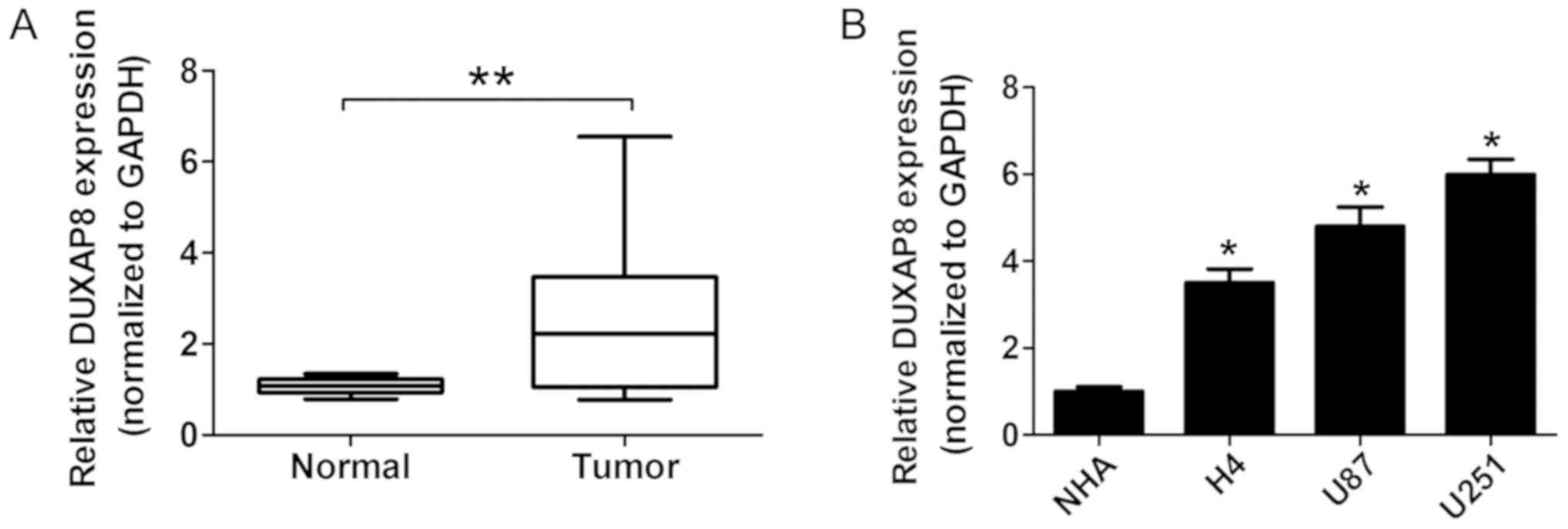
The expression of DUXAP8 in glioma tissues, compared
with that in normal tissues, was analyzed. The data revealed that
DUXAP8 expression was significantly upregulated in glioma tissues,
compared with that in normal tissues (Fig. 1A; P<0.05). Similarly, the data also
revealed that DUXAP8 expression was significantly upregulated in
glioma cells (U87, U251 and H4 cells), compared with normal NHA
cells (Fig. 1B; P<0.05).
Therefore, these results indicated that DUXAP8 expression is
upregulated in human glioma tissues and cells.
Upregulation of DUXAP8 expression is
associated with Karnofsky Performance Status (KPS), WHO grade and
poor prognosis of glioma
Furthermore, the mean expression of DUXAP8 (2.35
fold) was used as a cut-off value to divide patients into two
groups: High DUXAP8 expression and low DUXAP8 expression, with the
mean expression of DUXAP8 assigned to the high expression group.
The clinicopathological characteristics are presented in Table I. The association of DUXAP8 expression
with the clinicopathological characteristics was analyzed using χ2
test. The results indicated that increased DUXAP8 expression was
significantly associated with higher KPS (P=0.004) and advanced WHO
grade (P=0.001) in patients (Table
I). However, no significant association with sex, age and tumor
size was indicated (Table I;
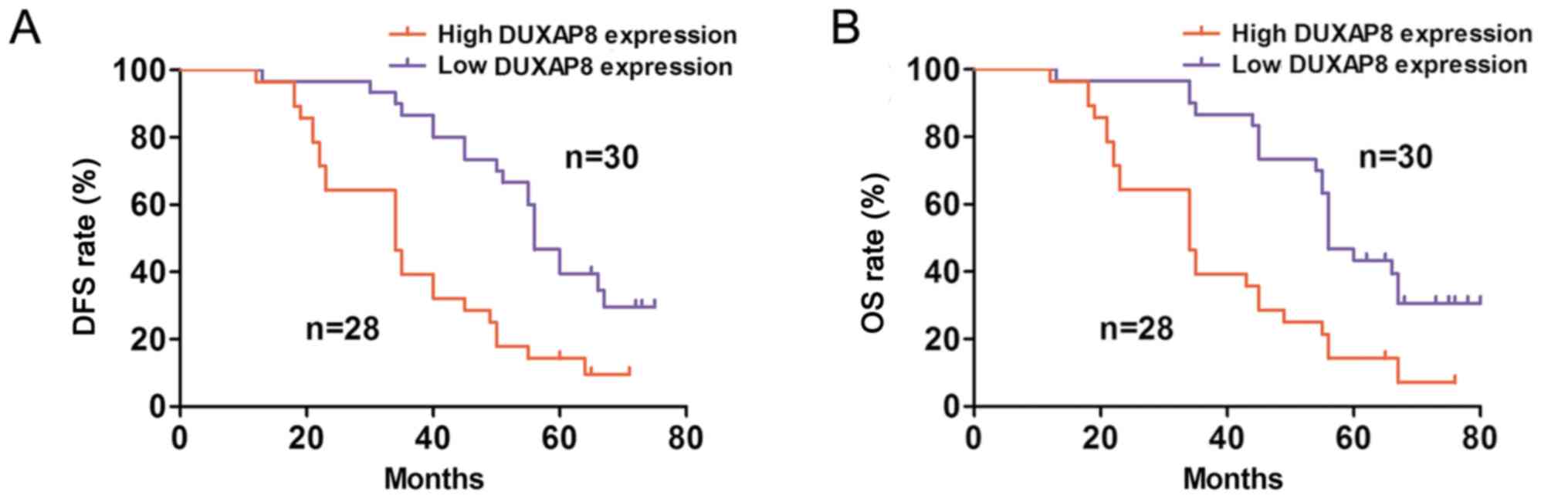
P>0.05). The survival plots were calculated by the Kaplan-Meier
methods and log-rank test. The results indicated that increased
DUXAP8 expression predicted poor disease-free survival (DFS; log
rank test=12.554; P<0.05) and overall survival (OS; log rank
test=13.374; P<0.05) rates, compared with reduced DUXAP8
expression groups (Fig. 2A and B).
Therefore, these results indicated that DUXAP8 may be a predictor
of glioma.
 | Table I.Association between DUXAP8 expression
and clinical features. |
Table I.
Association between DUXAP8 expression
and clinical features.
|
|
| DUXAP8
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Total cases,
n=58 | Low, n=30 | High, n=28 | P-value |
|---|
| Sex |
|
|
| 0.455 |
| Male | 36 | 20 | 16 |
|
|
Female | 22 | 10 | 12 |
|
| Age (years) |
|
|
| 0.300 |
| ≤55 | 31 | 18 | 13 |
|
|
>55 | 27 | 12 | 15 |
|
| Tumor size (cm) |
|
|
| 0.771 |
| ≤3 | 32 | 16 | 16 |
|
|
>3 | 26 | 14 | 12 |
|
| Tumor location |
|
|
| 0.457 |
|
Parenchyma | 40 | 22 | 18 |
|
|
Ventricle | 18 | 8 | 10 |
|
| Karnofsky performance
statusb |
|
|
| 0.004a |
| ≤80 | 32 | 22 | 10 |
|
|
>80 | 26 | 8 | 18 |
|
| WHO gradeb |
|
|
| 0.011a |
| I–II | 34 | 22 | 12 |
|
|
III–IV | 24 | 8 | 16 |
Knockdown of DUXAP8 expression
suppresses proliferation of glioma
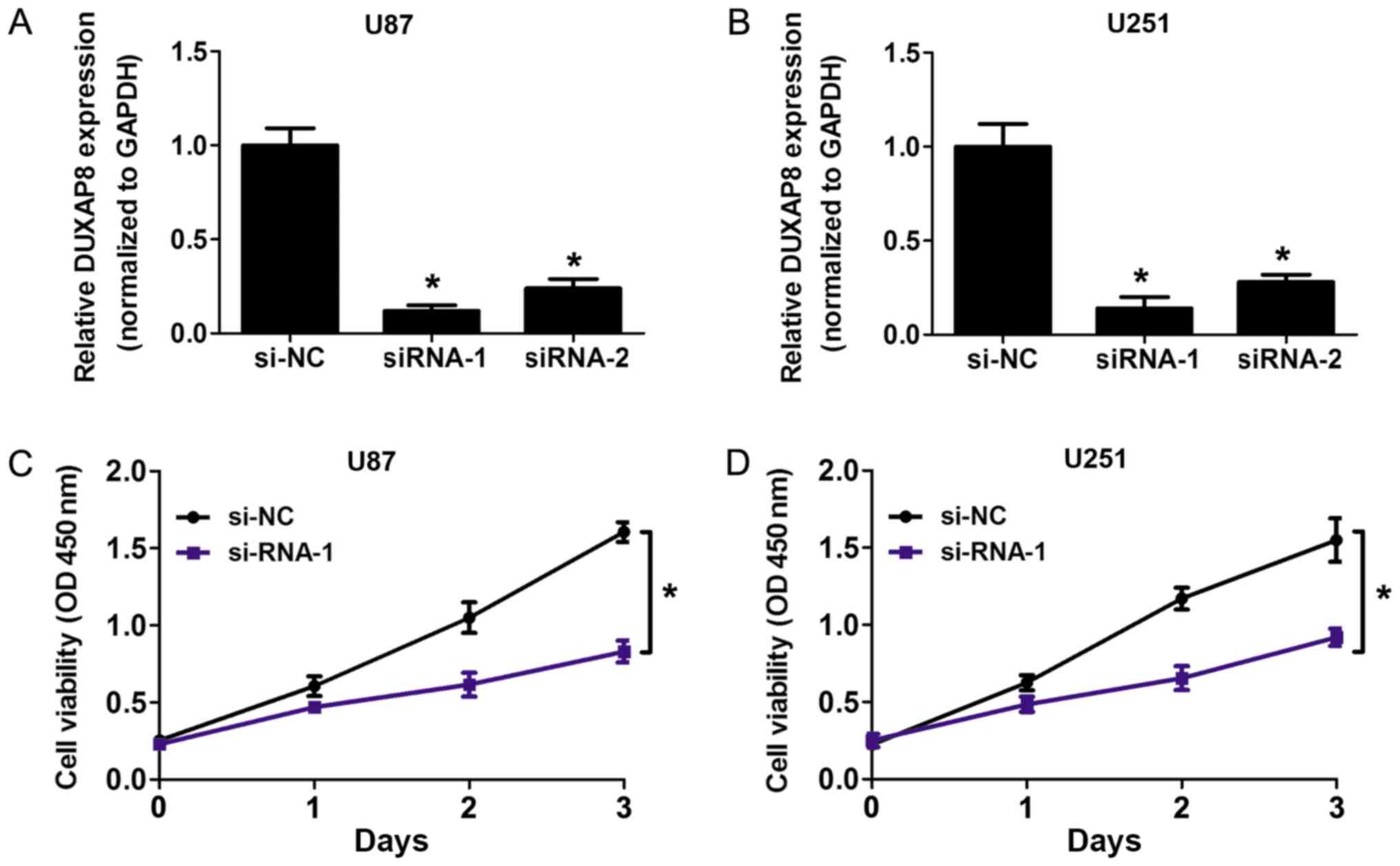
To examine the biological effects of DUXAP8
expression, a CCK8 assay and cell colony formation assay were
performed following DUXAP8 siRNA treatment. According to knockdown
efficiency in U87 and U251 cells, siRNA-1 was selected for the
following experiments (Fig. 3A and
B). The CCK8-assay results indicated that DUXAP8-knockdown
significantly inhibits proliferation, compared with the control
groups, in U87 and U251 cells (Fig. 3C
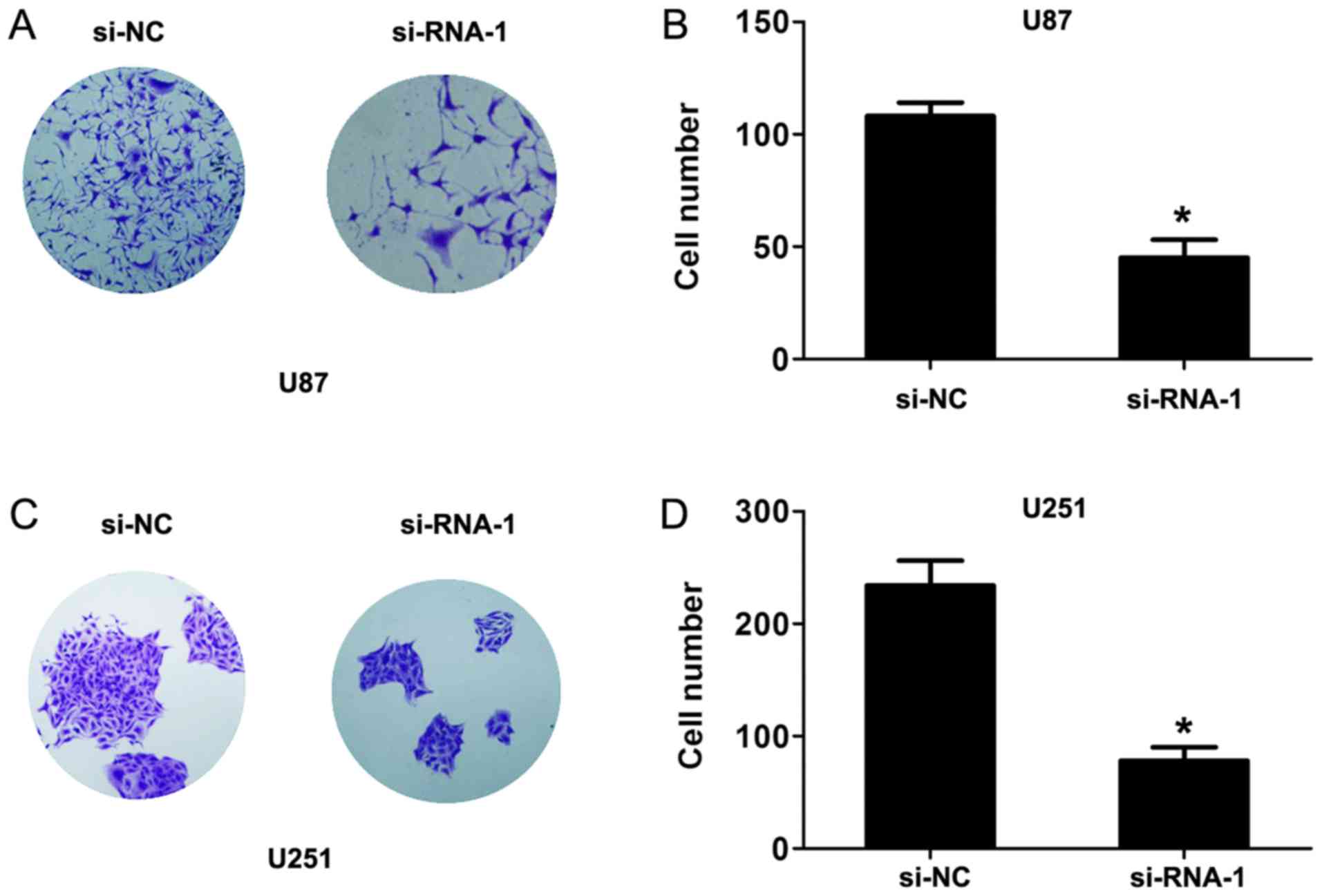
and D). Furthermore, the results of colony-formation assays
revealed that the clonogenic number is significantly decreased
following DUXAP8-knockdown in U87 and U251 cells (Fig. 4A-D). Therefore, these results
indicated that DUXAP8-knockdown suppresses the proliferation of
glioma.
Discussion
Pseudogenes were long identified to be
non-functional relics littering the genome (15). Previous studies revealed that the
involvement of pseudogenes in the pathogenesis and progression of
tumors, including the OCT4 pseudogene POU class 5 homeobox 1,
amplify and promote an aggressive phenotype in gastric cancer
(8,16). SUMO 1 pseudogene 3 was significantly
upregulated in gastric cancer and was significantly associated with
tumor size, differentiation, lymphatic metastasis and invasion
(8). In the present study, it was
revealed that DUXAP8 expression was significantly upregulated in
glioma tissues and cells, compared with that in normal tissues and
NHA cells. Increased DUXAP8 expression was significantly associated
with higher KPS scores and advanced WHO grade in patients.
Furthermore, the survival curves were calculated by the
Kaplan-Meier method and log-rank test. The results of the present
study indicated that increased DUXAP8 expression revealed a poor
outcome of patients, compared with reduced DUXAP8 expression
groups. Therefore, these results indicated that DUXAP8 may be
served as a potential predictor of glioma.
Pseudogene DUXAP8 was revealed to regulate tumor
progression in numerous tumors. For instance, the pseudogene DUXAP8
promotes non-small-cell lung cancer cell proliferation, and
invasion by epigenetically silencing early growth response 1 and
Ras homolog family member B (11).
Pseudogene DUXAP8 was significantly associated with overall
survival time of patients with renal cell carcinoma (RCC), and the
knockdown of DUXAP8 may impair RCC cells invasive ability in
vitro (17). Increased DUXAP8
promoted cell proliferation and invasion through epigenetically
silencing pleckstrin homology domain contain O1 expression by
binding with enhancer of zeste 2 polycomb repressive complex 2
subunit and SUZ12, polycomb repressive complex 2 subunit in gastric
cancer cells (12). The present study
demonstrated that knockdown of DUXAP8 expression inhibits the
proliferation and cell colony formation ability, which indicated
that DUXAP8 expression affects the proliferation ability in glioma
cells. Therefore, these results indicated that DUXAP8 is involved
in biological functions of glioma.
In conclusion, the results of the present study
first revealed that DUXAP8 expression is significantly upregulated
in glioma tissues and cells. Increased DUXAP8 expression predicted
a poor survival rate. Additionally, DUXAP8-knockdown significantly
inhibited proliferation in glioma. These results indicated for the
first time, to the best of our knowledge, that pseudogene DUXAP8
may be a potential biomarker and target of tumor treatment in
glioma. Future studies should investigate the molecular mechanism
for DUXAP8 in glioma progression.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, SH, MW and CW conceived and designed the study,
and drafted the manuscript. DX, XZ, SH, and MW collected, analyzed
and interpreted the experiment data, and revised the manuscript
critically for important intellectual content. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Second Hospital of Shandong University (Jinan, China). Written
informed consent was obtained from all patients in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Taylor LP: Diagnosis, treatment, and
prognosis of glioma: Five new things. Neurology. 75 (18 Suppl
1):S28–S32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lenting K, Verhaak R, Ter Laan M,
Wesseling P and Leenders W: Glioma: Experimental models and
reality. Acta Neuropathol. 133:263–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirst TC, Vesterinen HM, Conlin S, Egan
KJ, Antonic A, Lawson McLean A, Macleod MR, Grant R, Brennan PM,
Sena ES and Whittle IR: A systematic review and meta-analysis of
gene therapy in animal models of cerebral glioma: Why did promise
not translate to human therapy? Evid Based Preclin Med.
1:e000062014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei Y, Chang Z, Wu C, Zhu Y, Li K and Xu
Y: Identification of potential cancer-related pseudogenes in lung
adenocarcinoma based on ceRNA hypothesis. Oncotarget.
8:59036–59047. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiao-Jie L, Ai-Mei G, Li-Juan J and Jiang
X: Pseudogene in cancer: Real functions and promising signature. J
Med Gen. 52:17–24. 2015. View Article : Google Scholar
|
|
6
|
Harrison PM, Hegyi H, Balasubramanian S,
Luscombe NM, Bertone P, Echols N, Johnson T and Gerstein M:
Molecular fossils in the human genome: Identification and analysis
of the pseudogenes in chromosomes 21 and 22. Genome Res.
12:272–280. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jingsi T, Mingyao Y and Ying L: Functional
roles of pseudogenes in cancers. Yi Chuan. 37:8–16. 2015.PubMed/NCBI
|
|
8
|
Mei D, Song H, Wang K, Lou Y, Sun W, Liu
Z, Ding X and Guo J: Up-regulation of SUMO1 pseudogene 3 (SUMO1P3)
in gastric cancer and its clinical association. Med Oncol.
30:7092013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhan Y, Liu Y, Wang C, Lin J, Chen M, Chen
X, Zhuang C, Liu L, Xu W, Zhou Q, et al: Increased expression of
SUMO1P3 predicts poor prognosis and promotes tumor growth and
metastasis in bladder cancer. Oncotarget. 7:16038–16048. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang W, Li N, Hu J and Wang L: Inhibitory
effect of RNA-mediated knockdown of zinc finger protein 91
pseudogene on pancreatic cancer cell growth and invasion. Oncol
Lett. 12:1343–1348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei
C, Li W, He X and Lu KH: The Pseudogene DUXAP8 promotes
non-small-cell lung cancer cell proliferation and invasion by
epigenetically silencing EGR1 and RHOB. Mol Ther. 25:739–751. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma HW, Xie M, Sun M, Chen TY, Jin RR, Ma
TS, Chen QN, Zhang EB, He XZ, De W and Zhang ZH: The pseudogene
derived long noncoding RNA DUXAP8 promotes gastric cancer cell
proliferation and migration via epigenetically silencing PLEKHO1
expression. Oncotarget. 8:52211–52224. 2017.PubMed/NCBI
|
|
13
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pink RC, Wicks K, Caley DP, Punch EK,
Jacobs L and Carter DR: Pseudogenes: Pseudo-functional or key
regulators in health and disease? RNA. 17:792–798. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hayashi H, Arao T, Togashi Y, Kato H,
Fujita Y, De Velasco MA, Kimura H, Matsumoto K, Tanaka K, Okamoto
I, et al: The OCT4 pseudogene POU5F1B is amplified and promotes an
aggressive phenotype in gastric cancer. Oncogene. 34:199–208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu X, Xu Y, Shi C, Wang B, Yu X, Zou Y and
Hu T: A genome-wide comprehensively analyses of long noncoding RNA
profiling and metastasis associated lncRNAs in renal cell
carcinoma. Oncotarget. 8:87773–87781. 2017.PubMed/NCBI
|